Abstract
Pyridine nucleotides (PNs), such as NAD(H) and NADP(H), mediate electron transfer in many catabolic and anabolic processes. In general, NAD+ and NADP+ receive electrons to become NADH and NADPH by coupling with catabolic processes. These electrons are utilized for biologically essential reactions such as ATP production, anabolism and cellular oxidation-reduction (redox) regulation. Thus, in addition to ATP, NADH and NADPH could be defined as high-energy intermediates and “molecular units of currency” in energy transfer. We discuss the significance of PNs as energy/electron transporters and signal transducers, in regulating cell death and/or survival processes. In the first part of this review, we describe the role of NADH and NADPH as electron donors for NADPH oxidases (Noxs), glutathione (GSH) and thioredoxin (Trx) systems in cellular redox regulation. Noxs produce superoxide/hydrogen peroxide yielding oxidative environment, whereas GSH and Trx systems protect against oxidative stress. Then, we describe the role of NAD+ and NADH as signal transducers through NAD+ -dependent enzymes such as PARP-1 and Sirt1. PARP-1 is activated by damaged DNA in order to repair the DNA, which attenuates energy production through NAD+ consumption; Sirt1 is activated by an increased NAD+/NADH ratio in order to facilitate signal transduction for metabolic adaption, as well as stress responses. We conclude that PNs serve as an important interface for distinct cellular responses, including stress response, energy metabolism and cell survival/death.
Keywords: NADPH oxidases, Thioredoxin, Nampt, NAD+, sirtuins, PARP, apoptosis
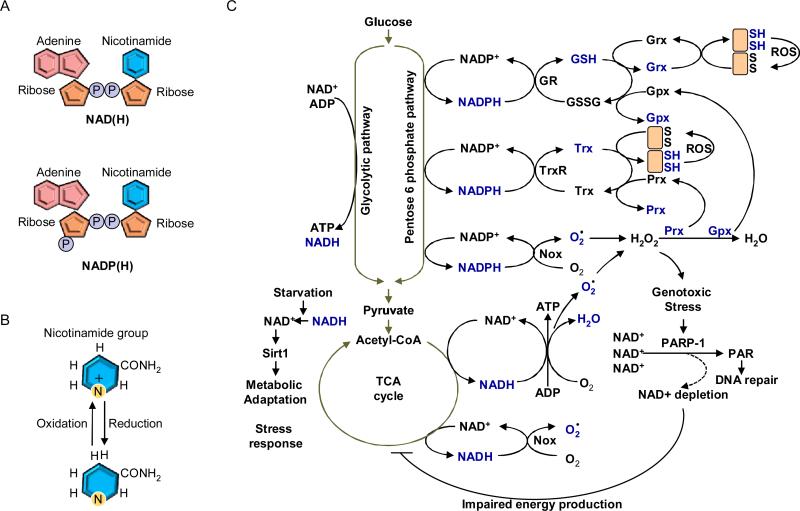
Pyridine is a basic heterocyclic organic compound with the chemical formula C5H5N. PNs are characterized by the presence of a pyridine derivative, such as nicotinamide, as a nitrogen base. For example, nicotinamide adenine dinucleotide (NAD+) is composed of two riboses and two nucleotides, including nicotinamide and adenine. Nicotinamide and adenine are connected to ribose with glycosidic linkage, and the ribose units bind each other through the phosphate groups (Figure 1A). NAD kinase phosphorylates 2’ of the ribose on the adenine side, which generates nicotinamide adenine dinucleotide phosphate (NADP+). Both NAD+ and NADP+ are subjected to two-electron reduction in the nicotinamide ring to be converted to NADH and NADPH, respectively (Figure 1B).
Figure 1.
(A) Schematic structures of NAD(H) and NADP(H). Adenine and nicotinamide connect to ribose unit. Those riboses connect each other through pyrophosphate. NADP(H) has a third phosphate group connected to the ribose unit of adenosine. (B) Oxidized and reduced forms of nicotinamide group. Nicotinamide group of NAD+ and NADP+ acquires two electrons to convert to NADH and NADPH, respectively. (C) Overview of the biological roles of PNs as electron carriers and signal mediators. NADP+ is reduced by the pentose phosphate pathway. The NADPH is utilized for scavenging ROS as well as reducing oxidized proteins through Trx and GSH systems. NAD+ is reduced by several catabolic pathways such as the glycolytic pathway and the tricarboxylic acid (TCA) cycle. The electron provided from NADH is transferred to the electron transport chain to convert ADP into ATP. Noxs utilize both NADH and NADPH as electron donors. Genotoxic stress including ROS, damages DNA, which is recognized by PARP-1. The highly activated PARP-1 facilitates poly ADP-ribosylation, an important step in DNA repair, resulting in depletion of NAD+. Energy production is impaired when NAD+ is depleted. Under starvation conditions, nutritional catabolism is attenuated, which results in an increased NAD+/NADH ratio resulting in activation of Sirt1. Grx: Glutharedoxin; Gpx: Gluthathione perodixdase; Prx: Peroxiredoxin.
The NAD(H) and NADP(H) are involved in many biological reactions as electron carriers. NAD(H) mainly participates in ATP production, while NADP(H) is utilized for modulating cellular reduction and oxidation (redox) status. Increasing lines of evidence suggest that the energy status and the redox status are the major mechanisms regulating survival and death of cardiovascular cell types. For example, the cellular level of remaining ATP is a crucial determinant for the cell to undergo either programmed cell death or necrosis in response to hypoxia. The level of oxidative stress regulated by PNs should affect the extent of cytochrome c release, mitochondrial permeability transition pore opening as well as endoplasmic reticulum stress, thereby serving the signaling mechanism mediating apoptosis and necrosis. Furthermore, NAD+ is used as a substrate for several enzymes, including poly(ADP-ribose) polymereases-1 (PARP-1) and sirtuins, whose targets and end-products critically regulate growth and death of cardiovascular cell types. Thus, the cellular level of PNs significantly affects survival and various forms of cell death through multiple mechanisms.
This review focuses on the roles of PNs in controlling cell death and survival in cardiovascular cells. In the first part, we discuss the role of PNs as electron donors for glutathione (GSH) and thioredoxin (Trx) systems as well as NAD(P)H oxidases (Noxs), key regulators of cellular redox states. In the second part, we discuss regulation of cellular levels of NAD+ and the role of NAD+ dependent enzymes, such as PARP-1 and sirtuins, which regulate cell survival and death through regulation of their target proteins as well as through consumption of NAD+.
Role of PNs in Cellular Redox Regulation as Electron Carriers
Excessive accumulation of reactive oxygen species (ROS), including superoxide (O2.), hydrogen peroxide (H2O2) and hydroxyl radical (OH.), is generally harmful to organisms because they react with cellular components, such as DNA, proteins and lipids. ROS are produced from various sources in cells, such as leakage from mitochondria, metabolic reactions, and irradiations 1. Oxidative damage of proteins and DNAs often triggers apoptotic cell death, while further production of ROS leads to mPTP opening, ATP depletion and eventual necrotic cell death 2. Increases in ROS are observed in response to a variety of stress in the heart, including ischemia/reperfusion (I/R) and hemodynamic overload, thereby mediating myocardial injury, hypertrophy and cardiac dysfunction 3. In order to protect against excessive ROS, cells possess an antioxidant systems, including glutathione (GSH) and thioredoxin (Trx), and reducing enzymes, such as superoxide dismutases and catalase 4.
NAD+ receives electrons to form NADH by coupling with several catabolic processes, such as the glycolytic pathway, β-oxidation and the TCA cycle. The free energy derived from NADH is utilized for conversion of ADP to ATP in mitochondria via the electron transport chain. NADP+ is converted to NADPH by the pentose phosphate pathway, which is an alternative glycolytic pathway. The electrons derived from NADPH are transferred to the GSH and Trx systems in order to reduce ROS and oxidized proteins. Thus, the cellular antioxidant machineries are maintained with energy provided by glucose catabolism through NADPH-mediated electron transport (Figure 1C). Importantly, however, electrons derived from both NADH and NADPH are also used to generate superoxide and hydrogen peroxide by NAD(P)H oxidases, which in turn stimulates oxidative stress 1. In the presence of oxidative stress, nitric oxide synthase (NOS), which normally produces NO by transfer of electrons from NADPH to O2- and oxidation of L-arginine to L-citrulline, produces superoxide due to uncoupling of the latter process 2. Mitochondrial electron transport chain in the failing heart produces more free radicals originated from NADH than that in the normal heart 3. Taken together, PNs play an important role in controlling the cellular redox status by serving as electron donors for both negative and positive regulators of oxidative stress.
NAD(P)H Oxidases (Noxs)
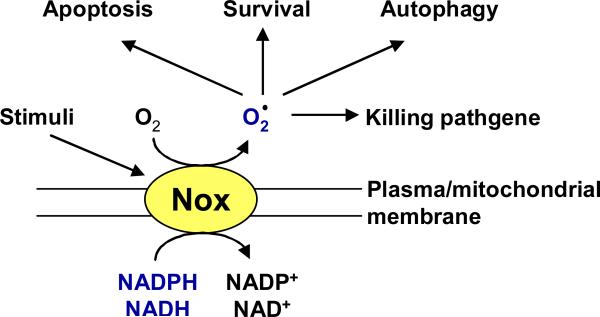
NADPH oxidases (Noxs) are transmembrane proteins that produce superoxide and/or hydrogen peroxide by transferring electrons provided by either NADH or NADPH to molecular oxygen. Noxs are either activated or upregulated by a variety of extracellular stimuli, including growth factors and cytokines. Thus, ROS production by Noxs is regulated acutely and chronically 6 (Figure 2). This is in contrast to electron leakage from mitochondria, which is a passive mechanism and does not appear to be regulated.
Figure 2.
Nox is activated by a wide variety of stimuli. The electrons derived from NADPH or NADH are transferred across the membrane to generate superoxide or hydrogen peroxide. The Nox derived ROS induce posttranslational modification of mitochondrial proteins and signaling molecules involved in cell survival, apoptosis or autophagy.
Currently, seven family members of Nox are identified which include Nox(s)1 to 5, Duox1 and Duox2 1. Among them, Nox2 and Nox4 are the major enzymes responsible for O2- production in CMs (Figure 2). Activation of Nox2 requires stimulus-induced membrane translocation of cytosolic regulatory subunits, including p47phox, p67phox, p40phox, and Rac1, a small GTPase 4. When cells are stimulated with agonists for G protein-coupled receptors, including the angiotensin II (Ang II) type 1 receptor (AT1-R), Rac1 and a ternary complex of p47phox, p67phox, and p40phox translocate to the membrane, where they form a functional complex with the Nox2-p22phox heterodimer, thereby initiating production of O2-5. Unlike Nox2, Nox4-mediated O2- generation does not require association with cytosolic factors, and Nox4 constitutively generates O2-6, 7.
The superoxide producing activity of Noxs is stimulated by various stresses in a regulated fashion, which in turn leads to cell death in cardiac myocytes. For example, mechanical stress activates Rac1, leading to NADPH oxidase activation in CMs 8. Angiotensin II (Ang II)-induced CM death is attenuated in p47phox knockout mice9. Although the identity of Nox remains to be clarified in these studies, regulation by the cytosolic factors suggests the involvement of the Nox2 isoform. Nox2 is activated by hyperglycemia in CMs, where the generated ROS promote apoptosis through activation of the c-Jun N-terminal kinase (JNK) 10.
Although the activity of Nox4 does not appear to be regulated by the cytosolic factors, expression of Nox4 is upregulated in response to aging, hypertrophic stimuli and heart failure 11. In neonatal rat CMs, overexpression of Nox4 primarily induces apoptosis, suggesting that the proapoptotic action of Nox4 is cell autonomous 11. Cardiac-specific overexpression of Nox4 enhances O2- production, mitochondrial dysfunction, as well as CM apoptosis in the middle-aged mouse heart in vivo12. Cardiac-specific Nox4 knockout mice show an ameliorated level of pressure overload-induced CM apoptosis and cardiac dysfunction 13. Taken together, Nox4-mediated ROS production promotes apoptosis in CMs.
Nox2 and Nox4 exhibit distinct subcellular localizations. Nox2 is expressed primarily on the plasma membrane, whereas Nox4 on intracellular membranes, including those in mitochondria, endoplasmic reticulum and nucleus 12, 13. Nox4 has a putative mitochondrial localization signal in the N-terminus domain, which is sufficient to direct Nox4 to the mitochondrial membrane 12. Consistently, oxidation of mitochondrial proteins in response to pressure overload is significantly attenuated in Nox4 KO mice. How Nox4 is inserted in the mitochondrial membrane remains to be shown. In our hands, Nox4 uses NADH more efficiently than NADPH as an electron donor 12, 13. However, other reports suggest that Nox4 directly generate H2O2 through a third cytosolic loop-dependent mechanism 14. Cardiac responses to pressure overload are quite different between Nox2 (systemic) KO mice and cardiac specific Nox4 KO mice 13, 15. Thus, ROS produced by Nox2 and Nox4 appear to function differently or in a cell type-dependent manner. The fact that Nox2 and Nox4 produce ROS at distinct subcellular localizations potentially explains the difference in their phenotype. In addition, Nox2 and Nox4 may produce distinct ROS, namely O2- vs H2O2, which could also contribute to the distinct cardiac effects between Nox2 and Nox4.
In addition to apoptotic signaling, Noxs are reported to prevent cell death through activation of cell survival signals, especially in cancer cells. Nox2 inhibitors induce cell death in epithelial ovarian cancer 16. Nox4 also prevents apoptosis, thereby acting as an oncoprotein in breast cancer cells 17 and pancreatic cancer cells 18. Interestingly, Nox4 can generate O2- in low O2 conditions. O2- produced by Nox4 during hypoxia inactivates proline hydroxylase and stabilizes HIF-1α, which in turn mediates cell survival and angiogenesis, both of which are protective against apoptosis. In fact, cardiac dysfunction induced by pressure overload was exacerbated due to insufficient angiogenesis in systemic Nox4 KO mice, suggesting that downregulation of Nox4 below physiological level could be detrimental in some experimental conditions 19, which might represent physiological functions of Noxs and may justify the fact that the heart express an active source of ROS in mitochondria.
GSH
GSH is a small antioxidant protein composed of tripeptide which is L-γ-glutamyl-L-cysteinyl-glycine. GSH exists in reduced (GSH) and oxidized (GSSG) forms. GSH carries electron provided by NADPH and donates a reducing equivalent (H+ e-) to reduce proteins with disulfide bonds. GSH is very abundant and exists in the mM order of concentration in cells. GSH is regenerated from GSSG in the presence of GSH reductase. GSH is synthesized through two ATP-dependent steps, namely the first step with glutamylcysteine-synthetase-(γ-GCS) and the second step with GSH synthase. GSH is essential for cell survival, as evidenced by the mortality rate of γ-GCS knockout mice, where the lack of this rate-limiting enzyme for GSH biosynthesis causes massive apoptotic cell death 20. Overexpression of γ-GCS inhibits TNFα- and UV-induced apoptosis in Hepa1 and HEK293 cells 21, 22. Increased GSH prevents apoptosis in murine hepatocytes and Cem cells, a leukemic T cell line23, 24, whereas GSH depletion, using buthionine sulfoximine (BSO), induces or potentiates apoptotic cell death in Cem cells 24. Interestingly, γ-GCS is a substrate of caspase 3 25. The cleavage of γ-GCS may have a role in facilitating the implementation of apoptosis.
The oxidation and reduction cycle between GSH and GSSG further couples with sub-antioxidant systems composed of glutathione-dependent peroxidase (Gpx) and glutaredoxin (Grx) (Figure 1C). These GSH-dependent antioxidant systems have reactive cysteine residues in the catalytic centers and function as carriers of electrons originated from PNs through GSH. Thus far, six family members of Gpx have been identified, namely Gpx1 to 6, and all Gpxs catalyze the reduction of H2O2 using electrons provided by GSH 26. Doxorubicin-induced CM death is exacerbated in Gpx1 knockout mice 27. Similarly, loss of Gpx2 increases apoptotic cell death in the intestine 28. Homozygous knockout of Gpx4 in mice results in embryonic lethality at E7.5 29. The Gpx4-null cell line is highly sensitive to oxidative stress 29. Mammalian cells have 4 Grxs including Grx1 to 3 and Grx5 30. Grx1 suppresses apoptosis-stimulating kinase 1 (ASK1) activation through direct interaction 31. In the presence of apoptotic stimuli, Grx1 is oxidized and dissociated from ASK1, thereby activating ASK1-induced apoptosis in MCF7 cells. Grx2 protects against oxidative stress induced by doxorubicin in HeLa cells 32.
Although the fact that excessive oxidative stress imposes many pathological influence in cardiovascular cell types has been established, excessive levels of an antioxidant or excessive reduction, termed reductive stress, has been recently recognized to be also toxic for the heart 33. αB-crystallin (CryAB) is a small heat shock protein abundantly expressed in CMs. An autosomal dominant missense mutation in CryAB (CryAB (R120G)) causes cataracts and desmin-related myopathy in humans. CryAB prevents aggregation of proteins inside CMs but proteins improperly unfold and form aggregates in the presence of CryAB(R120G) 34. In the presence of CryAB(R120G), glucose-6-phosphate dehydrogenase (G6PD), an enzyme producing NADPH, is upregulated, which in turn facilitate a conversion from GSSG to GSH. The enhancement of the cellular reducing power stimulates protein aggregation in CMs, since the reductive environment prevents correct protein folding in the ER 33, 35. Overexpression of heat shock protein 27 (Hsp27) also increased reductive stress and cardiomyopathy through increases in GSH/GSSG and upregulation of Gpx136. These results suggest that excessive production of the PN and increases in GSH/GSSG are detrimental for the heart. It is unclear, however, how dysregulation of chaperone leads to upregulation of G6PD and increases in GSH/GSSG. In addition, why the increase in NADPH leads to prominent reduction of GSH rather than reduction of Trx1 or induction of O2- is unknown. Furthermore, the destination of electrons originated from NADPH and GSH and their roles in mediating cardiomyopathy remains to be elucidated. It will be interesting to investigate posttranslational modification of cysteine residues of cardiac proteins in human desmin-related myopathy.
Trx System
Thioredoxin 1 (Trx1) is a 12KDa protein, which has conserved cysteine residues (Cys-32 and Cys-35) in the catalytic center. These two cysteine residues exist in a reduced state in the presence of NADPH, an electron donor, and Trx reductase1 (TrxR1). Trx1 reduces oxidized proteins by providing an electron. During this process, Trx1 transiently forms an inter-molecular disulfide bond with the target protein by transferring an electron from one of the conserved cysteine residues to the target protein. Trx1 then provides another electron from the other cysteine to reduce the intermolecular disulfide bond. This reaction is termed “thiol disulfide exchange reaction”. Importantly, electrons originated from NADPH are first provided to Trx1 and then to the oxidized target protein37. Trx1 has specific protein-protein interactions with intracellular signaling molecules and transcription factors. Thus, Trx1 is a carrier of electrons originated from the PN to deliver them to specific targets, which in turn exert a wide variety of function in cells (reviewed in 38, 39).
Trx1 was originally identified as an electron donor for ribonucleotide reductase which converts RNA to DNA in E.coli 37. Aside from Trx1, Grx1 also acts as an electron donor for ribonucleotide reductase in bacteria 40. In E.coli, the single gene mutant lacking either Trx1 or Grx1 is viable, but the double mutant is not viable in the minimum medium without reduced sulfate 41. In contrast to E.coli, single gene knockout of Trx1 in mice results in early embryonic lethality42. Thus, Trx1 is uniquely required for early differentiation and morphogenesis in mammalian cells.
Trx1 plays an important role in protecting against oxidative stress-induced cell death 43. Since the cellular concentration of GSH is approximately 1-10 mM 44, while that of Trx1 is at μM levels 45, it is unlikely that the protective effect of Trx1 is performed as a general free thiol buffer. Peroxiredoxins (Prxs) have been identified as Trx-dependent peroxidases 46. Trx1 acts as an electron donor for Prx, which converts H2O2 to H2O. During the identification and characterization of Prxs, it has been demonstrated that Trx1 itself does not effectively scavenge H2O2, but rather, Trx1-mediated peroxidase activity is highly dependent upon Prxs 46, 47. Since H2O2 is most abundant among ROS in cells, the insensitivity of Trx1 against H2O2 may be important for Trx1 to reduce its specific target proteins. This notion might be also supported by the fact that TrxR is even more resistant against oxidative inactivation compared to Trx1 48. Protein oxidation at specific cysteine residues alters the function or activity of target proteins. Trx1-mediated reduction of cysteine residues also exhibits specificity thereby acting as cell signaling mechanisms.
Several transcriptional regulators and other enzymes involved in cardiac hypertrophy and heart failure are directly reduced by Trx1. These include Redox factor-1 49, NE-F2 related factor (Nrf-2) 50, Nuclear factor-κB (NFκB) 51, p53 52, FoxO4 53 , Ras 54, methionine sulfoxide reductase 55, and class II histone deacetylases (HDACs) 11. Importantly, cysteine residues in these molecules are reduced by electrons originated from NADPH through Trx1. Since the major function of Trx1 in the heart is to suppress pathological hypertrophy and apoptosis, identifying the targets of Trx1 may allow us to identify additional key mediators of these pathological events in the heart 56. The role of Trx1-mediated posttranslational modification in mediating pathological hypertrophy and heart failure has been reviewed recently 38, 39.
Trx1 also mediates anti-apoptotic actions through other mechanisms as well. For example, Trx1 upregulates miroRNA-98/let-7, which in turn suppresses angiotensin II-induced hypertrophy and apoptosis in part though downregulation of cyclin D2 57. Trx1 also upregulates genes involved in both mitochondrial oxidative phosphorylation and the TCA cycle, thereby stimulating mitochondrial function in the heart 58. Trx1 directly interact with ASK1, a pro-apoptotic kinase, thereby inhibiting the kinase activity of ASK1 independently of cysteine modification.
Among Trx1-activating transcriptional factors, posttranslational redox regulation of NF-κB deserves further discussion. All NFκB family proteins possess a conserved motif, namely RXXRXRXXC. Oxidation of the cysteine residue attenuates its DNA-binding ability 59. Trx1 reduces the cysteine residue and enhances DNA binding of NFκB in the nucleus 51. In sharp contrast, the upstream signaling pathway leading to NFκB activation is activated by oxidative stress. ROS are produced by proinflammatory cytokines, such as TNFα, which leads to NFκB activation. Overexpression of Trx1 and treatment with N-acetyl cysteine, a plasma-membrane-permeable reducing agent, suppresses TNFκ-induced NFκB activation by inhibition of IKKs activation 60, 61. Thus, NFκB activation is enhanced by oxidative conditions in the cytosol and by reductive conditions in the nucleus. Interestingly, Trx1 translocates from the cytosol to the nucleus when stimulated by TNFα, Phorbol 12-myristate 13-acetete (PMA) and UVB 62. Forced expression of Trx1 in the nucleus enhances TNFα-induced NFκB activation 62. Thus, translocation of Trx1 from the cytosol to the nucleus facilitates NFκB activation by providing an oxidative environment in the cytosol and a reductive environment in the nucleus. Although the mechanism responsible for inhibition of IKK activation by cytosolic-Trx1 has not yet been determined, Trx1 may scavenge H2O2 through reduction of Prx1.
Thioredoxin 2 (Trx2)
Oxidative stress in mitochondria plays an important role in mediating mitochondrial dysfunction and cell death during aging and heart failure. Superoxide produced from molecular oxygen at electron transport chain or Nox4 is rapidly dismuted to hydrogen peroxide, whereas mitochondrial hydrogen peroxide is detoxified by the GSH and Trx2 systems. Trx2 is a member of the Trx family and is localized primarily in mitochondria. The GSH and Trx2 systems reduce H2O2 through Grxs 1/4 and peroxiredoxin3, respectively. Importantly the reducing activity of the GSH and Trx2 systems is maintained by GSH-reductase and TrxR2, respectively in the presence of NADPH. A recent study showed that energized mitochondria have more reduced Trx2 and that the reducing activity of Trx2 critically depends upon TrxR2 63.
Loss of Trx2 leads to apoptotic cell death in association with increased ROS levels, cleavage and activation of casepase-3 and caspase-9 in the DT40 cell line 64, suggesting that Trx2 is an essential gene for cell survival by suppressing mitochondrium-dependent cell death. Gene disruption of Trx2 in mice causes massive apoptotic cell death resulting in embryonic lethality 65. Cardiac specific deletion of TrxR2 exhibited dilated cardiomyopathy and early lethality 66, suggesting that the TrxR2-Trx2-Prx2 system is critically important in the postnatal heart as well. Suppression of mitochondrial oxidative stress by mitochondrially overexpressed catalase prevents pathological hypertrophy and aging in the heart. Since the level of endogenous catalase is low in mitochondria, intervention to upregulates endogenous mitochondrial Trx system could be an alternative method to prevent pathological events in the heart in elderly patients.
Competition of NADPH/NADH between Nox4 and Mitochondrial Enzyme
Currently, how distribution of electrons from the donor (NADPH/NADH) to the recipients (Noxs, Trx and GSH) is controlled is not well understood. A recent study showed, however, that Nox4 and G6PD are observed in the overlapping microdomains within nucleus and that G6PD controls Nox4 activity through nuclear production of NADPH in the liver, suggesting that G6PD and Nox4 are coupled locally 67. It is possible that consumption of PNs by Noxs may attenuate regeneration of Trx and GSH, thereby initiating a vicious cycle of oxidative stress, although a recent study showed that overexpression of Nox4 paradoxically increases GSH/GSSG in the heart 68.
The biological toxicity of O2- produced by Noxs is due to its capacity to inactivate the iron-sulfur cluster containing enzymes (which are critical in a wide variety of metabolic pathways, including the TCA cycle and the mitochondrial electron transport chain) 69, thereby liberating free iron in the cell, which can undergo Fenton chemistry and generate the highly reactive hydroxyl radical. In fact, a series of molecules in the TCA cycle are strongly oxidized in the mitochondrial fraction prepared from aging Nox4 overexpression hearts 12, and pressure overload-induced inhibition of aconitase activity was attenuated in Nox4 KO mice. Since Nox4 preferentially utilizes NADH as an electron donor 7, Nox4 may directly regulate the NADH/FADH2 generating enzymes in the TCA cycle by oxidizing them, thereby initiating regulatory feedback mechanisms controlling their O2- producing activity in mitochondria. In addition, consumption of NADH by Nox4 may interfere with electron transport and affect ATP synthesis in mitochondria during heart failure. Further investigation is needed to elucidate the local regulation of mitochondrial enzymes by PNs.
Regulation of Cell Survival and Death by NAD+-Dependent Enzymes
There are enzymes that consume NAD+, such as poly(ADP-ribose) polymerases (PARPs) and sirtuins. Among PARP family proteins, PARP-1 has a strong impact on NAD+ consumption. Highly activated PARP-1 leads to depletion of NAD+ pools in cells. Since NAD+ is required for ATP production, depletion of NAD+ attenuates ATP production, resulting in cell death. A major role of PARP-1 is repairing damaged DNA. Whether or not activation of Sirt1, a member of the sirtuin family, has strong impacts on the cellular level of NAD+ remains to be shown. In general, low nutrition and/or energy deficiency increase NAD+/NADH ratio where Sirt1 is activated. Sirt1 plays an important role in regulating cell survival and death and metabolic responses to caloric restriction and fasting. Activation of Sirt1 allows cells to alleviate the metabolic stress, and, thus, overconsumption of NAD+ may not take place. Taken together, PNs regulate cell survival and death by regulating the activity of the NAD+-dependent enzymes. The level of PNs could be reduced as a result of hyperactivation of the NAD+-dependent enzymes. The cellular level of NAD+ affects cell survival and death either by directly affecting the energy status or secondarily affecting other NAD+-dependent enzymes. In the following, we discuss 1) regulation of NAD+ in cells, 2) the function of Nampt, a key enzyme regulating the synthesis of NAD+, and 3) the function of NAD+-dependent enzymes, including PARP and Sirt1.
Regulation of NAD+
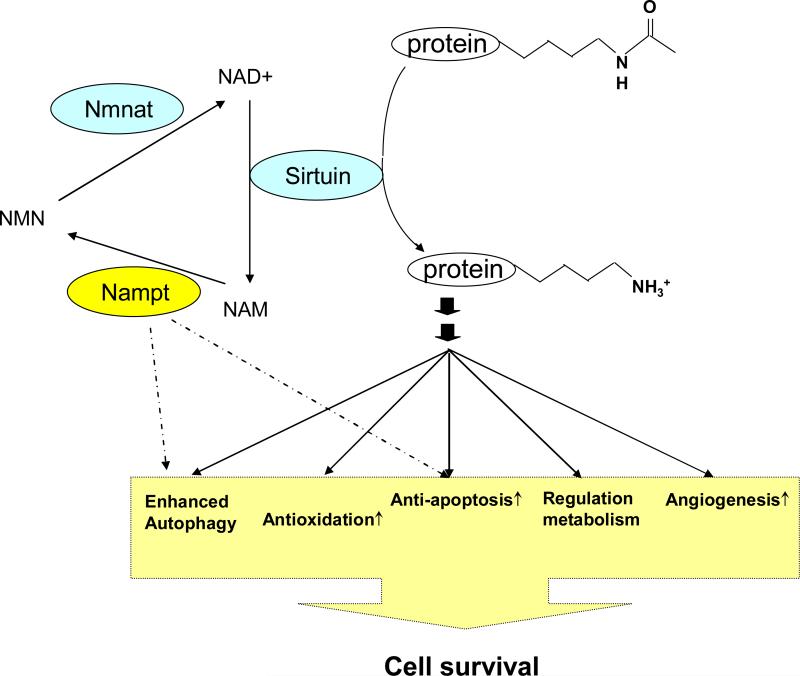
In mammals, NAD+ can be freshly synthesized from amino acids, including tryptophan or aspartic acid, via the de novo pathway 70 or taken up efficiently from the extracellular space 71. Importantly, NAD+ can also be re-synthesized from NAD+ metabolites through the salvage pathway 70. In the salvage pathway of NAD+ biosynthesis, Nampt was identified as a rate-limiting enzyme that converts nicotinamide into nicotinamide mononucleotide (NMN) 72, 73 (Figure 3). NMN is then directly synthesized into NAD+ by NMN adenylyltransferase (Nmnat) 74, 75. Nampt has been paid much attention because it increases the intracellular NAD+ level efficiently, and it has been proposed as a functional equivalent of pyrazinamidase/nicotinamidase 1 (PNC1), a master gene of longevity in yeast, in mammals. Upregulation of Nampt increases the cellular NAD+ level and enhances the activity of the catalytic domain of Sirt1 in mouse fibroblasts 73. It has been shown that there are large differences in the mitochondrial and cytosolic compartmentalization of NAD+ in different cell types 76. This implies that NAD+ concentration could be different among different compartments in the cell76. An isoform of Nmnat, termed NMNAT3, is enriched in mitochondria 77, suggesting that production of NAD+ may be regulated in a subcellular compartment.
Figure 3.
The salvage pathway of NAD+ biosynthesis facilitates cell survival through augmentation of sirtuin function. NAM, a natural inhibitor of sirtuin, is generated by sirtuin-mediated deacetylation of target proteins. Nampt converts from NAM to NMN, which is the rate limiting reaction in the salvage pathway of NAD+ biosynthesis.
Nampt
The gene that encodes for Nampt/PBEF was isolated from a human peripheral blood lymphocyte cDNA library termed pre-B-cell colony-enhancing factor (PBEF), because it was thought to be a cytokine that enhances the effect of interleukin-7 (IL-7) and stem cell factors (SCFs) on pre-B-cell colony formation 78. Later, on the basis of significant homology between PBEF and nadV, a prokaryotic nicotinamide phosphoribosyltranferase (NAmPRTase) from Haemophilus ducreyi, PBEF was identified as Nampt, an enzyme involved in NAD+ biosynthesis 72, 79. Visfatin, one of the adipocytokines, was also found to be an identical molecule as Nampt. Although it was originally proposed that Visfatin has insulin-mimetic effects in cultured cells, Nampt was found later to be a regulator of insulin secretion, rather than an insulin-mimetic, in pancreatic β-cells 80. Nampt is a highly conserved 52-kDa protein with no signal sequence, but it exists in most cells and tissues 78. In addition to its major role in intracellular NAD+ biosynthesis, Nampt also acts as an extracellular NAD+ biosynthetic enzyme whose secretion is observed in neutrophils, adipocytes and mesangial cells80.
Nampt is upregulated in neutrophils in response to a variety of inflammatory stimuli 81 in smooth muscle cells (SMCs) during maturation, and it lessens SMC apoptosis 82. Recently, Nampt was found to be upregulated in primary neonatal rat CMs under specific stresses (in vitro) and nutrient restriction (in vitro), and it was even upregulated in the liver tissue of fasting Sprague-Dawley rats 83. Nampt was also shown to protect against necrosis and apoptosis through mitochondrial NAD+ -dependent deacetylases Sirt3 and Sirt4 83. Moreover, PARP expression and NAD+ consumption were significantly increased in light of the genetic instability in tumors 84, and Nampt was shown to be upregulated in colorectal cancer 85. These findings suggest that Nampt is a stress-responsive protein that protects cells against injury and promotes cell survival. However, expression of Nampt in the heart was downregulated in response to some pathological stimuli in our study (in vivo) 86, and Dahl et al. also noted a decrease in the serum levels and hepatic mRNA levels of Nampt in nonalcoholic fatty liver disease 87. Hence, the regulation of Nampt is stimulus- as well as tissue-specific.
The NAD+ salvage pathway was recently found to be involved in circadian rhythm regulation through epigenetic control and chromatin remodeling 88, 89. The percent change of intracellular NAD+ concentration in liver tissue has a maximum of only 50%; however, it can accurately regulate the activity of Sirt1 in the circadian cycle. Furthermore, the transcription of Nampt may be modulated by the Sirt1-Clock:Bmal1 via a feedback loop. These pieces of evidence point out that the intracellular coupling of Nampt and Sirt1 must be intimate even without direct physical interaction.
Regulation of Oxidative Stress
In the human aortic endothelial cell (HAEC), Nampt can reduce cellular accumulation of ROS, an important cause of senescence, aging and cell death, in response to low and high glucose at least partly through sirtuin-dependent mechanisms. Overexpression of Nampt enhances proliferation, replicative lifespan and genomic stability in HAEC 90. Expression and activity of Nampt declined with senescence of human SMCs where Nampt protects against oxidative stress via enhanced Sirt1 activity and p53 degradation 91. Cardiac-specific overexpression of Nampt in transgenic mice increases NAD+ content in the heart reduces the size of myocardial infarction and apoptosis in response to prolonged ischemia and ischemia/reperfusion, which evoked a great impact on oxidative stress 86. Genetic disruption of angiotensin II type1 receptor (AT1R) promotes prolongation of lifespan in mice in vivo. The heart, aorta and kidneys from AT1R-/- mice displayed less oxidative damage than wild-type mice, which was associated with an increased number of mitochondria and upregulation of Nampt and Sirt3 in the kidney 92. Nampt can sustain intracellular NAD+ availability, even in mitochondria, through more efficient NAD+ production, when stresses induce consumption of NAD+ and ATP. It should be noted that there are some exceptions. Overexpression of Nampt increases its interactions with NADH dehydrogenase subunit 1, ferritin light chain, and interferon induced transmembrane 3, each of which are involved in oxidative stress and inflammation. Consequently, intracellular oxidative stress in human pulmonary vascular endothelial cells is increased, which may convey a mechanism of Nampt in the pathogenesis of acute lung injury 93. Regardless of whether it can universally decrease oxidative stress or not, Nampt indeed modulates oxidative activity.
Metabolic Effects
Nampt and NAD+ regulate the activity of sirtuins 73. Sirt1 can deacetylate many targets, thereby participating in a wide array of cellular processes 94. Some of its targets have metabolic effects, including PGC-1α, the FOXO family of transcription factors and acetyl-CoA synthetase 1 (AceCS1). Therefore, it is logically assumed that Nampt can modulate metabolic effects through the activity of Sirt1.
Overexpression of Nampt elicits a modest increase in aerobic glycolysis in response to high glucose (30mM) media, which could be blocked in the presence of either FK-866, a chemical inhibitor for Nampt, or sirtinol, an inhibitor for Sirt1,but it does not affect lactate production, glucose transport and oxidation in HAEC 90. Fatty acid β-oxidation was marginally increased in the low glucose medium (5mM). This data suggests a subtle but significant preference for ATP generation through aerobic glycolysis in Nampt-overexpressing HAEC, which may require Sirt1 activity. These findings are consistent with those on the effects of Sirt1, such as the increase of fatty acid oxidation in response to low glucose concentrations in skeletal muscles 95, and the inhibition of fat storage and the triggering of lipolysis in fat cells through suppression of the nuclear receptor PPAR-γ during calorie restriction 96.
The detrimental effect of elevated free fatty acids (FFAs) on insulin sensitivity could be normalized by thiazolidinediones (TZDs) via increased secretion of Nampt in patients with type 2 diabetes mellitus. Nampt secretion was blocked by synthetic fatty acids and by inhibition of phosphatidylinositol 3-kinase (PI3K) or Akt in human adipocytes 97. Nampt could regulate insulin secretion in β-cells through Nampt-mediated systemic NAD+ biosynthesis, suggesting vital roles for Nampt and NAD+ in the regulation of glucose metabolism 80. The synthesis and secretion of Nampt could be increased by a high level of glucose stimuli in cultured mesangial cells (MCs). Nampt treatment induces a rapid uptake of glucose via increased glucose transporter-1 (GLUT-1) protein expression in cellular membranes, which could be blocked by siRNA against insulin receptors and FK866 98. Furthermore, glucose restriction inhibits skeletal myoblast differentiation via the activated AMPK-Nampt-Sirt1 pathway 99. PPARα activation and glucose downregulated hepatic Nampt expression in vitro and in vivo 87. Nampt could be regarded as a sensor of intracellular energetic conditions in response to nutritional availability. These pieces of metabolic evidence have provided a possible molecular pathway connecting Nampt to cell survival and life extension.
Protection against Cell Death
Nampt is induced and secreted by peripheral blood mononuclear cells in response to inflammatory stimuli (LPS, IL-1β and TNF-α). Nampt inhibits apoptotic cell death in neutrophils, which was associated with reduced activity of caspases-8 and -3, but not of caspase-9. This indicates that the death receptor pathway was attenuated 81. Nampt protects against cell death (including necrosis and apoptosis) through mitochondrial NAD+-dependent deacetylases Sirt3 and Sirt4 83. Enzymatic activity of Nampt is required for its resistance to oxidative (genotoxic) stress where Nampt prevents apoptosis-inducing factor (AIF) delocalization from the mitochondrial innermembrane space in response to methyl-N′-nitro-N′-nitrosoguanidine, which is a PARP-1-dependent (caspase-independent) cell death. Nampt may modulate cell viability by affecting an earlier event preceding AIF release 100. Intracellular Nampt inhibited Huh7 hepatocytes apoptosis through NAD+ synthesis 87. However, we have shown that Nampt positively regulates survival and that downregulation of Nampt induces the release of cytochrome c from mitochondrial intermembrane space to the cytosol in cardiac myocytes, suggesting that the mitochondrial component of apoptosis is involved in cell death induced by Nampt downregulation 86. We also showed that Sirt1 downregulation could cause apoptotic cell death in CMs 101. Furthermore, there is no additive effect on increases in TUNEL-positive myocytes after downregulation of both Nampt and Sirt1, suggesting that downregulation of Nampt may induce cardiac myocyte apoptosis via a common mechanism such as downregulation of Sirt1 86. As a whole, this implies that the effect of Nampt inhibition on the mode of cell death is stimulus-specific and cell type-dependent, and that NAD+ plays an important role in controlling cell survival.
Not all anti-apoptotic effects of Nampt can be explained by NAD+ biosynthetic activity. For example, NMN, but not Nampt, protected macrophages from ER stress-induced apoptosis by activating an interlukin-6(IL-6)/STAT3 signaling pathway, indicating that the activation does not require Nampt enzymatic activity. Rather, it includes a two-step sequential process: rapid induction of IL-6 secretion, followed by IL-6-mediated activation of the prosurvival signal transducer STAT3 102. However, the mechanism by which NMN triggers IL-6 secretion is currently unknown.
Stimulation of Autophagy
Autophagy, an evolutionarily conserved process for the bulk degradation of cytoplasmic components, not only serves as a cell survival mechanism in starving cells but also mediates autophagic cell death in some conditions, such as I/R 103. Billington et al. showed that the autophagic mechanism could be modulated by NAD+71. Recently, we found that downregulation of Nampt inhibited autophagic flux in rat neonatal cardiac myocytes, which was reversed by the exogenous application of NAD+. Meanwhile, there were no additive effects on autophagic flux after downregulation of both Nampt and Sirt1 86. It was proposed that Nampt may affect the level of NADH, an reducing equivalent in cells, which in turn affects the activity of the electron transport chain on the lysosomal membrane. If the function of the electron transport chain is inhibited, lysosomal acidification and autophagosome-lysosome fusion as well as lysosomal degradation may be impaired 104. This implies that the regulation of autophagy by Nampt is mediated through NAD+ and/or Sirt1 (See below).
Angiogenesis
Xenotransplantation of Nampt-overexpressing SMCs into immunodeficient mice displayed an increased capacity for the transplanted SMCs to mature and intimately invest nascent endothelial channels 82, suggesting that Nampt stimulates angiogenesis. Hypoxia induces Nampt expression through hypoxia-inducible factor-1α in MCF7 breast cancer cells 105. Nampt promotes migration, invasion and tube formation in human endothelial cells via activation of extracellular signal-regulated kinase 1/2-dependent FGF-2 expression 106 and/or STAT3-dependent endothelial IL-6 induction 107. Nampt induces angiogenesis through endothelial VEGF and MMP2/9 production 108. Furthermore, MCP-1 is pivotal in modulating Nampt-induced angiogenesis via NF-κB and PI3K pathways, and autocrine/paracrine mechanisms (via the CCR2 receptor) 109. Augmenting Nampt activity enhances angiogenic response via Sirt1-mediated repression of FoxO1 in HAEC 90. Taken together, these results demonstrate that Nampt promotes angiogenesis, an important mechanism for cell survival.
Clinical Implication
Nampt/NAD+ promotes cell survival not only in normal cells, but also in cancer cells. FK866 effectively induces apoptotic cell death by gradual depletion of the intracellular NAD+ level in tumor cells (HepG2) 110. A strong anti-angiogenic potency in FK-866 was seen in a murine renal cell carcinoma model 111. Thus, it is in theory reasonable to inhibit Nampt to potentiate the chemotherapeutic effect of other antineoplastic agents.112. CHS-828 (a pyridyl cyanoguanidines), a potent chemoagent, also kills cancer cells by depleting NAD+ via inhibition of Nampt function and NF-κB signaling 113. Both of those drugs are under evaluation in clinical trials for cancer treatment.
Nampt is upregulated and promotes survival of neutrophils during immune and inflammatory responses in both human and animal models 81, and it can induce leukocyte adhesion to endothelial cells and the aortic endothelium through induction of cell adhesion molecules, including ICAM-1 and VCAM-1 114. Proteinuria, which is an important predictor of endothelial dysfunction in early diabetic nephropathy, was associated with altered circulating levels of Nampt and adiponectin 115. Nampt could be a novel marker for endothelial damage in renal transplantation 116. Nampt can be regarded as a proinflammatory molecule, and its inhibitor, FK866, can effectively reduce the severity of symptoms in mice with collagen induced arthritis 117. Macrophage plays a key role in obesity-associated pathophysiology 118. Several lines of evidence support a positive correlation of serum or plasma Nampt with obesity and diabetes 119, 120, suggesting a potentially important role of Nampt in the pathogenesis of obesity and type 2 diabetes.
PARP-1
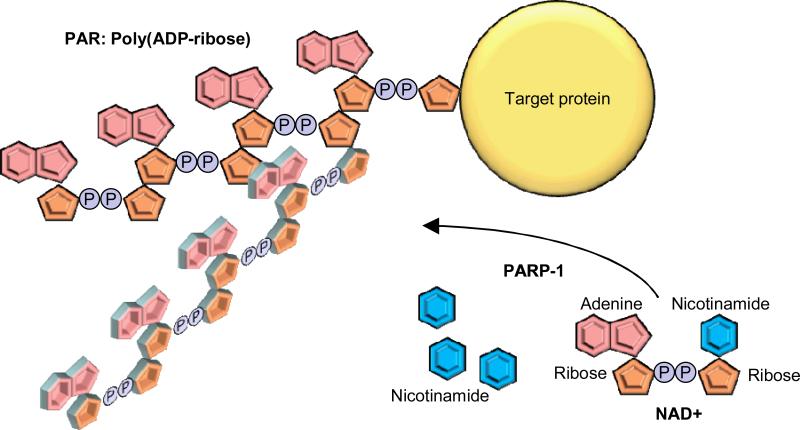
PARP-1 is an abundant nuclear protein, which polymerizes ADP-ribose on the target proteins, a reaction termed poly ADP-ribosylation (Figure 4) 121. The ADP-ribose units are derived from NAD+, and therefore, PARP-1 consumes NAD+. The enzymatic reaction also produces nicotinamide, a natural inhibitor for the PARP-1 and sirtuin families. Poly ADP ribose (PAR) polymers are attached to glutamate, aspartate or lysine residues on target proteins. PAR is negatively charged and acts as a posttranslational modifier. PARP-1 also produces free PAR polymers, which themselves act as a signal transducer. The significance of PARP-1 has been demonstrated in many cellular processes including DNA repair, genomic stability, cell death and survival, and transcriptional regulation. PARP-1 facilitates these functions through poly ADP-ribosylation, free PAR polymer formation, byproduct, protein-protein interaction and depletion of NAD+.
Figure 4.
Poly(ADP-ribosyl)ation (PAR) reaction. PARP-1 hydrolyses NAD+ to generate ADP-ribose units and nicotinamides. The ADP-ribose units are polymerized and attached to target proteins. The generated PAR could be linear or branched.
The Role of PARP-1 in Base Excision Repair (BER)
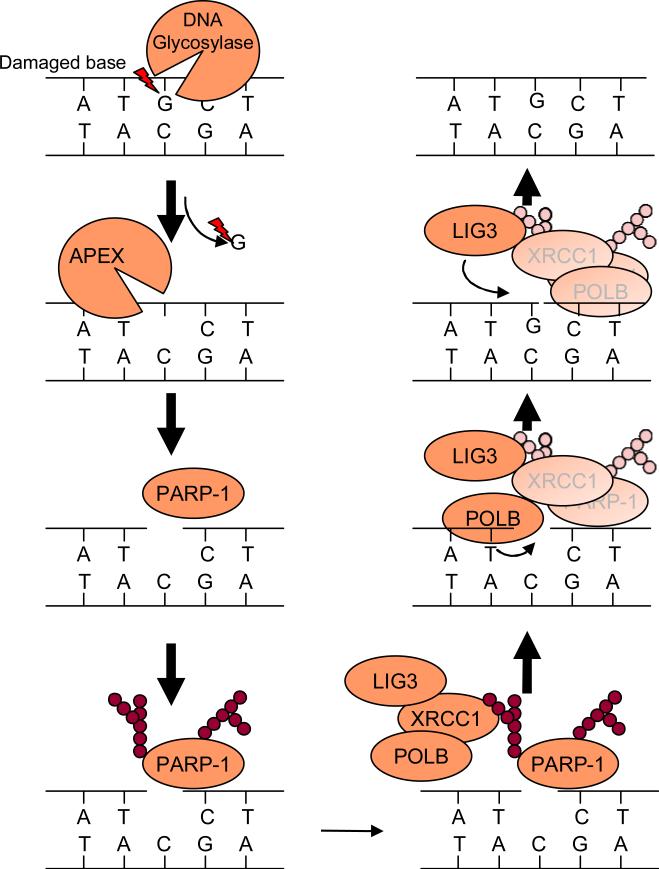
DNA damage is caused by multiple mechanisms, including oxidative damage and alkylation damage. DNA damage induces irregular pairing, which could eventually cause a mutation through incorporation of an incorrect nucleotide. The damaged DNA is repaired by a process termed base excision repair (BER) (Figure 5). In the first step of BER, damaged bases are recognized by DNA glycosylase and are removed by cleavage of an N-glycosidic bond. Next, Apurinic/apyrimidinic (AP) endonuclease cleaves the DNA backbone, thereby generating a single-strand DNA nick. PARP-1 recognizes the DNA nick as a single-strand break and facilitates poly ADP-ribosylation of target proteins. Foci of PAR can be detected within several minutes after hydrogen peroxide treatment or single-stranded DNA break induction 122. The PAR recruits X-ray repair cross-complementing 1 (XRCC1), an important scaffold protein for base excision repair. PARP-1 is subjected to self- poly ADP-ribosylation and binds to XRCC1 at the site of the single-strand DNA break 122. XRCC1 interacts with DNA polymerase II (polB) that fills the gap, and DNA ligase III that completes the repair process. PARP-1 knockout cells are highly sensitive to alkylating agents, toposiomerase inhibitors and γ-ray irradiation 123, consistent with the notion that PARP-1 plays an important role in mediating DNA repair. Double-strand DNA breaks are repaired by two distinct pathways: error-prone non homologous end-joining (NHEJ) and error-free homologous recombination (HR). The NHEJ process is initiated by the binding of the Ku70/80 heterodimer and DNA-activated protein kinase to the double-strand break end. Ku70 and Ku80 are subjected to PAR by PARP-1. The PAR decreases the affinity of Ku to the double-strand DNA break end, thereby inhibiting the NHEJ repair process 124. Thus, PARP-1 promotes HR by inhibition of NHEJ 125, since NHEJ and HR directly compete with each other 126.
Figure 5.
The involvement of PARP-1 in base excision repair (BER). Damaged bases are recognized and cleaved by appropriate DNA glycosidase. The cleavage of an N-glycosidic bond by DNA glycosidase results in generation of apurinic or apyrmidinic site (AP site). DNA AP endonuclease or DNA AP lyase cleave the DNA backbone to create a single-strand DNA nick. PARP-1 recognizes the DNA nick as a single-strand break. The DNA binding activates PARP-1 to poly ADP-ribosylate target proteins, such as histone and PARP-1 itself. XRCC1, an important scaffold protein for BER, is recruited to where PARP-1 facilitates PAR since XRCC1 has a high affinity for PAR. XRCC1 binds to and recruits DNA polymerase beta (POLB) and DNA ligase III (LIG3) to the DNA nick. Then POLB fills the GAP and LIG3 ligases.
PARP-1 Cleavage by Caspase-3
PARP-1 is a caspase-3 substrate. The caspase-dependent cleavage releases the N-terminal DNA binding domain from PARP-1 127. Since PARP-1 activity is highly dependent on DNA binding, the cleaved N-terminal fragment competitively inhibits the function of endogenous PARP-1 competing DNA binding with uncleaved PARP-1 128. The proteolytic inactivation of PARP-1 is achieved not only by breaking down intact PARP-1 to a non-functional form, but also by dominant negative inhibition by the cleaved product. Since PARP-1-mediated DNA repair is a crucial process for cell survival, the proteolytic inhibition of PARP-1 is likely important for ensuring apoptotic cell death. Indeed, PARP-1-induced BER is blocked by the N-terminal DNA-binding fragment of PARP-1 128. The PARP-1 inhibition is also important for preventing hyperactivation of PARP-1, which would otherwise result in depletion of cellular NAD+ as well as ATP. Apoptosis cannot be effectuated when cellular ATP levels are depleted, which results in necrosis 129. A specific DNase (CAD, caspase-activated DNase) that cleaves chromosomal DNA makes a complex with ICAD (inhibitor of CAD), which works as a specific chaperone for CAD. Caspase 3-cleaves ICAD and allows CAD to dissociate from ICAD and cleave chromosomal DNA. Caspase-mediated cleavage of PARP-1 may prevent hyperactivation of PARP-1 by fragmented DNA, which would induce necrosis. Indeed, the PARP-1 mutant, which cannot be cleaved by caspase-3, promotes necrotic cell death by TNF-α treatment 130.
Parthanatos
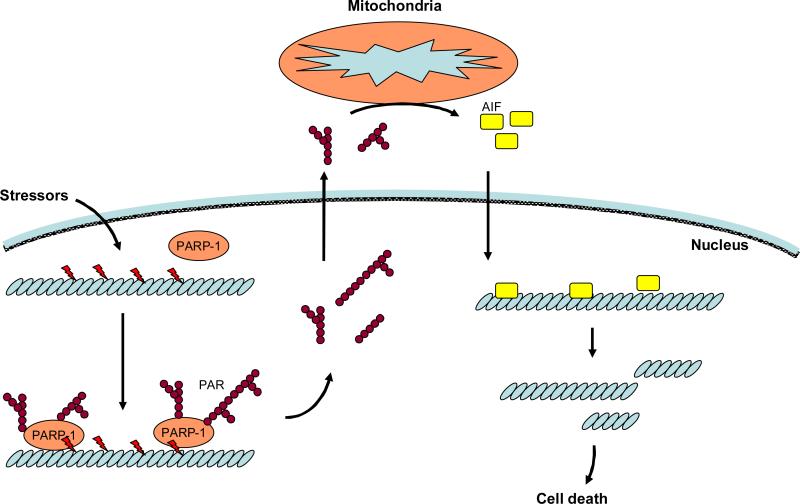
In response to low levels of genotoxic stress, PARP-1 promotes cell survival partly through DNA repair, while severe DNA damage triggers PARP-1 hyperactivation to induce cell death. At least two distinct mechanisms are proposed for PARP-1-induced cell death. Firstly, hyperactivation of PARP-1 results in ATP depletion, causing cells to undergo necrotic cell death. Secondly, PAR generation, stimulates AIF dependent programmed cell death, termed parthanatos (Figure 6). Parthanatos is induced by N-methl-D-asparate (NMDA) in neurons and is distinct from apoptosis and necrosis 131. Different from apoptosis, parthonatos does not form apoptotic bodies or induce small DNA fragmentation. The PARP-1-induced parthanatos cannot be prevented by pancaspase inhibitors, such as z-VAD-fmk, suggesting that parthanatos is a caspase-independent cell death program 132. Since hyperactivation of PARP-1 induces NAD+ depletion, PARP-1-induced cell death was presumed to result from intracellular energy depletion 129. However, PARP-1-induced cell death can take place even with a minor effect upon ATP content in some cases 133and, thus, ATP depletion may not be necessary for PARP-1-induced cell death. Instead, the essential role of PAR produced by PARP-1 has been proposed as the mechanism of cell death 131. The NMDA-induced cell death is attenuated by a neutralization antibody against PAR and by overexpression of PAR glycohydrolase, an enzyme that breaks PAR down. Exogenous PAR injection triggers AIF-dependent cell death. Thus, PAR itself is sufficient to induce cell death.
Figure 6.
PARP-1-induced caspase-independent cell death, termed parthanatos. Cell stressors, such as reactive oxygen species, cause DNA damage, which induces overactivation of PARP-1. PAR generated by PARP-1 activation translocates to the cytosol. Through direct interaction between PAR and mitochondria, apoptosis-inducing factor (AIF) is released from mitochondria and translocates to the nucleus. The AIF binds to DNA and induces large units of DNA fragmentation which is approximately 50 kbp.
The PAR generated by PARP-1 translocates from the nucleus to the cytosol. The PAR induces release of the AIF from mitochondria. The essential role of AIF in PARP-1-induced parthanatos has been proposed 132. Either knockdown of AIF or treatment of AIF neutralization antibody blocks PARP-1-induced cell death. Although a precise mechanism remains to be elucidated, direct interaction of PAR and mitochondria facilitates AIF release. Then, AIF translocates to the nucleus to promote DNA fragmentation; this fragmented DNA size is approximately 50 kbp, which is distinct from the small size of DNA fragmentation in apoptotic cell death. It remains unknown how nuclear AIF induces this large-scale DNA fragmentation.
Although PARP-1-mediated DNA repair is important for cell survival, NAD+ depletion and PAR promote either necrotic cell death or parthanatos. Whether PARP-1 induces cell survival or distinct forms of cell death depends upon the type of stress, the intensity of stress or the level of NAD+ consumption.
In the heart, pressure overload and I/R activates PARP-1 possibly through oxidative stress-induced DNA damage, which in turn induces translocation of AIF from mitochondria to the nucleus 134, 135 and consequent cell death. A PARP-1 inhibitor reduces AIF translocation, infarct size and cardiac dysfunction after I/R 134. Genetic deletion of PARP-1 in mice ameliorates I/R-induced cardiac and mitochondrial dysfunction 136. Downregulation of PARP-1 also prevents AIF translocation, cardiac hypertrophy and dysfunction in response to pressure overload 135. The beneficial effects of pharmacological and genetic inhibition of PARP-1 are also observed during cardiac dysfunction induced by septic shock 137, angiotensin II-induced cardiac hypertrophy and fibrosis 138, and doxorubicin-induced myocyte death and cardiac dysfunction 139, Interestingly, the beneficial effect of the ischemic preconditioning is abolished in PARP-1 knockout mice 140, suggesting that PARP-1 is protective in some experimental conditions. The function of PARP-1 in the heart critically depends upon the strength of the stress. For example, in the presence of mild to moderate stress, PARP-1 facilitates DNA repair to prevent further DNA damage and accumulation of mutations. At high levels of stress, cells undergo apoptotic cell death, where PARP-1 is subjected to proteolytic inactivation to restore NAD+ and ATP for effectuating apoptotic cell death. In the presence of more severe stress, hyperactivation of PAPR-1 facilitates necrotic cell death or parthanatos through depletion of NAD+/ATP.
Sirtuins
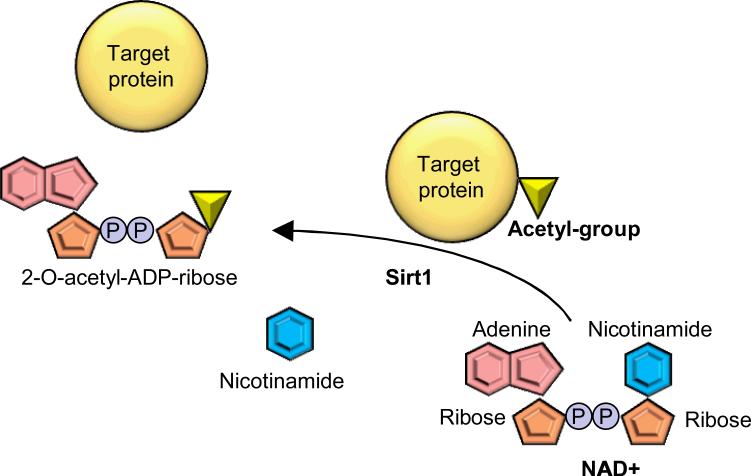
Since the function of sirtuins is discussed separately in this review series, here we discussed only briefly on their role in mediating cell survival/death. Sirt1 is an NAD+-dependent histone and protein deacetylase 141; an acetyl-group of target proteins is removed and conjugated to the ADP-ribose unit derived from NAD+. Nicotinamide is also produced as a byproduct in this enzymatic reaction (Figure 7). NADH acts as a competitive inhibitor of Sirt1, and therefore, an increased NAD+/NADH ratio is critical for Sirt1 activation 142. In mammalian cells, the NAD+/NADH ratio is increased by caloric restriction in several organs, which is associated with Sirt1 upregulation and activation 143. Sirtuin is also inhibited by nicotinamide, an NAD+ catabolite. Sirt1 is involved in a variety of biological reactions, including lipid and glucose metabolism, development, cell growth, immune reaction, autophagy, apoptosis and cancer development.
Figure 7.
Sirt1-mediated protein deacetylation. Sirt1 hydrolyses NAD+ to generate ADP-ribose units and nicotinamide. The acetyl group of the Sirt1 target proteins is removed and attached to the ADP-ribose to generate 2-O-acetyl-ADP-ribose.
Regulation of Apoptosis by Sirt1
In general, Sirt1 acts as an anti-apoptotic factor. Expression of Sirt1 is increased in both in vitro and in vivo model of cardiac hypertrophy 144. Sirt1 inhibition enhances cell death in cultured myocytes 144. Sirt1 is a negative regulator of the tumor suppressor protein p53 which induces apoptotic cell death in response to genotoxic stress. Acetylation of p53 leads to stabilization and activation of p53, which triggers apoptotic cell death and cell cycle arrest. Sirt1 deacetylates p53 thereby attenuating p53-mediated apoptotic cell death 145, 146. Sirt1 knockout in mice enhances radiation-induced acetylation of p53 in thymocytes 147. Interestingly, the Sirt1 knockout mice show developmental abnormalities in the heart, including septal and valvular defects 147. Cardiac-specific overexpression of Sirt1 prevents age-induced CM death in association with downregulation of p53 144. Sirt1 also regulates FoxO transcription factors, which involve in energy metabolism and cellular stress responses 148, 149. In response to oxidative stress and I/R, Sirt1 deacetylates FoxOs, which leads to nuclear translocation of FoxOs 86, 148. The nuclear FoxOs play an important role in resistance against ROS as well as apoptotic cell death. Cardiac-specific knockout of Sirt1 enhances apoptotic cell death in ischemia-reperfusion injury 150. The Sirt1 knockout mice show increased expression of proapoptotic proteins, such as Bax, decreased expression of anti-apoptotic proteins such as Bcl-xL, and decreased expression of anti-oxidants, such as Trx1 and MnSOD 150. Sirt1 promotes cell survival in the failing heart through upregulation of MnSOD151. Sirt1 also mediates ischemic preconditioning through lysine deacetylation of target proteins, such as p53, in the isolated perfused heart preparations 152. Sirt1 mediates cell survival through both transcriptional and posttranslational mechanisms.
Regulation of Autophagy by Sirt1
Increasing lines of evidence suggest that Sirt1 stimulates autophagy at baseline and in response to stress 153, 154. Perhaps, energy starvation increases NAD+/NADH, which in turn stimulates sirtuins, which could serve as a sensor for the energy status in cells. Glucose deprivation-induced autophagy is inhibited by knockdown of Sirt1, whereas overexpression of Sirt1 is sufficient to induce autophagy 155. The Sirt1-induced autophagy is mediated by FoxO1 155. Under glucose deprivation, FoxO1 is deacetylated and activated by Sirt1. A critical target gene of FoxO1 includes Rab7, which is a GTP-binding protein that promotes autophagosome-lysosomal fusion 154. Cardiac specific knockout of FoxO1 shows impaired cardiac function under starvation conditions, suggesting that Sirt1/FoxO1-mediated autophagy plays an important role in maintaining cardiac function under starvation conditions 154. It is interesting to note that Sirt1, FoxO and autophagy are all involved in lifespan extension and stress resistance in lower organisms. Together with the fact that Nampt also regulates autophagy in the heart through an NAD+-Sirt1-dependent mechanisms suggest that the PN-regulated signaling pathway is a fundamentally important mechanism controlling survival and death of cardiomyocytes and that autophagy takes part of it.
Antagonizing Effect between PARP-1 and Sirt1
PARP-1 and Sirt1 functionally antagonize one another. Both enzymes generate nicotinamide, a natural inhibitor for both Sirt1 and PARP-1. In response to DNA damage, PARP-1 localizes to the site of DNA damage, which leads to local PAR production, NAD+ depletion and nicotinamide generation. The local NAD+ depletion and nicotinamide generation induces histone acetylation through Sirt1 inhibition, which results in chromatin decondensation 156. This decondensated chromatin structure may be important for recruiting the DNA repair machinery and facilitating the repair process. In the failing heart, hyperactivation of PARP-1, is associated with NAD+ depletion and Sirt1 inactivation 138. Sirt1 function is enhanced when PARP-1 is inhibited. PARP-1 knockout mice show increased mitochondrial function, oxygen consumption and energy expenditure in a Sirt1-dependent manner 157.
On the other hand, Sirt1 activation results in reduced PARP-1 activity, whereas hydrogen peroxide-induced PARP-1 activation is enhanced in Sirt1 knockout cells 158. Loss of Sirt1 potentiates hydrogen peroxide-induced AIF nuclear translocation and cell death 158. Sirt1 directly inhibits PARP-1 through PARP-1 deacetylation 159. Acetylated PARP-1 shows high enzymatic activity independently of DNA damage. Sirt1 inhibits PARP-1-induced cell death through direct deacetylation of PARP-1 159. Sirt1 also attenuates PARP-1 promoter activity 159 and downregulates PARP-1 expression. In contrast to PARP-1's localization to active promoters, combined analyses of ChIP on ChIP and gene expression profile revealed that Sirt1 mainly localizes on inactive promoters 160. During hydrogen peroxide–induced DNA damage, PARP-1 interacts with chromatin whereas Sirt1 is released from chromatin 160. Such reciprocal interaction with chromatin between PARP-1 and Sirt1 may have a role in the DNA repair process and gene transcription. These results demonstrate a crucial link between Sirt1 and PARP-1 in stress response.
Conclusions
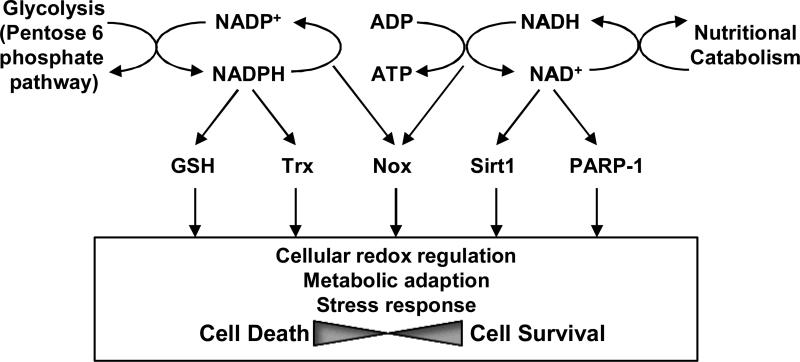
In summary, PNs are high-energy intermediates and crucial cofactors that protect against multiple stresses such as ROS and starvation, in many cardiovascular cell types (Figure 8). The functions of PNs are mediated primarily through the GSH and Trx systems, Noxs, Sirt1 and PARP1. During the past decade, a dramatic advancement was seen in the elucidation of the cellular functions of these PN-dependent molecules. As we summarized in this review, all of these molecules clearly play important roles in controlling cellular survival and death in cardiovascular cell types through regulation of cellular redox, energy metabolism and stress responses. However, there are still many outstanding questions. For example, the local coupling mechanism between NADPH/NADH and redox enzymes, namely GSH and Trx1/2, and that between Nampt and NAD+-dependent enzymes, including PARP-1 and sirtuins, remain to be elucidated. Although the source of PN production and the PN-dependent enzymes appear to be localized closely in subcellular compartments, essentially nothing is known regarding how they are coupled functionally in both physiological and pathological conditions. For example, all electrons utilized by the Trx and GSH systems are derived primarily from the pentose phosphate pathway via NADPH at physiological conditions. However, the source of PNs may be altered during pathological conditions. In the same token, more studies are needed to clarify how the activity of one PN-dependent enzyme affects the other PN-dependent ones. Whether activation Noxs could affect the other PNs-dependent process, including reduction of GSH/Trx and ATP synthesis through the mitochondrial electron transport chain remains to be clarified. More studies are needed to clarify the functional interaction between PARP-1 and Sirt1 in the heart since both PARP-1 and Sirt1profoundly affect cardiac function in the failing heart. Finally, more studies are needed to elucidate the functional significance of NAD+ production by Nampt in various cell types, subcellular compartments, as well as extracellular spaces, and the function of the sirtuin family members, using genetically altered mouse models. It is likely that such studies should allow us to identify a novel function of PNs in cardiovascular cell types.
Figure 8.
Schematic representation of PNs. NADH and NADPH are high-energy intermediates for the transfer of energy from nutritional catabolism to cellular redox regulation and ATP production coupled with electron transfer. NADPH is mainly used for maintaining cellular redox status, whereas NADH is used for ATP production. NAD+ is also used for providing ADP-ribose units for Sirt1 and PARP1, which is essential for the function of these enzymes. Although PNs have essential roles in cell survival, PARP-1 and Nox could be cell death inducers.
Acknowledgement
We thank Christopher Brady for critical reading of the manuscript.
Funding Sources
This work was supported in part by U.S. Public Health Service Grants HL59139, HL67724, HL69020, HL91469, HL102738, AG27211 and the Foundation of Leducq Transatlantic Network of Excellence
List of abbreviations
- AceCS1
Acetyl-CoA synthetase 1
- AIF
Apoptosis-inducing factor
- AMPK
5’adenosine monophosphate-activated protein kinase
- Ang II
Angiotensin II
- AP
Apurinic/apyrimidinic
- ASK1
Apoptosis signal regulating kinase 1
- AT1R
Ang II type 1 receptor
- Bcl-xL
B-cell lymphoma-extra large
- BER
Base excision repair
- BMAL1
brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like 1
- BSO
Buthionine sulfoximine
- CAD
Caspase-activated DNase
- CCR2
Chemokine (C-C motif) receptor 2
- ChIP
Chromatin immunoprecipitation
- CM
Cardiomyocytes
- CryAB
αB-crystallin
- ER
Endoplasmic reticulum
- FAD
Flavin adenine dinucleotide
- FGF
Fibroblast growth factor
- FoxO
Forkhead box protein O
- γ-GCS
γ-Glutamylcysteine-synthetase
- GLUT-1
Glucose transporter-1
- GSH
Glutathione
- GSSG
Glutathione disulfide
- Gpx
Glutathione dependent peroxidase
- Grx
Glutaredoxin
- G6PD
Glucose-6-phosphate dehydrogenase
- HAEC
Human aortic endothelial cell
- HDAC
Histone deacetylase
- HR
Homologous recombination
- Hsp
Heat shock protein
- ICAM-1
Inter-Cellular Adhesion Molecule 1
- IKK
IκB kinase
- IL
Interleukin
- IR
Ischemia/reperfusion
- JNK
c-Jun N-terminal kinase
- KO
Knockout
- LPS
Lipopolysaccharide
- MAPK
Mitogen-activated protein kinase
- MC
Mesangial cell
- MCP-1
Monocyte chemotactic protein-1
- MMP
Matrix metalloproteinase
- MnSOD
Manganase superoxide dismutase
- mPTP
Mitochondrial permeability transition pore
- NAmPRTase
Nicotinamide phosphoribosyltranferase
- Nampt
Nicotinamide phosphoribosyltransferase
- NFκB
Nuclear factor-κB
- NHEJ
Non homologous end-joining
- NMDA
N-methl-D-asparate
- NMN
Nicotinamide mononucleotide
- NMNAT
Nicotinamide mononucleotide adenylyltransferase
- NO
Nitrogen oxide
- Nox
NADPH oxidase
- PAR
Poly ADP ribose
- PARP-1
Poly [ADP-ribose] polymerase 1
- PBEF
Pre-B-cell colony-enhancing factor
- PGC-1α
PPAR-gamma coactivator-1α
- Phox
Phagocytic oxidase
- PI3K
Phosphatidylinositol 3-kinase
- PMA
Phorbol 12-myristate 13-acetete
- PN
Pyridine nucleotide
- PNC1
Pyrazinamidase/nicotinamidase 1
- PPAR
Peroxisome proliferator activated receptor
- Prx
Peroxiredoxin
- Rab7
Ras(Rat sarcoma)-related in brain
- ROS
Reactive oxygen species
- Sirt1
Silent information regulator 1
- SMC
Smooth muscle cell
- STAT
Signal transducer and activator of transcription
- TCA
Tricarboxylic acid
- TNFα
Tumor necrosis factor-α
- Trx
Thioredoxin
- TrxR
Thioredoxin reductase
- VCAM-1
Vascular cell adhesion protein 1
- VEGF
Vascular endothelial growth factor
- UV
Ultra violet
- XRCC1
X-ray repair cross-complementing 1
Footnotes
Disclosures
None
References
- 1.Maejima Y, Kuroda J, Matsushima S, Ago T, Sadoshima J. Regulation of myocardial growth and death by NADPH oxidase. J Mol Cell Cardiol. 2011;50:408–416. doi: 10.1016/j.yjmcc.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moens AL, Ketner EA, Takimoto E, Schmidt TS, O'Neill CA, Wolin MS, Alp NJ, Channon KM, Kass DA. Bi-modal dose-dependent cardiac response to tetrahydrobiopterin in pressure-overload induced hypertrophy and heart failure. J Mol Cell Cardiol. 2011 doi: 10.1016/j.yjmcc.2011.05.017. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ide T, Tsutsui H, Kinugawa S, Utsumi H, Kang D, Hattori N, Uchida K, Arimura K, Egashira K, Takeshita A. Mitochondrial electron transport complex I is a potential source of oxygen free radicals in the failing myocardium. Circ Res. 1999;85:357–363. doi: 10.1161/01.res.85.4.357. [DOI] [PubMed] [Google Scholar]
- 4.Abo A, Pick E, Hall A, Totty N, Teahan CG, Segal AW. Activation of the NADPH oxidase involves the small GTP-binding protein p21rac1. Nature. 1991;353:668–670. doi: 10.1038/353668a0. [DOI] [PubMed] [Google Scholar]
- 5.Ago T, Kuribayashi F, Hiroaki H, Takeya R, Ito T, Kohda D, Sumimoto H. Phosphorylation of p47phox directs phox homology domain from SH3 domain toward phosphoinositides, leading to phagocyte NADPH oxidase activation. Proc Natl Acad Sci U S A. 2003;100:4474–4479. doi: 10.1073/pnas.0735712100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambasta RK, Kumar P, Griendling KK, Schmidt HH, Busse R, Brandes RP. Direct interaction of the novel Nox proteins with p22phox is required for the formation of a functionally active NADPH oxidase. J Biol Chem. 2004;279:45935–45941. doi: 10.1074/jbc.M406486200. [DOI] [PubMed] [Google Scholar]
- 7.Shiose A, Kuroda J, Tsuruya K, Hirai M, Hirakata H, Naito S, Hattori M, Sakaki Y, Sumimoto H. A novel superoxide-producing NAD(P)H oxidase in kidney. J Biol Chem. 2001;276:1417–1423. doi: 10.1074/jbc.M007597200. [DOI] [PubMed] [Google Scholar]
- 8.Aikawa R, Nagai T, Tanaka M, Zou Y, Ishihara T, Takano H, Hasegawa H, Akazawa H, Mizukami M, Nagai R, Komuro I. Reactive oxygen species in mechanical stress-induced cardiac hypertrophy. Biochem Biophys Res Commun. 2001;289:901–907. doi: 10.1006/bbrc.2001.6068. [DOI] [PubMed] [Google Scholar]
- 9.Erickson JR, Joiner ML, Guan X, Kutschke W, Yang J, Oddis CV, Bartlett RK, Lowe JS, O'Donnell SE, Aykin-Burns N, Zimmerman MC, Zimmerman K, Ham AJ, Weiss RM, Spitz DR, Shea MA, Colbran RJ, Mohler PJ, Anderson ME. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133:462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuo WW, Wang WJ, Lin CW, Pai P, Lai TY, Tsai CY. NADPH oxidase-derived superoxide anion-induced apoptosis is mediated via the JNK-dependent activation of NF-kappaB in cardiomyocytes exposed to high glucose. J Cell Physiol. 2011 doi: 10.1002/jcp.22847. (in press) [DOI] [PubMed] [Google Scholar]
- 11.Ago T, Liu T, Zhai P, Chen W, Li H, Molkentin JD, Vatner SF, Sadoshima J. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 12.Ago T, Kuroda J, Pain J, Fu C, Li H, Sadoshima J. Upregulation of Nox4 by hypertrophic stimuli promotes apoptosis and mitochondrial dysfunction in cardiac myocytes. Circ Res. 2010;106:1253–1264. doi: 10.1161/CIRCRESAHA.109.213116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuroda J, Ago T, Matsushima S, Zhai P, Schneider MD, Sadoshima J. NADPH oxidase 4 (Nox4) is a major source of oxidative stress in the failing heart. Proc Natl Acad Sci U S A. 2010;107:15565–15570. doi: 10.1073/pnas.1002178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takac I, Schroder K, Zhang L, Lardy B, Anilkumar N, Lambeth JD, Shah AM, Morel F, Brandes RP. The E-loop is involved in hydrogen peroxide formation by the NADPH oxidase Nox4. J Biol Chem. 2011;286:13304–13313. doi: 10.1074/jbc.M110.192138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grieve DJ, Byrne JA, Siva A, Layland J, Johar S, Cave AC, Shah AM. Involvement of the nicotinamide adenosine dinucleotide phosphate oxidase isoform Nox2 in cardiac contractile dysfunction occurring in response to pressure overload. J Am Coll Cardiol. 2006;47:817–826. doi: 10.1016/j.jacc.2005.09.051. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Z, Fletcher NM, Ali-Fehmi R, Diamond MP, Abu-Soud HM, Munkarah AR, Saed GM. Modulation of redox signaling promotes apoptosis in epithelial ovarian cancer cells. Gynecol Oncol. 2011;122:418–423. doi: 10.1016/j.ygyno.2011.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham KA, Kulawiec M, Owens KM, Li X, Desouki MM, Chandra D, Singh KK. NADPH oxidase 4 is an oncoprotein localized to mitochondria. Cancer Biol Ther. 2010;10:223–231. doi: 10.4161/cbt.10.3.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edderkaoui M, Nitsche C, Zheng L, Pandol SJ, Gukovsky I, Gukovskaya AS. NADPH oxidase activation in pancreatic cancer cells is mediated through Akt-dependent up-regulation of p22phox. J Biol Chem. 2011;286:7779–7787. doi: 10.1074/jbc.M110.200063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang M, Brewer AC, Schroder K, Santos CX, Grieve DJ, Wang M, Anilkumar N, Yu B, Dong X, Walker SJ, Brandes RP, Shah AM. NADPH oxidase-4 mediates protection against chronic load-induced stress in mouse hearts by enhancing angiogenesis. Proc Natl Acad Sci U S A. 2010;107:18121–18126. doi: 10.1073/pnas.1009700107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalton TP, Chen Y, Schneider SN, Nebert DW, Shertzer HG. Genetically altered mice to evaluate glutathione homeostasis in health and disease. Free Radic Biol Med. 2004;37:1511–1526. doi: 10.1016/j.freeradbiomed.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 21.Botta D, Franklin CC, White CC, Krejsa CM, Dabrowski MJ, Pierce RH, Fausto N, Kavanagh TJ. Glutamate-cysteine ligase attenuates TNF-induced mitochondrial injury and apoptosis. Free Radic Biol Med. 2004;37:632–642. doi: 10.1016/j.freeradbiomed.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 22.Fan Y, Wu D, Jin L, Yin Z. Human glutamylcysteine synthetase protects HEK293 cells against UV-induced cell death through inhibition of c-Jun NH2-terminal kinase. Cell Biol Int. 2005;29:695–702. doi: 10.1016/j.cellbi.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Cazanave S, Berson A, Haouzi D, Vadrot N, Fau D, Grodet A, Letteron P, Feldmann G, El-Benna J, Fromenty B, Robin MA, Pessayre D. High hepatic glutathione stores alleviate Fas-induced apoptosis in mice. J Hepatol. 2007;46:858–868. doi: 10.1016/j.jhep.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 24.Friesen C, Kiess Y, Debatin KM. A critical role of glutathione in determining apoptosis sensitivity and resistance in leukemia cells. Cell Death Differ. 2004;11(Suppl 1):S73–85. doi: 10.1038/sj.cdd.4401431. [DOI] [PubMed] [Google Scholar]
- 25.Franklin CC, Krejsa CM, Pierce RH, White CC, Fausto N, Kavanagh TJ. Caspase-3-Dependent Cleavage of the Glutamate-L-Cysteine Ligase Catalytic Subunit during Apoptotic Cell Death. Am J Pathol. 2002;160:1887–1894. doi: 10.1016/S0002-9440(10)61135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei XG, Cheng WH, McClung JP. Metabolic regulation and function of glutathione peroxidase-1. Annu Rev Nutr. 2007;27:41–61. doi: 10.1146/annurev.nutr.27.061406.093716. [DOI] [PubMed] [Google Scholar]
- 27.Gao J, Xiong Y, Ho YS, Liu X, Chua CC, Xu X, Wang H, Hamdy R, Chua BH. Glutathione peroxidase 1-deficient mice are more susceptible to doxorubicin-induced cardiotoxicity. Biochim Biophys Acta. 2008;1783:2020–2029. doi: 10.1016/j.bbamcr.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Florian S, Krehl S, Loewinger M, Kipp A, Banning A, Esworthy S, Chu FF, Brigelius-Flohe R. Loss of GPx2 increases apoptosis, mitosis, and GPx1 expression in the intestine of mice. Free Radic Biol Med. 2010;49:1694–1702. doi: 10.1016/j.freeradbiomed.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, Motta L, Richardson A, Prolla TA. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 30.Lillig CH, Berndt C, Holmgren A. Glutaredoxin systems. Biochim Biophys Acta. 2008;1780:1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Song JJ, Rhee JG, Suntharalingam M, Walsh SA, Spitz DR, Lee YJ. Role of glutaredoxin in metabolic oxidative stress. Glutaredoxin as a sensor of oxidative stress mediated by H2O2. J Biol Chem. 2002;277:46566–46575. doi: 10.1074/jbc.M206826200. [DOI] [PubMed] [Google Scholar]
- 32.Lillig CH, Lonn ME, Enoksson M, Fernandes AP, Holmgren A. Short interfering RNA-mediated silencing of glutaredoxin 2 increases the sensitivity of HeLa cells toward doxorubicin and phenylarsine oxide. Proc Natl Acad Sci U S A. 2004;101:13227–13232. doi: 10.1073/pnas.0401896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajasekaran NS, Connell P, Christians ES, Yan LJ, Taylor RP, Orosz A, Zhang XQ, Stevenson TJ, Peshock RM, Leopold JA, Barry WH, Loscalzo J, Odelberg SJ, Benjamin IJ. Human alpha B-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130:427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vicart P, Caron A, Guicheney P, Li Z, Prevost MC, Faure A, Chateau D, Chapon F, Tome F, Dupret JM, Paulin D, Fardeau M. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Osinska H, Klevitsky R, Gerdes AM, Nieman M, Lorenz J, Hewett T, Robbins J. Expression of R120G-alphaB-crystallin causes aberrant desmin and alphaB-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89:84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Min X, Li C, Benjamin IJ, Qian B, Ding Z, Gao X, Yao Y, Ma Y, Cheng Y, Liu L. Involvement of Reductive Stress in the Cardiomyopathy in Transgenic Mice With Cardiac-Specific Overexpression of Heat Shock Protein 27. Hypertension. 2010;55:1412–1417. doi: 10.1161/HYPERTENSIONAHA.109.147066. [DOI] [PubMed] [Google Scholar]
- 37.Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 38.Ago T, Sadoshima J. Thioredoxin1 as a negative regulator of cardiac hypertrophy. Antioxid Redox Signal. 2007;9:679–687. doi: 10.1089/ars.2007.1529. [DOI] [PubMed] [Google Scholar]
- 39.Ago T, Sadoshima J. Thioredoxin and ventricular remodeling. J Mol Cell Cardiol. 2006;41:762–773. doi: 10.1016/j.yjmcc.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmgren A. Glutaredoxin from Escherichia coli and calf thymus. Methods Enzymol. 1985;113:525–540. doi: 10.1016/s0076-6879(85)13071-5. [DOI] [PubMed] [Google Scholar]
- 41.Russel M, Holmgren A. Construction and characterization of glutaredoxin-negative mutants of Escherichia coli. Proc Natl Acad Sci U S A. 1988;85:990–994. doi: 10.1073/pnas.85.4.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsui M, Oshima M, Oshima H, Takaku K, Maruyama T, Yodoi J, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse thioredoxin gene. Dev Biol. 1996;178:179–185. doi: 10.1006/dbio.1996.0208. [DOI] [PubMed] [Google Scholar]
- 43.Masutani H, Ueda S, Yodoi J. The thioredoxin system in retroviral infection and apoptosis. Cell Death Differ. 2005;12(Suppl 1):991–998. doi: 10.1038/sj.cdd.4401625. [DOI] [PubMed] [Google Scholar]
- 44.Franco R, Cidlowski JA. Apoptosis and glutathione: beyond an antioxidant. Cell Death Differ. 2009;16:1303–1314. doi: 10.1038/cdd.2009.107. [DOI] [PubMed] [Google Scholar]
- 45.Das KC, White CW. Detection of thioredoxin in human serum and biological samples using a sensitive sandwich ELISA with digoxigenin-labeled antibody. J Immunol Methods. 1998;211:9–20. doi: 10.1016/s0022-1759(97)00163-4. [DOI] [PubMed] [Google Scholar]
- 46.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 47.Netto LES, Chae HZ, Kang SW, Rhee SG, Stadtman ER. Removal of hydrogen peroxide by thiol-specific antioxidant enzyme (TSA) is involved with its antioxidant properties. TSA possesses thiol peroxidase activity. J Biol Chem. 1996;271:15315–15321. doi: 10.1074/jbc.271.26.15315. [DOI] [PubMed] [Google Scholar]
- 48.Starke DW, Chen Y, Bapna CP, Lesnefsky EJ, Mieyal JJ. Sensitivity of protein sulfhydryl repair enzymes to oxidative stress. Free Radic Biol Med. 1997;23:373–384. doi: 10.1016/s0891-5849(97)00009-9. [DOI] [PubMed] [Google Scholar]
- 49.Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. Embo J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen JM, Watson WH, Jones DP. Compartmentation of Nrf-2 redox control: regulation of cytoplasmic activation by glutathione and DNA binding by thioredoxin-1. Toxicol Sci. 2004;82:308–317. doi: 10.1093/toxsci/kfh231. [DOI] [PubMed] [Google Scholar]
- 51.Hayashi T, Ueno Y, Okamoto T. Oxidoreductive regulation of nuclear factor kappa B. Involvement of a cellular reducing catalyst thioredoxin. J Biol Chem. 1993;268:11380–11388. [PubMed] [Google Scholar]
- 52.Ueno M, Masutani H, Arai RJ, Yamauchi A, Hirota K, Sakai T, Inamoto T, Yamaoka Y, Yodoi J, Nikaido T. Thioredoxin-dependent redox regulation of p53-mediated p21 activation. J Biol Chem. 1999;274:35809–35815. doi: 10.1074/jbc.274.50.35809. [DOI] [PubMed] [Google Scholar]
- 53.Dansen TB, Smits LM, van Triest MH, de Keizer PL, van Leenen D, Koerkamp MG, Szypowska A, Meppelink A, Brenkman AB, Yodoi J, Holstege FC, Burgering BM. Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat Chem Biol. 2009;5:664–672. doi: 10.1038/nchembio.194. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto M, Yang G, Hong C, Liu J, Holle E, Yu X, Wagner T, Vatner SF, Sadoshima J. Inhibition of endogenous thioredoxin in the heart increases oxidative stress and cardiac hypertrophy. J Clin Invest. 2003;112:1395–1406. doi: 10.1172/JCI17700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lowther WT, Brot N, Weissbach H, Honek JF, Matthews BW. Thiol-disulfide exchange is involved in the catalytic mechanism of peptide methionine sulfoxide reductase. Proc Natl Acad Sci U S A. 2000;97:6463–6468. doi: 10.1073/pnas.97.12.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oka S, Ago T, Kitazono T, Zablocki D, Sadoshima J. The role of redox modulation of class II histone deacetylases in mediating pathological cardiac hypertrophy. J Mol Med (Berl) 2009;87:785–791. doi: 10.1007/s00109-009-0471-2. [DOI] [PubMed] [Google Scholar]
- 57.Yang Y, Ago T, Zhai P, Abdellatif M, Sadoshima J. Thioredoxin 1 negatively regulates angiotensin II-induced cardiac hypertrophy through upregulation of miR-98/let-7. Circ Res. 2011;108:305–313. doi: 10.1161/CIRCRESAHA.110.228437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ago T, Yeh I, Yamamoto M, Schinke-Braun M, Brown JA, Tian B, Sadoshima J. Thioredoxin1 upregulates mitochondrial proteins related to oxidative phosphorylation and TCA cycle in the heart. Antioxid Redox Signal. 2006;8:1635–1650. doi: 10.1089/ars.2006.8.1635. [DOI] [PubMed] [Google Scholar]
- 59.Kumar S, Rabson AB, Gelinas C. The RxxRxRxxC motif conserved in all Rel/kappa B proteins is essential for the DNA-binding activity and redox regulation of the v-Rel oncoprotein. Mol Cell Biol. 1992;12:3094–3106. doi: 10.1128/mcb.12.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oka S, Kamata H, Kamata K, Yagisawa H, Hirata H. N-acetylcysteine suppresses TNF-induced NF-kappaB activation through inhibition of IkappaB kinases. FEBS Lett. 2000;472:196–202. doi: 10.1016/s0014-5793(00)01464-2. [DOI] [PubMed] [Google Scholar]
- 61.Takeuchi J, Hirota K, Itoh T, Shinkura R, Kitada K, Yodoi J, Namba T, Fukuda K. Thioredoxin inhibits tumor necrosis factor- or interleukin-1-induced NF-kappaB activation at a level upstream of NF-kappaB-inducing kinase. Antioxid Redox Signal. 2000;2:83–92. doi: 10.1089/ars.2000.2.1-83. [DOI] [PubMed] [Google Scholar]
- 62.Hirota K, Murata M, Sachi Y, Nakamura H, Takeuchi J, Mori K, Yodoi J. Distinct roles of thioredoxin in the cytoplasm and in the nucleus. A two-step mechanism of redox regulation of transcription factor NF-kappaB. J Biol Chem. 1999;274:27891–27897. doi: 10.1074/jbc.274.39.27891. [DOI] [PubMed] [Google Scholar]
- 63.Stanley BA, Sivakumaran V, Shi S, Macdonald I, Lloyd D, Watson WH, Aon MA, Paolocci N. Thioredoxin reductase 2 is essential for keeping low levels of H2O2 emission from isolated heart mitochondria. J Biol Chem. 2011 doi: 10.1074/jbc.M111.284612. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka T, Hosoi F, Yamaguchi-Iwai Y, Nakamura H, Masutani H, Ueda S, Nishiyama A, Takeda S, Wada H, Spyrou G, Yodoi J. Thioredoxin-2 (TRX-2) is an essential gene regulating mitochondria-dependent apoptosis. Embo J. 2002;21:1695–1703. doi: 10.1093/emboj/21.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nonn L, Williams RR, Erickson RP, Powis G. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol Cell Biol. 2003;23:916–922. doi: 10.1128/MCB.23.3.916-922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, Schneider M, Beck H, Hatzopoulos AK, Just U, Sinowatz F, Schmahl W, Chien KR, Wurst W, Bornkamm GW, Brielmeier M. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol Cell Biol. 2004;24:9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spencer NY, Yan Z, Boudreau RL, Zhang Y, Luo M, Li Q, Tian X, Shah AM, Davisson RL, Davidson B, Banfi B, Engelhardt JF. Control of hepatic nuclear superoxide production by glucose 6-phosphate dehydrogenase and NADPH oxidase-4. J Biol Chem. 2011;286:8977–8987. doi: 10.1074/jbc.M110.193821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brewer AC, Murray TV, Arno M, Zhang M, Anilkumar NP, Mann GE, Shah AM. Nox4 regulates Nrf2 and glutathione redox in cardiomyocytes in vivo. Free Radic Biol Med. 2011;51:205–215. doi: 10.1016/j.freeradbiomed.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bulteau AL, Ikeda-Saito M, Szweda LI. Redox-dependent modulation of aconitase activity in intact mitochondria. Biochemistry. 2003;42:14846–14855. doi: 10.1021/bi0353979. [DOI] [PubMed] [Google Scholar]
- 70.Revollo JR, Grimm AA, Imai S. The regulation of nicotinamide adenine dinucleotide biosynthesis by Nampt/PBEF/visfatin in mammals. Curr Opin Gastroenterol. 2007;23:164–170. doi: 10.1097/MOG.0b013e32801b3c8f. [DOI] [PubMed] [Google Scholar]
- 71.Billington RA, Genazzani AA, Travelli C, Condorelli F. NAD depletion by FK866 induces autophagy. Autophagy. 2008;4:385–387. doi: 10.4161/auto.5635. [DOI] [PubMed] [Google Scholar]
- 72.Rongvaux A, Shea RJ, Mulks MH, Gigot D, Urbain J, Leo O, Andris F. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur J Immunol. 2002;32:3225–3234. doi: 10.1002/1521-4141(200211)32:11<3225::AID-IMMU3225>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 73.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 74.Emanuelli M, Carnevali F, Saccucci F, Pierella F, Amici A, Raffaelli N, Magni G. Molecular cloning, chromosomal localization, tissue mRNA levels, bacterial expression, and enzymatic properties of human NMN adenylyltransferase. J Biol Chem. 2001;276:406–412. doi: 10.1074/jbc.M008700200. [DOI] [PubMed] [Google Scholar]
- 75.Schweiger M, Hennig K, Lerner F, Niere M, Hirsch-Kauffmann M, Specht T, Weise C, Oei SL, Ziegler M. Characterization of recombinant human nicotinamide mononucleotide adenylyl transferase (NMNAT), a nuclear enzyme essential for NAD synthesis. FEBS Lett. 2001;492:95–100. doi: 10.1016/s0014-5793(01)02180-9. [DOI] [PubMed] [Google Scholar]
- 76.Alano CC, Tran A, Tao R, Ying W, Karliner JS, Swanson RA. Differences among cell types in NAD(+) compartmentalization: a comparison of neurons, astrocytes, and cardiac myocytes. J Neurosci Res. 2007;85:3378–3385. doi: 10.1002/jnr.21479. [DOI] [PubMed] [Google Scholar]
- 77.Nikiforov A, Dolle C, Niere M, Ziegler M. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J Biol Chem. 2011;286:21767–21778. doi: 10.1074/jbc.M110.213298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Samal B, Sun Y, Stearns G, Xie C, Suggs S, McNiece I. Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol. 1994;14:1431–1437. doi: 10.1128/mcb.14.2.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin PR, Shea RJ, Mulks MH. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J Bacteriol. 2001;183:1168–1174. doi: 10.1128/JB.183.4.1168-1174.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Revollo JR, Korner A, Mills KF, Satoh A, Wang T, Garten A, Dasgupta B, Sasaki Y, Wolberger C, Townsend RR, Milbrandt J, Kiess W, Imai S. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab. 2007;6:363–375. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jia SH, Li Y, Parodo J, Kapus A, Fan L, Rotstein OD, Marshall JC. Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J Clin Invest. 2004;113:1318–1327. doi: 10.1172/JCI19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.van der Veer E, Nong Z, O'Neil C, Urquhart B, Freeman D, Pickering JG. Pre-B-cell colony-enhancing factor regulates NAD+-dependent protein deacetylase activity and promotes vascular smooth muscle cell maturation. Circ Res. 2005;97:25–34. doi: 10.1161/01.RES.0000173298.38808.27. [DOI] [PubMed] [Google Scholar]
- 83.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 85.Van Beijnum JR, Moerkerk PT, Gerbers AJ, De Bruine AP, Arends JW, Hoogenboom HR, Hufton SE. Target validation for genomics using peptide-specific phage antibodies: a study of five gene products overexpressed in colorectal cancer. Int J Cancer. 2002;101:118–127. doi: 10.1002/ijc.10584. [DOI] [PubMed] [Google Scholar]
- 86.Hsu CP, Oka S, Shao D, Hariharan N, Sadoshima J. Nicotinamide phosphoribosyltransferase regulates cell survival through NAD+ synthesis in cardiac myocytes. Circ Res. 2009;105:481–491. doi: 10.1161/CIRCRESAHA.109.203703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dahl TB, Haukeland JW, Yndestad A, Ranheim T, Gladhaug IP, Damas JK, Haaland T, Loberg EM, Arntsen B, Birkeland K, Bjoro K, Ulven SM, Konopski Z, Nebb HI, Aukrust P, Halvorsen B. Intracellular nicotinamide phosphoribosyltransferase protects against hepatocyte apoptosis and is down-regulated in nonalcoholic fatty liver disease. J Clin Endocrinol Metab. 2011;95:3039–3047. doi: 10.1210/jc.2009-2148. [DOI] [PubMed] [Google Scholar]
- 88.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Borradaile NM, Pickering JG. Nicotinamide phosphoribosyltransferase imparts human endothelial cells with extended replicative lifespan and enhanced angiogenic capacity in a high glucose environment. Aging Cell. 2009;8:100–112. doi: 10.1111/j.1474-9726.2009.00453.x. [DOI] [PubMed] [Google Scholar]
- 91.van der Veer E, Ho C, O'Neil C, Barbosa N, Scott R, Cregan SP, Pickering JG. Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J Biol Chem. 2007;282:10841–10845. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
- 92.Benigni A, Corna D, Zoja C, Sonzogni A, Latini R, Salio M, Conti S, Rottoli D, Longaretti L, Cassis P, Morigi M, Coffman TM, Remuzzi G. Disruption of the Ang II type 1 receptor promotes longevity in mice. J Clin Invest. 2009;119:524–530. doi: 10.1172/JCI36703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang LQ, Adyshev DM, Singleton P, Li H, Cepeda J, Huang SY, Zou X, Verin AD, Tu J, Garcia JG, Ye SQ. Interactions between PBEF and oxidative stress proteins--a potential new mechanism underlying PBEF in the pathogenesis of acute lung injury. FEBS Lett. 2008;582:1802–1808. doi: 10.1016/j.febslet.2008.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hsu CP, Odewale I, Alcendor RR, Sadoshima J. Sirt1 protects the heart from aging and stress. Biol Chem. 2008;389:221–231. doi: 10.1515/BC.2008.032. [DOI] [PubMed] [Google Scholar]
- 95.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. Embo J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Haider DG, Mittermayer F, Schaller G, Artwohl M, Baumgartner-Parzer SM, Prager G, Roden M, Wolzt M. Free fatty acids normalize a rosiglitazone-induced visfatin release. Am J Physiol Endocrinol Metab. 2006;291:E885–890. doi: 10.1152/ajpendo.00109.2006. [DOI] [PubMed] [Google Scholar]
- 98.Song HK, Lee MH, Kim BK, Park YG, Ko GJ, Kang YS, Han JY, Han SY, Han KH, Kim HK, Cha DR. Visfatin: a new player in mesangial cell physiology and diabetic nephropathy. Am J Physiol Renal Physiol. 2008;295:F1485–1494. doi: 10.1152/ajprenal.90231.2008. [DOI] [PubMed] [Google Scholar]
- 99.Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rongvaux A, Galli M, Denanglaire S, Van Gool F, Dreze PL, Szpirer C, Bureau F, Andris F, Leo O. Nicotinamide phosphoribosyl transferase/pre-B cell colony-enhancing factor/visfatin is required for lymphocyte development and cellular resistance to genotoxic stress. J Immunol. 2008;181:4685–4695. doi: 10.4049/jimmunol.181.7.4685. [DOI] [PubMed] [Google Scholar]
- 101.Alcendor RR, Kirshenbaum LA, Imai S, Vatner SF, Sadoshima J. Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ Res. 2004;95:971–980. doi: 10.1161/01.RES.0000147557.75257.ff. [DOI] [PubMed] [Google Scholar]
- 102.Li Y, Zhang Y, Dorweiler B, Cui D, Wang T, Woo CW, Brunkan CS, Wolberger C, Imai S, Tabas I. Extracellular Nampt promotes macrophage survival via a nonenzymatic interleukin-6/STAT3 signaling mechanism. J Biol Chem. 2008;283:34833–34843. doi: 10.1074/jbc.M805866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 104.Hsu CP, Hariharan N, Alcendor RR, Oka S, Sadoshima J. Nicotinamide phosphoribosyltransferase regulates cell survival through autophagy in cardiomyocytes. Autophagy. 2009;5:1229–1231. doi: 10.4161/auto.5.8.10275. [DOI] [PubMed] [Google Scholar]
- 105.Bae SK, Kim SR, Kim JG, Kim JY, Koo TH, Jang HO, Yun I, Yoo MA, Bae MK. Hypoxic induction of human visfatin gene is directly mediated by hypoxia-inducible factor-1. FEBS Lett. 2006;580:4105–4113. doi: 10.1016/j.febslet.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 106.Bae YH, Bae MK, Kim SR, Lee JH, Wee HJ, Bae SK. Upregulation of fibroblast growth factor-2 by visfatin that promotes endothelial angiogenesis. Biochem Biophys Res Commun. 2009;379:206–211. doi: 10.1016/j.bbrc.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 107.Kim J-Y, Bae Y-H, Bae M-K, Kim S-R, Park H-J, Wee H-J, Bae S-K. Visfatin through STAT3 activation enhances IL-6 expression that promotes endothelial angiogenesis. Biochimica et Biophysica Acta (BBA) -Molecular Cell Research. 2009;1793:1759–1767. doi: 10.1016/j.bbamcr.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 108.Adya R, Tan BK, Punn A, Chen J, Randeva HS. Visfatin induces human endothelial VEGF and MMP-2/9 production via MAPK and PI3K/Akt signalling pathways: novel insights into visfatin-induced angiogenesis. Cardiovasc Res. 2008;78:356–365. doi: 10.1093/cvr/cvm111. [DOI] [PubMed] [Google Scholar]
- 109.Adya R, Tan BK, Chen J, Randeva HS. Pre-B cell colony enhancing factor (PBEF)/visfatin induces secretion of MCP-1 in human endothelial cells: role in visfatin-induced angiogenesis. Atherosclerosis. 2009;205:113–119. doi: 10.1016/j.atherosclerosis.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 110.Hasmann M, Schemainda I. FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res. 2003;63:7436–7442. [PubMed] [Google Scholar]
- 111.Drevs J, Loser R, Rattel B, Esser N. Antiangiogenic potency of FK866/K22.175, a new inhibitor of intracellular NAD biosynthesis, in murine renal cell carcinoma. Anticancer Res. 2003;23:4853–4858. [PubMed] [Google Scholar]
- 112.Pogrebniak A, Schemainda I, Azzam K, Pelka-Fleischer R, Nussler V, Hasmann M. Chemopotentiating effects of a novel NAD biosynthesis inhibitor, FK866, in combination with antineoplastic agents. Eur J Med Res. 2006;11:313–321. [PubMed] [Google Scholar]
- 113.Olesen UH, Christensen MK, Bjorkling F, Jaattela M, Jensen PB, Sehested M, Nielsen SJ. Anticancer agent CHS-828 inhibits cellular synthesis of NAD. Biochem Biophys Res Commun. 2008;367:799–804. doi: 10.1016/j.bbrc.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 114.Kim SR, Bae YH, Bae SK, Choi KS, Yoon KH, Koo TH, Jang HO, Yun I, Kim KW, Kwon YG, Yoo MA, Bae MK. Visfatin enhances ICAM-1 and VCAM-1 expression through ROS-dependent NF-kappaB activation in endothelial cells. Biochim Biophys Acta. 2008;1783:886–895. doi: 10.1016/j.bbamcr.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 115.Yilmaz MI, Saglam M, Qureshi AR, Carrero JJ, Caglar K, Eyileten T, Sonmez A, Cakir E, Oguz Y, Vural A, Yenicesu M, Stenvinkel P, Lindholm B, Axelsson J. Endothelial dysfunction in type-2 diabetics with early diabetic nephropathy is associated with low circulating adiponectin. Nephrol Dial Transplant. 2008;23:1621–1627. doi: 10.1093/ndt/gfm828. [DOI] [PubMed] [Google Scholar]
- 116.Yilmaz MI, Saglam M, Carrero JJ, Qureshi AR, Caglar K, Eyileten T, Sonmez A, Oguz Y, Aslan I, Vural A, Yenicesu M, Stenvinkel P, Lindholm B, Axelsson J. Normalization of endothelial dysfunction following renal transplantation is accompanied by a reduction of circulating visfatin/NAMPT. A novel marker of endothelial damage? Clin Transplant. 2009;23:241–248. doi: 10.1111/j.1399-0012.2008.00921.x. [DOI] [PubMed] [Google Scholar]
- 117.Busso N, Karababa M, Nobile M, Rolaz A, Van Gool F, Galli M, Leo O, So A, De Smedt T. Pharmacological inhibition of nicotinamide phosphoribosyltransferase/visfatin enzymatic activity identifies a new inflammatory pathway linked to NAD. PLoS ONE. 2008;3:e2267. doi: 10.1371/journal.pone.0002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Haider DG, Holzer G, Schaller G, Weghuber D, Widhalm K, Wagner O, Kapiotis S, Wolzt M. The adipokine visfatin is markedly elevated in obese children. J Pediatr Gastroenterol Nutr. 2006;43:548–549. doi: 10.1097/01.mpg.0000235749.50820.b3. [DOI] [PubMed] [Google Scholar]
- 120.Berndt J, Kloting N, Kralisch S, Kovacs P, Fasshauer M, Schon MR, Stumvoll M, Bluher M. Plasma visfatin concentrations and fat depot-specific mRNA expression in humans. Diabetes. 2005;54:2911–2916. doi: 10.2337/diabetes.54.10.2911. [DOI] [PubMed] [Google Scholar]
- 121.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 122.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li B, Navarro S, Kasahara N, Comai L. Identification and biochemical characterization of a Werner's syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J Biol Chem. 2004;279:13659–13667. doi: 10.1074/jbc.M311606200. [DOI] [PubMed] [Google Scholar]
- 125.Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, Zhao GY, Saberi A, Masutani M, Adachi N, Koyama H, de Murcia G, Takeda S. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. Embo J. 2006;25:1305–1314. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. Embo J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 128.D'Amours D, Sallmann FR, Dixit VM, Poirier GG. Gain-of-function of poly(ADP-ribose) polymerase-1 upon cleavage by apoptotic proteases: implications for apoptosis. J Cell Sci. 2001;114:3771–3778. doi: 10.1242/jcs.114.20.3771. [DOI] [PubMed] [Google Scholar]
- 129.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci U S A. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Los M, Mozoluk M, Ferrari D, Stepczynska A, Stroh C, Renz A, Herceg Z, Wang ZQ, Schulze-Osthoff K. Activation and caspase-mediated inhibition of PARP: a molecular switch between fibroblast necrosis and apoptosis in death receptor signaling. Mol Biol Cell. 2002;13:978–988. doi: 10.1091/mbc.01-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 133.Fossati S, Cipriani G, Moroni F, Chiarugi A. Neither energy collapse nor transcription underlie in vitro neurotoxicity of poly(ADP-ribose) polymerase hyper-activation. Neurochem Int. 2007;50:203–210. doi: 10.1016/j.neuint.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 134.Choudhury S, Bae S, Ke Q, Lee JY, Kim J, Kang PM. Mitochondria to nucleus translocation of AIF in mice lacking Hsp70 during ischemia/reperfusion. Basic Res Cardiol. 2011;106:397–407. doi: 10.1007/s00395-011-0164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xiao CY, Chen M, Zsengeller Z, Li H, Kiss L, Kollai M, Szabo C. Poly(ADP-Ribose) polymerase promotes cardiac remodeling, contractile failure, and translocation of apoptosis-inducing factor in a murine experimental model of aortic banding and heart failure. J Pharmacol Exp Ther. 2005;312:891–898. doi: 10.1124/jpet.104.077164. [DOI] [PubMed] [Google Scholar]
- 136.Zhou HZ, Swanson RA, Simonis U, Ma X, Cecchini G, Gray MO. Poly(ADP-ribose) polymerase-1 hyperactivation and impairment of mitochondrial respiratory chain complex I function in reperfused mouse hearts. Am J Physiol Heart Circ Physiol. 2006;291:H714–723. doi: 10.1152/ajpheart.00823.2005. [DOI] [PubMed] [Google Scholar]
- 137.Soriano FG, Nogueira AC, Caldini EG, Lins MH, Teixeira AC, Cappi SB, Lotufo PA, Bernik MM, Zsengeller Z, Chen M, Szabo C. Potential role of poly(adenosine 5'-diphosphate-ribose) polymerase activation in the pathogenesis of myocardial contractile dysfunction associated with human septic shock. Crit Care Med. 2006;34:1073–1079. doi: 10.1097/01.CCM.0000206470.47721.8D. [DOI] [PubMed] [Google Scholar]
- 138.Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J Biol Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- 139.Pacher P, Liaudet L, Bai P, Virag L, Mabley JG, Hasko G, Szabo C. Activation of poly(ADP-ribose) polymerase contributes to development of doxorubicin-induced heart failure. J Pharmacol Exp Ther. 2002;300:862–867. doi: 10.1124/jpet.300.3.862. [DOI] [PubMed] [Google Scholar]
- 140.Liaudet L, Yang Z, Al-Affar EB, Szabo C. Myocardial ischemic preconditioning in rodents is dependent on poly (ADP-ribose) synthetase. Mol Med. 2001;7:406–417. [PMC free article] [PubMed] [Google Scholar]
- 141.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 145.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 146.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 147.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 149.Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hsu CP, Zhai P, Yamamoto T, Maejima Y, Matsushima S, Hariharan N, Shao D, Takagi H, Oka S, Sadoshima J. Silent information regulator 1 protects the heart from ischemia/reperfusion. Circulation. 2011;122:2170–2182. doi: 10.1161/CIRCULATIONAHA.110.958033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tanno M, Kuno A, Yano T, Miura T, Hisahara S, Ishikawa S, Shimamoto K, Horio Y. Induction of manganese superoxide dismutase by nuclear translocation and activation of SIRT1 promotes cell survival in chronic heart failure. J Biol Chem. 2010;285:8375–8382. doi: 10.1074/jbc.M109.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nadtochiy SM, Redman E, Rahman I, Brookes PS. Lysine deacetylation in ischaemic preconditioning: the role of SIRT1. Cardiovasc Res. 2011;89:643–649. doi: 10.1093/cvr/cvq287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 Plays an Essential Role in Mediating Starvation-Induced Autophagy in Cardiac Myocytes. Circ Res. 2011;107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, Wang D, Feng J, Yu L, Zhu WG. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 156.Kruszewski M, Szumiel I. Sirtuins (histone deacetylases III) in the cellular response to DNA damage--facts and hypotheses. DNA Repair (Amst) 2005;4:1306–1313. doi: 10.1016/j.dnarep.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 157.Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissier-de Murcia J, Auwerx J. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P. Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle. 2006;5:873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- 159.Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Birukov KG, Samant S, Hottiger MO, Gupta MP. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol Cell Biol. 2009;29:4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]