Abstract
Pyridine nucleotides are abundant soluble coenzymes and they undergo reversible oxidation and reduction in several biological electron-transfer reactions. They are comprised of two mononucleotides, adenosine monophosphate and nicotinamide mononucleotide, and are present as oxidized and reduced nicotinamide adenine dinucleotides in their unphosphorylated (NAD+ and NADH) and phosphorylated (NADP+ and NADPH) forms. In the past, pyridine nucleotides were considered to be primarily electron-shuttling agents involved in supporting the activity of enzymes that catalyze oxidation-reduction reactions. However, it has recently been demonstrated that pyridine nucleotides and the balance between the oxidized and reduced forms play a wide variety of pivotal roles in cellular functions as important interfaces, beyond their coenzymatic activity. These include maintenance of redox status, cell survival and death, ion channel regulation, and cell signaling under normal and pathological conditions. Furthermore, targeting pyridine nucleotides could potentially provide therapeutically useful avenues for treating cardiovascular diseases. This review series will highlight the functional significance of pyridine nucleotides and underscore their physiological role in cardiovascular function and their clinical relevance to cardiovascular medicine.
Keywords: pyridine nucleotides, NAD(H), NADP(H), metabolism, redox, cardiovascular disease
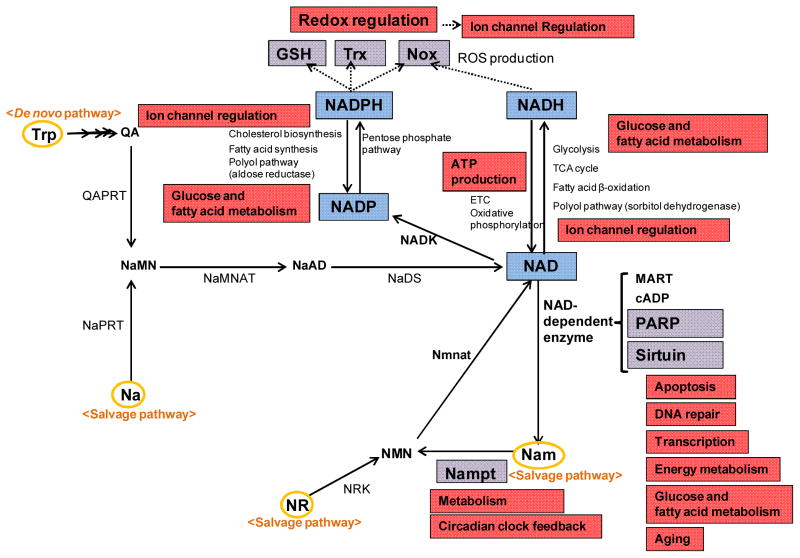
Pyridine nucleotides are small molecules comprised of two mononucleotides, adenosine monophosphate (AMP) and nicotinamide mononucleotide (NMN). They consist of oxidized and reduced nicotinamide adenine dinucleotides in their unphosphorylated (NAD+ or NADH) and phosphorylated (NADP+ or NADPH) forms. Arthur Harden first discovered the existence of a high molecular weight ferment (enzyme) and a low molecular weight co-ferment (coenzyme), while Hans von Euler-Chelpin isolated the coenzyme and later described the character of NAD+ as being comprised of two mononucleotides, namely, AMP and NMN. These investigators won a Nobel Prize in 1929 for their investigations into the fermentation of sugar and fermentative enzymes. Subsequently, Otto Warburg, another Nobel laureate, provided evidence for the functional significance of NAD+, which serves to transfer hydrogen, and discovered another coenzyme, NADP+ 1. Since then, it has become increasingly clear that these small molecules, termed pyridine nucleotides, are involved in a wide variety of cellular functions, including energy production, metabolism, reduction-oxidation (redox) reactions, cell survival and death, transcriptional regulation, and protein modification. Furthermore, interest in the field of pyridine nucleotides has been dramatically stimulated in the past decade by the discoveries that 1) the sirtuin family proteins, the key regulators of energy metabolism and lifespan, utilize NAD+ as a co-factor2, 3, 2) nicotinamide phosphoribosyltransferase (Nampt), an enzyme of the salvage pathway, is a key enzyme for NAD+ synthesis4, 5 3) NADPH provides electrons to both oxidases and reductases, thereby stimulating oxidative and reductive stress6, and 4) pyridine nucleotides regulate ion channels7, 8 (Figure).
Figure. Schematic of pyridine nucleotides biosynthesis and their functions.
Pyridine nucleotides are involved in a wide variety of cellular functions, including energy production, metabolism, redox reactions, survival/death, and ion channels under normal and pathological conditions. Trp, tryptophan; Na, nicotinic acid; QA, quinolinic acid; NaMN, nicotinic acid mononucleotide; Nam, nicotinamide; NMN, nicotinamide mononucleotide; NR, nicotinamide riboside; Nampt, nicotinamide phosphoribosyltransferase; Nmnat, Nam/Na mononucleotide adenylyl transferase; NADK, NAD kinase; NaPRT, Na phosphoribosyltransferase; QAPRT, quinolate phosphoribosyltransferase; NaMNAT, nicotinic acid mononucleotide adenylyltransferase; NaAD, nicotinic acid adenine dinucleotide; NaDS, NAD synthase; Trx, thioredoxin; Nox, NADPH oxidase; GSH, glutathione; Redox, reduction-oxidation; ROS, reactive oxygen species; PARP, poly(ADP-ribose) polymerases; MART, mono-ADP-ribose transferase; cADP, cyclic ADP-ribose synthase; ETC, electron transport chain.
Generally, pyridine nucleotides have been shown to serve 1) as coenzymes for various dehydrogenase enzymes to regulate cellular metabolism, 2) as electron carriers to synthesize adenosine triphosphate (ATP) in mitochondria via the electron transport chain (ETC) and oxidative phosphorylation, 3) as electron donors for glutathione (GSH), thioredoxin (Trx), and NADPH oxidases (Noxs) to regulate the redox status, 4) as donors of adenosine diphosphate (ADP)-ribose moieties in ADP-ribosylation reactions, 5) as substrates for NAD+-dependent enzymes, such as mono-ADP-ribose transferases (MARTs), poly(ADP-ribose) polymerases (PARPs), cyclic ADP-ribose synthases (cADPs) and sirtuins, and 6) as redox regulators that modify ion channel function. As one can speculate from these findings, pyridine nucleotides play an important role in regulating a wide variety of functions in the heart and other cardiovascular tissues. Therefore, understanding the role of pyridine nucleotides has become increasingly important in the field of cardiovascular medicine. We therefore put forward a review series to summarize our current knowledge of the cardiovascular function of pyridine nucleotides. The goal of this editorial is to provide brief highlights on the cardiovascular role of pyridine nucleotides under both physiological and pathological conditions as discussed in individual review articles of this series.
Biosynthesis of pyridine nucleotides
How are pyridine nucleotides generated in cardiomyocytes? First of all, NAD+ and NADH are interchangeable through oxidation/reduction reactions, as are NADP+ and NADPH. Since NAD+ can be converted to NADP+ by NAD kinase (NADK), describing how NAD+ is synthesized would be most informative. The biosynthesis of NAD+ occurs through two major pathways: a de novo pathway and a salvage pathway9, 10 (Figure). The de novo pathway (the Preiss-Handler pathway) begins with tryptophan to generate quinolinic acid (QA). QA is converted to nicotinic acid mononucleotide (NaMN) by quinolate phosphoribosyltransferase (QAPRT). NaMN is then adenylylated by nicotinic acid mononucleotide adenylyltransferase (NaMNAT) to produce nicotinic acid adenine dinucleotide (NaAD), which is subsequently amidated to NAD+ by NAD synthase (NaDS).
On the other hand, the salvage pathway regenerates NAD+ from nicotinic acid (Na), nicotinamide (Nam), or nicotinamide riboside (NR) present in the metabolites of NAD+ or dietary sources. Na is converted to NaMN through Na phosphoribosyltransferase (NaPRT). Nam is converted to NMN by Nampt, and NMN is subsequently adenylylated by Nam/Na mononucleotide adenylyl transferase (Nmnat) to form NAD+. NR is used to generate NMN by NR kinase (NRK). Among the components of the salvage pathway, Nam is the major precursor of the NAD+ synthesizing pathways in mammals, and Nampt is the rate-limiting enzyme4. Thus, Nam and Nampt are the most important components of the NAD+ synthesizing pathways in mammalian cells. Importantly, in the heart, the expression of Nampt is regulated by stress11. Furthermore, the levels of Nampt and NAD+ display circadian oscillations that are regulated by the core clock machinery in mice12, 13. While the expression of Nampt is regulated by the circadian transcription factor CLOCK, Nampt in turn negatively regulates the core circadian clock machinery CLOCK/BMAL1 through NAD+/Sirt1. How NAD+ levels are regulated through changes in Nampt expression in response to stress and how this mechanism is influenced by the clock genes remain to be elucidated. Na and Nam are collectively called niacin or vitamin B3. Vitamins taken up through diet can be precursors of pyridine nucleotide biosynthesis through the salvage pathway. Nevertheless, it remains to be elucidated whether supplementation of vitamins or precursors of NAD+ can maintain or improve NAD+ levels during stress, thereby influencing the energy metabolism and other cellular functions14.
Cellular metabolism
Pyridine nucleotides play an important role in regulating energy metabolism. The heart requires a high level of energy, in the form of ATP, nutrients, lipids, carbohydrates, and amino acids, to continue its pumping and maintain a constant protein turnover. The predominant energy source in the adult heart is fatty acids, used for fatty acid β-oxidation that produces NADH, the reduced form of flavin adenine dinucleotide (FADH2), and acetyl-CoA. However, in the failing heart, the energy source shifts from fatty acids to carbohydrates, for glycolysis15, 16. The expression of the enzymes involved in fatty acid β-oxidation is regulated at the level of transcription by nuclear receptor transcription factors, such as peroxisome proliferator-activated receptor (PPAR) and PPARγ co-activator 1 α (PGC-1α)17. In the failing myocardium, fatty acid β-oxidation is downregulated, which is accompanied by an increase in glucose uptake and glycolysis. During glycolysis, the tricarboxylic acid (TCA) cycle, and fatty acid β-oxidation, NAD+ is utilized as a coenzyme to produce NADH. During ATP synthesis through the mitochondrial ETC and oxidative phosphorylation, NADH is utilized as a hydride donor to generate a proton motive force across the inner mitochondrial membrane. NADPH is produced in the pentose phosphate pathway, and is re-oxidized as a coenzyme of aldose reductase in the polyol pathway. While aldose reductase utilizes NADPH to reduce the reactive products of lipid peroxidation under normal conditions18, excessive activation of the polyol pathway in diabetes reduces the amount of NADPH, which, in turn, induces oxidative stress. NADPH is also utilized as a coenzyme for de novo fatty acid synthesis and in the biosynthesis of cholesterol. Because NAD+/NADH and NADP+/NADPH play a central role in energy production and metabolism as electron carriers and coenzymes, an imbalance in the ratios of the oxidized and reduced forms affects the energy level and cellular metabolism in the heart. Under normal conditions, NADH and acetyl-CoA, which are the products of fatty acid β-oxidation, inhibit carbohydrate oxidation through phosphorylation/inhibition of pyruvate dehydrogenase (PDH). Thus, carbohydrate oxidation is regulated by PDH activity, and the ratios of NADH/NAD+ and acetyl-CoA/free CoA play a central role in regulating PDH activity15. The regulation of cellular metabolism by pyridine nucleotides, the metabolic changes during ischemia, ischemic preconditioning, ischemia and reperfusion, pulmonary artery hypertension, and heart failure, and the effect of the changes in enzymatic activities upon cardiac function will be reviewed by Gary Lopaschuk and his colleagues.
Oxidative and reductive stress
The reduced forms of pyridine nucleotides, NADH and NADPH, not only serve as electron donors to produce ATP (mainly by NADH), but also regulate the cellular redox status (mainly by NADPH) through Noxs, GSH and Trx. Electrons provided by NADPH/NADH are transferred to either molecular oxygen to produce O2− by Nox, or to GSH or Trx whose cysteine residues have been oxidized in the oxidative environment. Uncoupled NO synthase can also produce O2− using electrons obtained from NADPH.
The function of Noxs is unique in that these enzymes purposefully produce reactive oxygen species (ROS), which damage proteins and DNA, causing apoptotic cell death and mitochondrial permeability transition pore (mPTP) opening, ATP depletion and necrotic cell death6. Nox2 and Nox4 are the major Noxs in the heart. Nox4 is primarily expressed on intracellular membranes, including mitochondrial membranes in cardiomyocytes, while Nox2 is preferentially expressed on the plasma membrane19, 20. The expression of Nox2/4 is upregulated during cardiac hypertrophy and heart failure. However, the role of Noxs in hypertrophy is not clear. Whereas systemic deletion of Nox4 exacerbates load-induced hypertrophy in mice21, cardiac-specific deletion of Nox4 inhibits it20. Until recently, it was believed that Noxs are unlikely to be expressed in mitochondria. However, both ROS production in mitochondria and cardiomyocyte apoptosis in response to pressure overload are ameliorated in Nox4 cardiac-specific knockout mice. Given this, together with the presence of a mitochondrial localization signal in Nox4, it is likely that Nox4 is expressed in mitochondria. The fact that Nox4 consumes pyridine nucleotides for O2− production raises an intriguing possibility that Nox4 and the ETC may compete with one another for pyridine nucleotides. It should be noted that whether Nox4 preferentially uses NADH or NADPH as a substrate has been debated. Overexpression of Nox4 in cardiomyocytes in vitro and in vivo induces cardiomyocyte apoptosis rather than hypertrophy, suggesting that Nox4 stimulates cardiomyocyte apoptosis through ROS production and mitochondrial dysfunction19. On the other hand, Nox4 can also act as a sensor to generate O2− as a signaling molecule in the presence of hypoxia, suggesting that Nox4 also possesses an important physiological function6. Further investigations are required to understand these pathological and physiological roles of Nox in greater detail.
In contrast to Nox, GSH and Trx reduce ROS and oxidized proteins, thereby protecting against oxidative stress22, 23. GSH-dependent antioxidant systems, composed of GSH-dependent peroxidase (Gpx) and glutaredoxin (Grx), have been shown to protect cells against H2O2. Although the intracellular concentration of Trx1 is not as high as that of GSH (sub-μM vs mM range)24, Trx1 is able to reduce key proteins through direct/indirect protein-protein interactions, thereby exerting a significant influence on a wide variety of cellular functions23, 25–27. Trx1 is upregulated in response to ischemia/reperfusion (I/R), and overexpression of Trx1 is cardioprotective against I/R by 1) reducing ROS and oxidized proteins, 2) modulating signaling proteins and transcription factors, 3) upregulating mitochondrial proteins, 4) inhibiting expression of proinflammatory cytokines, and 5) preventing the oxidative stress-induced downregulation of the K+v4 channel through modification of the ion channel26.
The cellular source of electrons for reducing thiols is NADPH, which is produced primarily through glucose-6-phosphate dehydrogenase (G6PD), a cytosolic enzyme in the pentose phosphate pathway. The way in which G6PD couples to Nox, GSH and Trx1 is not well understood. For example, stimulation of G6PD appears to enhance O2− production through Nox in failing hearts28. It can also induce hyper-production of GSH and “reductive stress”, which can, in turn, lead to cardiac dysfunction29. It is unclear why Nox and antioxidants are not equally affected under these conditions. It is possible that the activity of Nox, GSH and Trx1 might be regulated locally and their functions compartmentalized. When one considers the function of these molecules, it is important to elucidate how they regulate posttranslational oxidative modification of specific targets. For example, Trx1 prevents pathological hypertrophy through reduction of class II histone deacetylases (HDACs)30, 31. The regulation of Nox, GSH and Trx1 by pyridine nucleotides will be summarized by Jun Sadoshima and his colleagues.
NADH not only acts as an electron donor in the mitochondrial ETC to produce an electrochemical gradient for ATP synthesis but it also serves as a source of electrons by itself or after conversion into NADPH in mitochondria. NADP+ is synthesized from NAD+ by NADK, while NADPH is converted from NADH by three major enzymes in the matrix of mitochondria: NAD(P) transhydrogenase, the NADP+-dependent isocitrate dehydrogenase (IDH-NADP+) and malic enzyme32. The IDH-NADP+ and malic enzyme are dependent upon TCA cycle intermediates. Thus, respiration and the activity of mitochondrial antioxidants are linked at the level of NADH. Therefore, the state of oxidative phosphorylation affects the activity of nearly all antioxidants. The proper balances in the NADH/NAD+ and NADPH/NADP+ ratios are also essential for the regulation of the metabolism and redox state in the heart. The role of pyridine nucleotides in the energy metabolism and their interaction with the antioxidant pathways will be reviewed by Brian O’Rourke.
NAD+-dependent enzymes and their functions
Pyridine nucleotides also act as substrates for NAD+-dependent enzymes, such as PARPs and sirtuins. Both proteins induce posttranslational modifications of target proteins, thereby regulating key cellular functions such as DNA repair and transcription. However, excessive activation of the NAD+-dependent enzymes causes depletion of NAD+, thereby secondarily disturbing other NAD+-dependent functions in cells. PARPs polymerize ADP-ribose derived from NAD+ onto target proteins in the nucleus. Although the primary function of PARPs is to regulate single-strand DNA break repair through poly-ADP ribosylation, they also affect cell death through consumption of NAD25, 33, 34 and stimulation of AIF-induced cell death, termed parthanatos35. PARP-1 is hyperactivated in response to cardiac stress, including I/R, pressure overload, and ROS. Inhibition of PARP-1 protects the heart from I/R injury and pressure overload through multiple mechanisms, including inhibition of the depletion of NAD+ and ATP, inhibition of mitochondrial-to-nuclear translocation of AIF, and inhibition of mitochondrial complex I dysfunction.36, 37 Hyperactivation of PARP-1 depletes nuclear NAD+, thereby inhibiting transcription of sirtuin-dependent genes, such as mitochondrial genes, and stimulating proapoptotic genes, such as p53. Although the catalytic activity of PARP-1, namely, the stimulation of poly-ADP ribosylation of target proteins, also appears to stimulate myocardial injury, the direct involvement of enzymatic targets of PARP-1 in myocardial injury remains to be shown. While excessive activation of PARP-1 causes depletion of ATP, PARP-1 can be cleaved by caspase-3, which serves as a negative feedback mechanism to prevent further activation of PARP-1.
Sirtuins are mammalian homologues of the silent information regulator (Sir) proteins in yeast. The Sir 1–4 proteins were initially discovered as gene silencers of the silent mating loci and telomerases. Sirtuins are evolutionarily conserved from bacteria to humans, and they comprise 7 isoforms, sirtuins 1–7, that serve as NAD+-dependent deacetylases (Sirt1–3 and 5–7) and as an ADP-ribosyltransferase (Sirt4). Their cellular localizations are different; Sirt1, 6 and 7 are predominantly localized in the nucleus, although Sirt1 is also located in the cytosol in some situations, Sirt2 is preferentially localized in the cytosol, and Sirt3–5 are predominantly localized in the mitochondrial matrix.
Sirtuins are involved in transcriptional regulation, glucose and fatty acid metabolism, apoptosis, energy metabolism, and DNA repair2. Sirt1, the most characterized isoform in the sirtuin family, deacetylates transcription factors, such as PPARγ, PPARα, PGC-1α, and the forkhead box subgroup O (FoxO) family, to induce insulin secretion, gluconeogenesis and fatty acid β-oxidation. Importantly, Sirt1 is activated by energy stress and in turn activates metabolic adaptation. For example, Sirt1 works in parallel with AMP-dependent kinase (AMPK)-mediated phosphorylation to activate PGC-1α during caloric restriction and exercise. Interestingly, a cross-talk exists between Sirt1 and AMPK where AMPK stimulates Sirt1 with or without upregulation of Nampt, whereas Sirt1 stimulates AMPK though deacetylation of LKB1, an upstream regulator (reviewed in 38). Sirt1 also controls apoptosis and inflammation through deacetylation of p53 and NF-kB, respectively39.
In the heart, mild to modest upregulation of Sirt1 prevents aging-induced cardiomyopathy40. However, the issue of whether endogenous Sirt1 controls aging in the heart requires further investigation. Just as other molecular mechanisms extending lifespan often confer stress resistance to the organism, Sirt1 protects the heart from I/R by inducing preconditioning effects or directly inhibiting cell death during ischemia and reperfusion41, 42. The deacetylation of FoxOs plays an important role in mediating the Sirt1 protective effect. The Sirt1-FoxO axis also protects the heart from nutrient starvation and prolonged ischemia through activation of autophagy43. Importantly, however, upregulation of Sirt1 during pressure overload facilitates downregulation of genes involved in the energy metabolism and cardiac contraction by suppressing transcription through the estrogen-related receptor element (ERRE), where upregulation of Sirt1 and PPARα competitively excludes binding of estrogen-related receptor α (ERRα) to the ERRE44. Thus, the effect of Sirt1 upon the heart is stimulus-dependent.
Sirt3 knockout mice developed cardiac hypertrophy and interstitial fibrosis at 8 weeks of age and showed more severe cardiac hypertrophy in response to angiotensin II and pressure overload than control mice. The overexpression of Sirt3 protected the heart against hypertrophic stimuli by upregulating MnSOD and catalase via FoxO3a45. However, whether endogenous Sirt3 regulates hypertrophy through its nuclear targets remains to be elucidated. Alternatively, Sirt3 may inhibit cardiac hypertrophy by deacetylating mitochondrial targets, such as cyclophilin D46. In fact, the function of many mitochondrial genes is regulated by acetylation of lysine residues. Sirt3 deacetylates proteins involved in the metabolism of the mitochondrial matrix, including acetyl-CoA synthase 2. Recently, general control of amino acid synthesis–like 1 (GCN5L1) has been identified as the protein acetyltransferase in mitochondria47. How the balance of protein acetylation in mitochondrial proteins is regulated appears to be a hot topic in the field.
Polyphenol compounds, including resveratrol, and small-molecule compounds mimicking the function of resveratrol have been shown to extend the lifespan of mice fed a high fat diet. The major premise is that these compounds mimic the effect of dietary restriction, currently the most established intervention to prevent age-related diseases and extend lifespan in many species, from yeast to primates, by stimulating the action of sirtuins. However, whether Sirt1 has a major effect upon lifespan extension in lower organisms and whether resveratrol and other small-molecule compounds mediate lifespan extension through Sirt1 in mammals have been debated extensively48. Regardless of the underlying mechanism, if caloric restriction mimetics prevent both aging and stress-induced myocardial injury, they will become promising candidates for cardiovascular therapy. Maha Abdellatif will review recent progress in the study of sirtuins.
Pyridine nucleotide ion channel interaction
The state of redox couples of pyridine nucleotides, namely, NADH/NAD+ and NADPH/NADP+, also plays an important role in regulating ion channel activities. For example, the β-subunits of the voltage- gated (Kv) channels bind to pyridine nucleotides, and intracellular changes in the redox ratio of these nucleotides affect the inactivation or activation characteristics of these channels7. Other K channels, such as the KATP channels and the KNa channels, have also been shown to respond to changes in intercellular levels of pyridine nucleotides49, 50, although the physiological significance of these regulatory mechanisms has not been fully assessed. An intriguing study also shows that the cystic fibrosis transmembrane conductance regulator (CFTR) channels (Cl− channels) are regulated by the pyridine nucleotide redox potential51, 52. This suggests that CFTR has the ability to sense the energy and redox status of the cell through pyridine nucleotides. In addition to direct binding to channel protein or associated subunits, pyridine nucleotides also regulate the activity/conductance of ion channels through ROS- or protein kinase-dependent posttranslational modifications. In the heart, the elevated intracellular NADH results in a rapid reduction in the cardiac Na+ current (Ina) through downregulation of sodium channels (Nav1.5), which is associated with ventricular tachycardia8. Since the NADH/NAD+ ratio may increase during I/R, this enhances mitochondrial ROS, thereby predisposing the heart to fatal arrhythmia through decreases in Na+ conductance 53. An increase in the cytosolic NADH/NAD+ ratio also inhibits cardiac sarcoplasmic reticulum Ca2+ release channels in cardiomyocytes54. It should be noted that an accurate assessment of the NADH/NAD+ ratio in subcellular compartments with standard metabolomic methods is challenging. Further investigation is necessary to clarify how the redox status of NADH/NAD+ is regulated in pathophysiological conditions in the heart. Oxidative stress increases ryanodine receptor calcium leakage55, and scavenging of ROS with antioxidants has been reported to decrease the incidence of arrhythmias56. Thus, the regulation of pyridine nucleotides and consequent changes in the redox status in cells play an important role in regulating several ion channels. This regulatory axis could link changes in cell energetics to excitability, allowing the cell to mount an integrated response to altered states of metabolism or redox potential. Perturbation in this mode of regulation may be associated with fatal arrhythmia and the progression of heart failure. Regulation of ion channels by pyridine nucleotides will be reviewed by Aruni Bhatnagar and his colleagues.
Summary
We here described that pyridine nucleotides act as electron carriers, enzyme substrates, and redox regulators. They have been recognized as important regulators of energy production, metabolism, redox reactions, survival/death, and ion channels under normal and pathological conditions. The deregulation of pyridine nucleotides is intimately involved in the pathogenesis of heart failure, I/R injury, arrhythmia, and age-associated abnormality in the heart. Importantly, both the metabolic status of cells and the redox status of the subcellular compartment regulated by pyridine nucleotides secondarily affect the function of mitochondria and ion channels. Changes in the activity of sirtuins and PARPs in the nucleus affect expression of genes through transcriptional as well as epigenetic mechanisms. Furthermore, cellular mechanisms regulated by pyridine nucleotides appear to be linked to one another, since they are widely used as co-substrates in many enzymatic processes. For example, NAD+ may be used not only by sirtuins and PARP but also by the glycolysis/TCA cycles. NADH may be competitively used by the ETC and mitochondrial Nox, and also by GSH/Trx2 after it is converted to NADPH in mitochondria. Nox, GSH and Trx might compete for electrons provided by NADPH. More investigations are required to clarify the interactions among multiple cellular functions through competition for pyridine nucleotides. In order to elucidate how changes in the level of pyridine nucleotides affect the function of cardiomyocytes, systematic approaches globally assessing the effects of pyridine nucleotides are needed. We hope that this review series serves as a convenient introduction to the understanding of the complex functions of pyridine nucleotides in cardiovascular tissues under both physiological and pathological conditions.
Acknowledgments
We thank Daniela Zablocki and Christopher D. Brady for critical reading of the manuscript.
Funding Sources
This work was supported in part by U.S. Public Health Service Grants HL59139, HL67724, HL69020, HL91469, HL102738, AG27211 and the Foundation of Leducq Transatlantic Network of Excellence.
List of Abbreviations
- AIF
apoptosis inducible factor
- AMPK
AMP-dependent protein kinase
- cADP
cyclic ADP-ribose synthase
- CFTR
cystic fibrosis transmembrane conductance regulator
- ERRα
estrogen-related receptor α
- ERRE
estrogen-related receptor element
- ETC
electron transport chain
- FDH2
flavin adenine dinucleotide
- FoxO
forkhead box subgroup O
- G6PD
glucose-6-phosphate dehydrogenase
- GCN5L1
general control of amino acid synthesis–like 1
- Gpx
GSH-dependent peroxidase
- Grx
glutaredoxin
- GSH
glutathione
- HDAC
histone deacetylases
- I/R
ischemia/reperfusion
- IDH
the NADP+-dependent isocitrate dehydrogenase
- MART
mono-ADP-ribose transferases
- mPTP
mitochondrial permeability transition pore
- NaAD
nicotinic acid adenine dinucleotide
- NAD+
oxidized nicotinamide adenine dinucleotide
- NADH
nicotinamide adenine dinucleotide
- NADK
NAD kinase
- Na
nicotinic acid
- NaDS
NAD synthase
- Nam
nicotinamide
- NaMN
nicotinic acid mononucleotide
- NaMNAT
nicotinic acid mononucleotide adenylyltransferase
- Nampt
nicotinamide phosphoribosyltransferase
- NaPRT
Na phosphoribosyltransferase
- NMN
nicotinamide mononucleotide
- Nmnat
Nam/Na mononucleotide adenylyl transferase
- Nox
NADPH oxidases
- NR
nicotinamide riboside
- NRK
NR kinase
- PARP
poly(ADP-ribose) polymerase
- PDH
pyruvate dehydrogenase
- PGC-1α
PPARγ co-activator 1α
- PPAR
peroxisome proliferator-activated receptor
- QA
quinolinic acid
- QART
quinolate phosphoribosyltransferase
- ROS
reactive oxygen species
- Sir
silent information regulator
- TCA
tricarboxylic acid
Footnotes
Disclosures
None
References
- 1.Berger F, Ramirez-Hernandez MH, Ziegler M. The new life of a centenarian: signalling functions of NAD(P) Trends Biochem Sci. 2004;29:111–118. doi: 10.1016/j.tibs.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 5.Imai S. Dissecting systemic control of metabolism and aging in the NAD World: the importance of SIRT1 and NAMPT-mediated NAD biosynthesis. FEBS Lett. 2011;585:1657–1662. doi: 10.1016/j.febslet.2011.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maejima Y, Kuroda J, Matsushima S, Ago T, Sadoshima J. Regulation of myocardial growth and death by NADPH oxidase. J Mol Cell Cardiol. 2011;50:408–416. doi: 10.1016/j.yjmcc.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tipparaju SM, Saxena N, Liu SQ, Kumar R, Bhatnagar A. Differential regulation of voltage-gated K+ channels by oxidized and reduced pyridine nucleotide coenzymes. Am J Physiol Cell Physiol. 2005;288:C366–376. doi: 10.1152/ajpcell.00354.2004. [DOI] [PubMed] [Google Scholar]
- 8.Liu M, Sanyal S, Gao G, Gurung IS, Zhu X, Gaconnet G, Kerchner LJ, Shang LL, Huang CL, Grace A, London B, Dudley SC., Jr Cardiac Na+ current regulation by pyridine nucleotides. Circ Res. 2009;105:737–745. doi: 10.1161/CIRCRESAHA.109.197277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends Biochem Sci. 2007;32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Pollak N, Dolle C, Ziegler M. The power to reduce: pyridine nucleotides--small molecules with a multitude of functions. Biochem J. 2007;402:205–218. doi: 10.1042/BJ20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu CP, Oka S, Shao D, Hariharan N, Sadoshima J. Nicotinamide phosphoribosyltransferase regulates cell survival through NAD+ synthesis in cardiac myocytes. Circ Res. 2009;105:481–491. doi: 10.1161/CIRCRESAHA.109.203703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai S. “Clocks” in the NAD World: NAD as a metabolic oscillator for the regulation of metabolism and aging. Biochim Biophys Acta. 2010;1804:1584–1590. doi: 10.1016/j.bbapap.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005;85:1093–1129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- 16.Lionetti V, Stanley WC, Recchia FA. Modulating fatty acid oxidation in heart failure. Cardiovasc Res. 2011;90:202–209. doi: 10.1093/cvr/cvr038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopaschuk GD, Ussher JR, Folmes CD, Jaswal JS, Stanley WC. Myocardial fatty acid metabolism in health and disease. Physiol Rev. 2010;90:207–258. doi: 10.1152/physrev.00015.2009. [DOI] [PubMed] [Google Scholar]
- 18.Srivastava SK, Ramana KV, Bhatnagar A. Role of aldose reductase and oxidative damage in diabetes and the consequent potential for therapeutic options. Endocr Rev. 2005;26:380–392. doi: 10.1210/er.2004-0028. [DOI] [PubMed] [Google Scholar]
- 19.Ago T, Kuroda J, Pain J, Fu C, Li H, Sadoshima J. Upregulation of Nox4 by hypertrophic stimuli promotes apoptosis and mitochondrial dysfunction in cardiac myocytes. Circ Res. 2010;106:1253–1264. doi: 10.1161/CIRCRESAHA.109.213116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuroda J, Ago T, Matsushima S, Zhai P, Schneider MD, Sadoshima J. NADPH oxidase 4 (Nox4) is a major source of oxidative stress in the failing heart. Proc Natl Acad Sci U S A. 2010;107:15565–15570. doi: 10.1073/pnas.1002178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Brewer AC, Schroder K, Santos CX, Grieve DJ, Wang M, Anilkumar N, Yu B, Dong X, Walker SJ, Brandes RP, Shah AM. NADPH oxidase-4 mediates protection against chronic load-induced stress in mouse hearts by enhancing angiogenesis. Proc Natl Acad Sci U S A. 2010;107:18121–18126. doi: 10.1073/pnas.1009700107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickinson DA, Forman HJ. Cellular glutathione and thiols metabolism. Biochem Pharmacol. 2002;64:1019–1026. doi: 10.1016/s0006-2952(02)01172-3. [DOI] [PubMed] [Google Scholar]
- 23.Ago T, Sadoshima J. Thioredoxin and ventricular remodeling. J Mol Cell Cardiol. 2006;41:762–773. doi: 10.1016/j.yjmcc.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franco R, Cidlowski JA. Apoptosis and glutathione: beyond an antioxidant. Cell Death Differ. 2009;16:1303–1314. doi: 10.1038/cdd.2009.107. [DOI] [PubMed] [Google Scholar]
- 25.Ago T, Sadoshima J. Thioredoxin1 as a negative regulator of cardiac hypertrophy. Antioxid Redox Signal. 2007;9:679–687. doi: 10.1089/ars.2007.1529. [DOI] [PubMed] [Google Scholar]
- 26.Matsushima S, Zablocki D, Sadoshima J. Application of recombinant thioredoxin1 for treatment of heart disease. J Mol Cell Cardiol. 2011;51:570–573. doi: 10.1016/j.yjmcc.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oka S, Ago T, Kitazono T, Zablocki D, Sadoshima J. The role of redox modulation of class II histone deacetylases in mediating pathological cardiac hypertrophy. J Mol Med. 2009;87:785–791. doi: 10.1007/s00109-009-0471-2. [DOI] [PubMed] [Google Scholar]
- 28.Gupte SA, Levine RJ, Gupte RS, Young ME, Lionetti V, Labinskyy V, Floyd BC, Ojaimi C, Bellomo M, Wolin MS, Recchia FA. Glucose-6-phosphate dehydrogenase-derived NADPH fuels superoxide production in the failing heart. J Mol Cell Cardiol. 2006;41:340–349. doi: 10.1016/j.yjmcc.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Rajasekaran NS, Connell P, Christians ES, Yan LJ, Taylor RP, Orosz A, Zhang XQ, Stevenson TJ, Peshock RM, Leopold JA, Barry WH, Loscalzo J, Odelberg SJ, Benjamin IJ. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130:427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- 31.Erickson JR, Joiner ML, Guan X, Kutschke W, Yang J, Oddis CV, Bartlett RK, Lowe JS, O’Donnell SE, Aykin-Burns N, Zimmerman MC, Zimmerman K, Ham AJ, Weiss RM, Spitz DR, Shea MA, Colbran RJ, Mohler PJ, Anderson ME. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133:462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aon MA, Cortassa S, O’Rourke B. Redox-optimized ROS balance: a unifying hypothesis. Biochim Biophys Acta. 2010;1797:865–877. doi: 10.1016/j.bbabio.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jagtap P, Szabo C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4:421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 34.Abd Elmageed ZY, Naura AS, Errami Y, Zerfaoui M. The poly(ADP-ribose) polymerases (PARPs): new roles in intracellular transport. Cell Signal. 2012;24:1–8. doi: 10.1016/j.cellsig.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp Neurol. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou HZ, Swanson RA, Simonis U, Ma X, Cecchini G, Gray MO. Poly(ADP-ribose) polymerase-1 hyperactivation and impairment of mitochondrial respiratory chain complex I function in reperfused mouse hearts. Am J Physiol Heart Circ Physiol. 2006;291:H714–723. doi: 10.1152/ajpheart.00823.2005. [DOI] [PubMed] [Google Scholar]
- 37.Choudhury S, Bae S, Ke Q, Lee JY, Kim J, Kang PM. Mitochondria to nucleus translocation of AIF in mice lacking Hsp70 during ischemia/reperfusion. Basic Res Cardiol. 2011;106:397–407. doi: 10.1007/s00395-011-0164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, Ido Y. AMPK and SIRT1: a long-standing partnership? Am J Physiol Endocrinol Metab. 298:E751–760. doi: 10.1152/ajpendo.00745.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto T, Sadoshima J. Protection of the heart against ischemia/reperfusion by silent information regulator 1. Trends Cardiovasc Med. 2011;21:27–32. doi: 10.1016/j.tcm.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 41.Nadtochiy SM, Redman E, Rahman I, Brookes PS. Lysine deacetylation in ischaemic preconditioning: the role of SIRT1. Cardiovasc Res. 2011;89:643–649. doi: 10.1093/cvr/cvq287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu CP, Zhai P, Yamamoto T, Maejima Y, Matsushima S, Hariharan N, Shao D, Takagi H, Oka S, Sadoshima J. Silent information regulator 1 protects the heart from ischemia/reperfusion. Circulation. 2010;122:2170–2182. doi: 10.1161/CIRCULATIONAHA.110.958033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 Plays an Essential Role in Mediating Starvation-Induced Autophagy in Cardiac Myocytes. Circ Res. 2010;107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oka S, Alcendor R, Zhai P, Park JY, Shao D, Cho J, Yamamoto T, Tian B, Sadoshima J. PPARalpha-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway. Cell Metab. 2011;14:598–611. doi: 10.1016/j.cmet.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hafner AV, Dai J, Gomes AP, Xiao CY, Palmeira CM, Rosenzweig A, Sinclair DA. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging (Albany NY) 2010;2:914–923. doi: 10.18632/aging.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott I, Webster BR, Li JH, Sack MN. Identification of a molecular component of the mitochondrial acetyltransferase programme: a novel role for GCN5L1. Biochem J. 2012;443:655–661. doi: 10.1042/BJ20120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baur JA. Resveratrol, sirtuins, and the promise of a DR mimetic. Mech Ageing Dev. 2010;131:261–269. doi: 10.1016/j.mad.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tokube K, Kiyosue T, Arita M. Openings of cardiac KATP channel by oxygen free radicals produced by xanthine oxidase reaction. Am J Physiol. 1996;271:H478–489. doi: 10.1152/ajpheart.1996.271.2.H478. [DOI] [PubMed] [Google Scholar]
- 50.Tamsett TJ, Picchione KE, Bhattacharjee A. NAD+ activates KNa channels in dorsal root ganglion neurons. J Neurosci. 2009;29:5127–5134. doi: 10.1523/JNEUROSCI.0859-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stutts MJ, Gabriel SE, Price EM, Sarkadi B, Olsen JC, Boucher RC. Pyridine nucleotide redox potential modulates cystic fibrosis transmembrane conductance regulator Cl- conductance. J Biol Chem. 1994;269:8667–8674. [PubMed] [Google Scholar]
- 52.Harrington MA, Kopito RR. Cysteine residues in the nucleotide binding domains regulate the conductance state of CFTR channels. Biophys J. 2002;82:1278–1292. doi: 10.1016/S0006-3495(02)75484-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu M, Liu H, Dudley SC., Jr Reactive oxygen species originating from mitochondria regulate the cardiac sodium channel. Circ Res. 2010;107:967–974. doi: 10.1161/CIRCRESAHA.110.220673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zima AV, Copello JA, Blatter LA. Effects of cytosolic NADH/NAD(+) levels on sarcoplasmic reticulum Ca(2+) release in permeabilized rat ventricular myocytes. J Physiol. 2004;555:727–741. doi: 10.1113/jphysiol.2003.055848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belevych AE, Terentyev D, Viatchenko-Karpinski S, Terentyeva R, Sridhar A, Nishijima Y, Wilson LD, Cardounel AJ, Laurita KR, Carnes CA, Billman GE, Gyorke S. Redox modification of ryanodine receptors underlies calcium alternans in a canine model of sudden cardiac death. Cardiovasc Res. 2009;84:387–395. doi: 10.1093/cvr/cvp246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cho J, Won K, Wu D, Soong Y, Liu S, Szeto HH, Hong MK. Potent mitochondria-targeted peptides reduce myocardial infarction in rats. Coron Artery Dis. 2007;18:215–220. doi: 10.1097/01.mca.0000236285.71683.b6. [DOI] [PubMed] [Google Scholar]