Abstract
The truncated brain-derived neurotrophic factor (BDNF) receptors (truncated TrkB [TrkB-TK−] and sarc homology containing TrkB [TrkB-Shc]) are alternative transcripts of the full-length TrkB receptor (TrkB-TK+) that produce isoforms capable of binding to BDNF but not being able to mediate the classic neurotrophic response via tyrosine kinase signaling. We hypothesized that in the dorsolateral prefrontal cortex (DLPFC) of people with schizophrenia, truncated TrkB receptors (TK− and Shc) would be altered and may contribute to deficits in BDNF function. Using a large cohort of controls and schizophrenics (n = 72/72), we measured mRNA expression of the full-length TrkB receptor, TrkB-TK+ and the truncated TrkB receptors, TrkB-TK− and TrkB-Shc, by quantitative real-time polymerase chain reaction and protein expression by western blotting. We found highly significant increases in mRNA expression of both truncated TrkB receptor isoforms in people with schizophrenia. When we examined the full-length TrkB-TK+:truncated TrkB ratios, we observed significant decreases in schizophrenia both on the mRNA and protein level. We found a slight reduction in TrkB-TK+ mRNA and a significant reduction in TrkB-TK+ protein expression in schizophrenia, which was evident in females. No gender-specific changes were found for the truncated TrkB receptors. Diagnostic changes in TrkB-TK+ mRNA and protein may be subtle and/or gender-specific, whereas changes in TrkB-TK− and TrkB-Shc expression are robust and may generalize to both males and females with schizophrenia. Increased truncated TrkB receptors may contribute to reduced overall BDNF/tyrosine receptor kinase B (TrkB) signaling and lead to reduced neuronal plasticity in the DLPFC in schizophrenia suggesting that therapies aimed at ameliorating neurotrophin deficits may need to consider blocking excessive truncated TrkB function.
Keywords: dorsolateral prefrontal cortex, postmortem, BDNF, Trk, gene expression
Introduction
In schizophrenia, people show cognitive deficits linked to abnormalities in the function of the dorsolateral prefrontal cortex (DLPFC).1 While multiple lines of evidence support the idea of altered neuronal function in schizophrenia, a definitive mechanism for this neuronal deficit is elusive. The neurotrophin, brain-derived neurotrophic factor (BDNF), is consistently decreased in the DLPFC of people with schizophrenia (in at least 4 postmortem brain cohorts).2–4 Neurotrophins are potent regulators of neuronal maturation, differentiation, survival, plasticity, and function5–8; and their effects are mediated through high-affinity tropomyosin receptor kinase receptors (Trks).9,10 People with schizophrenia not only show reduced expression of BDNF but they also demonstrate reductions in full-length TrkB receptors (TrkB-TK+).3,11 Moreover, reductions in TrkB-TK+ were speculated to be more deleterious to interneuron health and viability than BDNF reductions alone.3 However, the function of tyrosine receptor kinase B (TrkB) receptors depends not only on the abundance of TrkB-TK+ but also on the levels of other TrkB splice variants known to be expressed in the human DLPFC.12
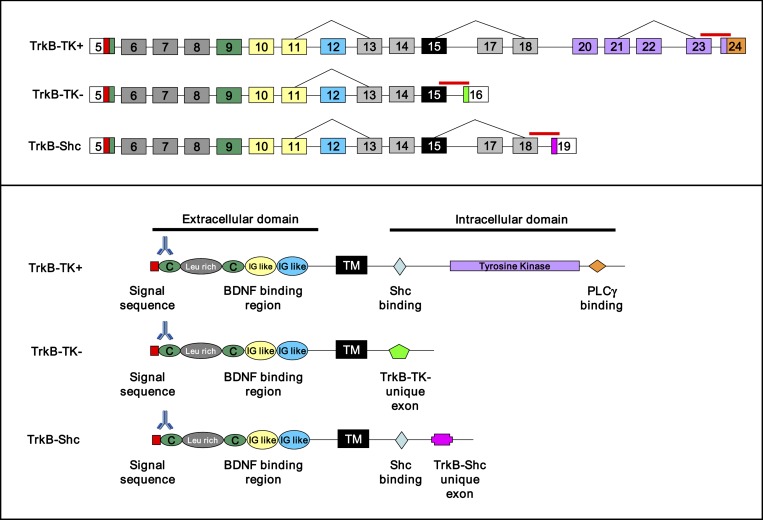
The full-length (TrkB-TK+) and truncated TrkB receptors (TrkB-TK− and sarc homology containing TrkB [TrkB-Shc]) are generated by alternative splicing and are the 3 best characterized and most abundant TrkB isoforms in the brain13 (figure 1). The high-affinity TrkB-TK+ contains a catalytic tyrosine kinase domain, is expressed almost exclusively in neurons within the human prefrontal cortex,11 and is necessary for activating second messenger signaling and mediating the neurotrophic effects of BDNF.9,14 TrkB-TK− lacks the catalytic tyrosine kinase domain but contains a short isoform-specific cytoplasmic domain.15 It is expressed in both neurons (pyramidal and interneurons) and glia (astrocytes)16–18 and in glia, TrkB-TK− may inhibit neuronal neurotrophin signaling by sequestering or trapping neurotrophins to prevent binding.19,20 Alternatively, TrkB-TK− has been shown in vitro cell culture to act as a dominant-negative receptor by forming inactive heterodimers with TrkB-TK+ to prevent neurotrophin signaling.21,22 Considering that neurons can co-express both TrkB-TK+ and TrkB-TK−,23 this event may occur in vivo. Thus, the cellular response to BDNF has been proposed to depend on the relative levels of the TrkB-TK+ and TrkB-TK− isoforms.21,24 The other truncated TrkB receptor isoform, TrkB-Shc, contains a sarc homolgy binding site (Shc) in the juxtamembrane domain similar to TrkB-TK+; however, it contains a unique truncated carboxy-terminus which also lacks the tyrosine kinase domain and can act as a negative regulator of TrkB-TK+.13 Previously, we found that levels of TrkB-TK− mRNA are robust and at least equivalent to TrkB-TK+ mRNA levels in the human cortex by Northern Blotting25 suggesting that TrkB-TK− may contribute significantly to cortical neurotrophin response. Thus, efforts to stimulate BDNF or TrkB-TK+ signaling in schizophrenia may be limited without knowing if alterations in truncated TrkB receptor isoforms also exist in the diseased state.
Fig. 1.
TrkB gene structure and organization. Upper panel: coding regions of the 3 best characterized tyrosine receptor kinase B (TrkB) transcripts from 5′ to 3′, full-length TrkB (TrkB-TK+), truncated TrkB (TrkB-TK−), and sarc homology containing TrkB (TrkB-Shc). Nomenclature is derived from Luberg et al12. Target regions of the Taqman primer/probes used for qPCR are indicated by solid lines above exons. Bottom panel: functional domains of the TrkB proteins from N terminus to C terminus. The target region of the TrkB antibody used for western blotting is indicated by the antibody symbol above the protein domains. TM, transmembrane domain.
Although the truncated TrkB isoforms are critical in determining the cellular response to BDNF, no study has been published that specifically examines the mRNA and/or protein expression of the truncated TrkB receptors, (TrkB-TK−/TrkB-Shc) in schizophrenia. In this current study, we determined whether mRNA and/or protein expression of the TrkB receptors, TrkB-TK+, TrkB-TK−, and TrkB-Shc or their ratios of expression, are altered in the DLPFC of people with schizophrenia. We hypothesized that in addition to decreased TrkB-TK+, people with schizophrenia may also have alterations in truncated TrkB receptor expression, either of which may interfere with normal neurotrophin signaling.
Methods
Brain Cohort
The combined cohort used in this study comprises the Tissue Resource Centre (TRC) cohort (Sydney, Australia) (n = 37 controls, n = 37 schizophrenics) and the Stanley Medical Research Institute’s (SMRI) brain “array” collection (MD) (n = 35 controls, n = 35 schizophrenics). Cases with schizophrenia were matched to healthy controls for both cohorts. For pair matching (TRC), the following guidelines were used: (1) age at death (within 10 y), (2) brain pH (within 0.59 pH units), and (3) postmortem interval (PMI) (within 22 h). Demographic data for the combined cohort are listed in table 1. The overall quality of the RNA was high (average RNA integrity number [RIN] >7). The diagnostic groups did not differ according to age, brain pH, PMI, RIN, or brain weight (all P > .06). Anatomical dissections for the DLPFC and detailed demographics for each cohort have been published.26–28 For the 2 different brain cohorts, the manner in which the schizophrenia cases were diagnosed (all meeting Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, criteria for schizophrenia) and the protocols used for tissue collection, characterization, and RNA extraction were similar.26–29
Table 1.
Brain Cohort Demographics
| Control n = 72 | Schizophrenia n = 72 | t value | P value | |
| Age | 47.8 ± 1.43 (18–78) | 47.1 ± 1.47 (19–75) | 0.34 | .74 |
| Gender | 16F/56M | 22F/50M | ||
| pH | 6.63 ± 0.03 (5.84–7.15) | 6.55 ± 0.03 (5.69–7.09) | 1.87 | .06 |
| PMI | 27 ± 1.42 (6.50–58) | 29.9 ± 1.72 (5.0–80) | −1.28 | .20 |
| RIN | 7.78 ± 0.09 (6.0–9.70) | 7.85 ± 0.10 (6.20–9.60) | −0.53 | .59 |
| Brain weight (g) | 1445.2 ± 16.1 (1120–1900) | 1417.6 ± 16.6 (1020–1700) | 1.20 | .23 |
| Age of onset (y) | 22.5 ± 0.72 (9–40) | |||
| DOI (y) | 24.5 ± 1.47 (1–52) | |||
| Lifetime APD use | 3952352 ± 809053.2 |
Note: Cohort is a combination of the Tissue Resource Centre (TRC) (n = 37/37) and Stanley Medical Research Institute (SMRI) (n = 35/35) cohorts. Demographics are presented as mean values ±SEM. The lowest and highest values of each variable are listed in brackets. Race: TRC cohort: 2 Asians, 72 Caucasians. SMRI cohort: 1 Native American, 1 African American, 1 Hispanic, 67 Caucasians. M, male; F, female; PMI, postmortem interval; RIN, RNA integrity number; DOI, duration of illness; APD, antipsychotic drug.
RNA Extraction and cDNA Synthesis
Total RNA was extracted from ∼300 mg of frozen DLPFC tissue using the TRIZOL Reagent method (Life Technologies Inc, Grand Island, NY) as described.30 RIN was assessed with high-resolution capillary electrophoresis (Agilent Technologies, Palo Alto, CA). Three aliquots of 3 μg RNA were reverse transcribed to synthesize cDNA by random hexamer priming using the SuperScript First-Strand Synthesis System (Life Technologies Inc) as described and the 3 aliquots were then pooled.30
Quantitative Real-Time Polymerase Chain Reaction
TrkB mRNA levels from cDNA were measured by Taqman (primers/probes) quantitative real-time polymerase chain reaction (qPCR) using an ABI Prism 7900HT Fast Real-Time PCR System in a 384-well format as described.31 The most optimal housekeeping genes were determined and the 3 that overlapped between the 2 cohorts were used to calculate the geometric means as previously described.26,32 The housekeeping genes used included β-actin (ACTB) (Cat#hs99999903-m1), ubiquitin-C (UBC) (Cat#hs00824723-m1), and TATA box-binding protein (TBP) (Cat#hs00427620-m1). The TrkB transcripts were targeted by the following ABI Taqman primers/probes (Applied Biosystems): TrkB-TK+ (Cat#hs01093098-m1), TrkB-TK− (Cat#hs01093110-m1), and TrkB-Shc (custom probe Cat#AIVI3IH) (figure 1). Samples (9 ng cDNA) were run in triplicate with an 8-point standard curve using serial dilutions of pooled cDNA (from all cases) under standard cycling conditions.31 No template and no reverse-transcription controls produced no signal. PCR data was obtained with the Sequence Detector Software (SDS version 2.0, Applied Biosystems). TrkB mRNA levels from each subject in the 2 cohorts were normalized by the geometric mean of housekeepers measured from the same subject. There was no significant difference between the housekeeping genes or geometric mean between controls and schizophrenics (all P > .15) (see online supplementary material for figure S1and Results 1 for details).
Western Blotting
Protein extraction and western blotting were conducted as described.4,31 Briefly, samples of equal protein (10 μg) were analyzed by 10% sodium dodecyl sulfate polyacrylamide (SDS-PAGE) gels or 12% Bis-Tris gels (BioRad). Internal controls of pooled DLPFC homogenates were run as a correction for gel to gel variation. The primary antibody used to detect human TrkB protein was from Cell Signaling Technology (diluted 1:3000; 4°C; overnight; Cat#4603) (figure 1). β-Actin (Chemicon International, Cat#MAB1501) was probed as a protein loading control. Bands were visualized on a Chemidoc Imaging System (BioRad) and quantitated by densitometry using Quantity One 1-D Analysis Software v4.6.5 (BioRad). TrkB protein levels were determined after normalizing with β-actin.
Statistical Analysis
We excluded qPCR measurement errors (TRC cohort: no more than 3/group removed; none were removed from the SMRI cohort). Population outliers (±2 SDs) were excluded from both mRNA and protein analyses (in all assays for both cohorts, ∼6% were outliers). Statistical analyses were conducted (StatSoft Inc, 2000, STATISTICA for Windows) with 2-tailed unpaired tests to assess mRNA expression of the qPCR housekeeping genes and geometric mean between controls and schizophrenics. One-tailed unpaired t tests were conducted if a directional hypothesis was employed. Pearson’s Product Moment correlations were conducted to determine if any relationship existed between measured target molecules and demographic variables (see online supplementary material for Results 2). Where a relationship existed, ANCOVA was calculated. Pearson’s Product Moment correlations were also conducted to assess the relationship between mRNA and protein targets. To determine the main effect of diagnosis and gender and the diagnosis × gender interaction effect on TrkB mRNA and protein expression and receptor ratios, 2-way ANOVA was conducted followed by Fisher Least Significant Difference (LSD) post hoc analysis. A P < .05 was considered statistically significant.
Results
mRNA Expression of TrkB Receptors
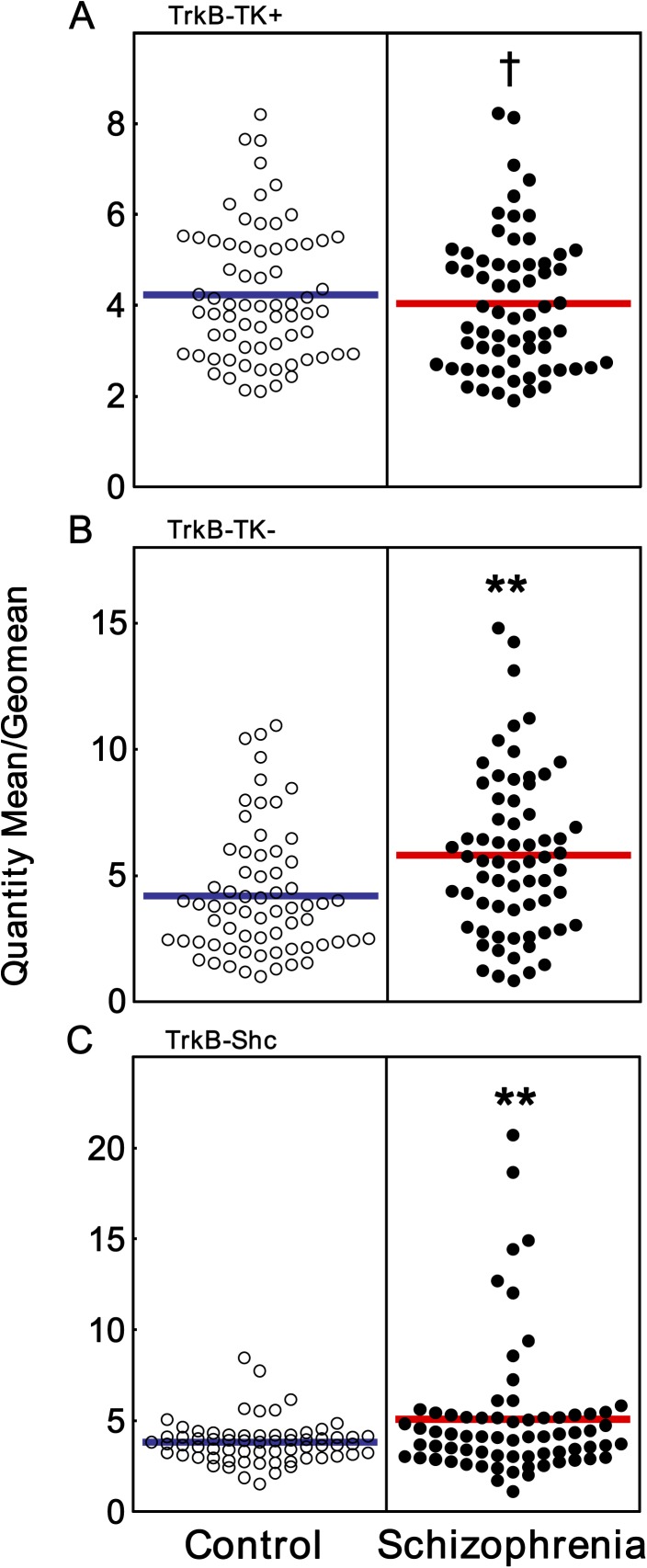
We found that TrkB-TK− and TrkB-Shc mRNAs were highly and significantly elevated (38% and 33% increase, repsectively) in the DLPFC of people with schizophrenia (ANCOVA with pH, diagnosis main effect: TrkB-TK−: F = 5.33, df = 1, 130, P = .02; ANOVA diagnosis main effect: TrkB-Shc: F = 7.61, df = 1, 132, P = .007) (figure 2B and C). ANCOVA analysis of TrkB-TK+ mRNA expression covarying for age, PMI, and RIN revealed a trend toward decreased expression in people with schizophrenia (ANCOVA: F = 3.63, df = 1, 129, P = .059) (figure 2A), but unlike the large increase in mRNAs encoding the truncated TrkB receptors, the magnitude of the decrease in TrkB-TK+ was relatively small (<10% decrease).
Fig. 2.
TrkB mRNA expression in the DLPFC of people with schizophrenia. Expression of (A) full-length TrkB (TrkB-TK+), (B) truncated TrkB (TrkB-TK−), and (C) sarc homology containing TrkB (TrkB-Shc) mRNAs (control white circles, schizophrenics black circles) were measured by qPCR. Data are expressed as quantity mean/geomean of individual subjects. **P < .01, † denotes a trend toward significance following ANCOVA (covarying for age, PMI, and RIN [P = .059]).
Protein Expression of TrkB Receptors
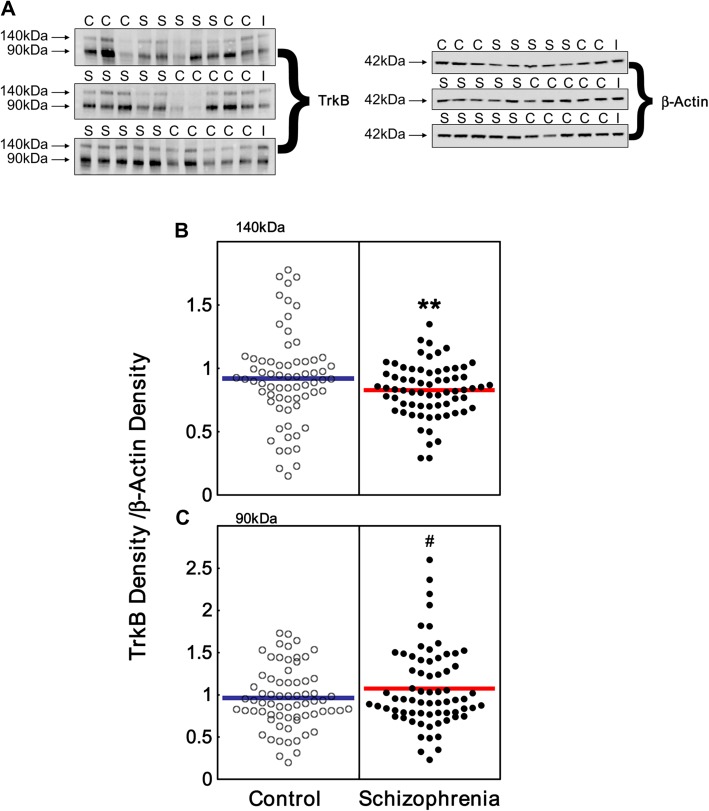
We found immunoreactive TrkB bands migrating at ∼140 kDa and ∼90 kDa in all human DLPFC samples (figure 3A) with the smaller band being more intense. The ∼140 kDa band corresponding to TrkB-TK+ protein showed a significant reduction in schizophrenia (10% decrease; ANOVA diagnosis main effect: F = 10.4, df = 1, 132, P = .002) (figure 3B).
Fig. 3.
TrkB protein expression in the dorsolateral prefrontal cortex (DLPFC) of people with schizophrenia. (A) Representative western blots showing control (C) and schizophrenia (S) cases. We identified 2 bands in all samples, one corresponding to full-length TrkB (TrkB-TK+) protein (∼140 kDa) and one corresponding to truncated TrkB proteins (∼90 kDa). β-Actin was probed as a loading control and a pooled internal control was run (I). (B–C) Bands were quantitated by densitometry. (B) Expression of TrkB-TK+ (∼140 kDa) and (C) truncated TrkB (∼90 kDa) (control white circles, schizophrenics black circles) are expressed as TrkB density/β-actin density of individual subjects. **P < .005, # denotes a trend toward significance P = .057.
The smaller TrkB immunopositive bands migrating at ∼90 kDa was large and diffuse. This is likely because TrkB-TK− and TrkB-Shc are similar in size and both migrate at ∼90 kDa. A trend toward increased expression was observed in the ∼90 kDa TrkB protein in people with schizophrenia compared with controls (12% increase; unpaired t test: t = 1.59, df = 135, P = .057 one-tailed, figure 3C). β-actin levels did not vary between controls and schizophrenics (P > .05).
The mRNA and protein levels of all 3 TrkB isoforms across individual subjects in the study were tested for correlation. We found a significant negative correlation between the 90 kDa truncated TrkB protein and TrkB-TK+ mRNA (r=−.38, P = .00001). No other significant correlations were observed between TrkB mRNA and protein.
mRNA and Protein Ratios Between Full-Length and Truncated TrkB Receptor Expression
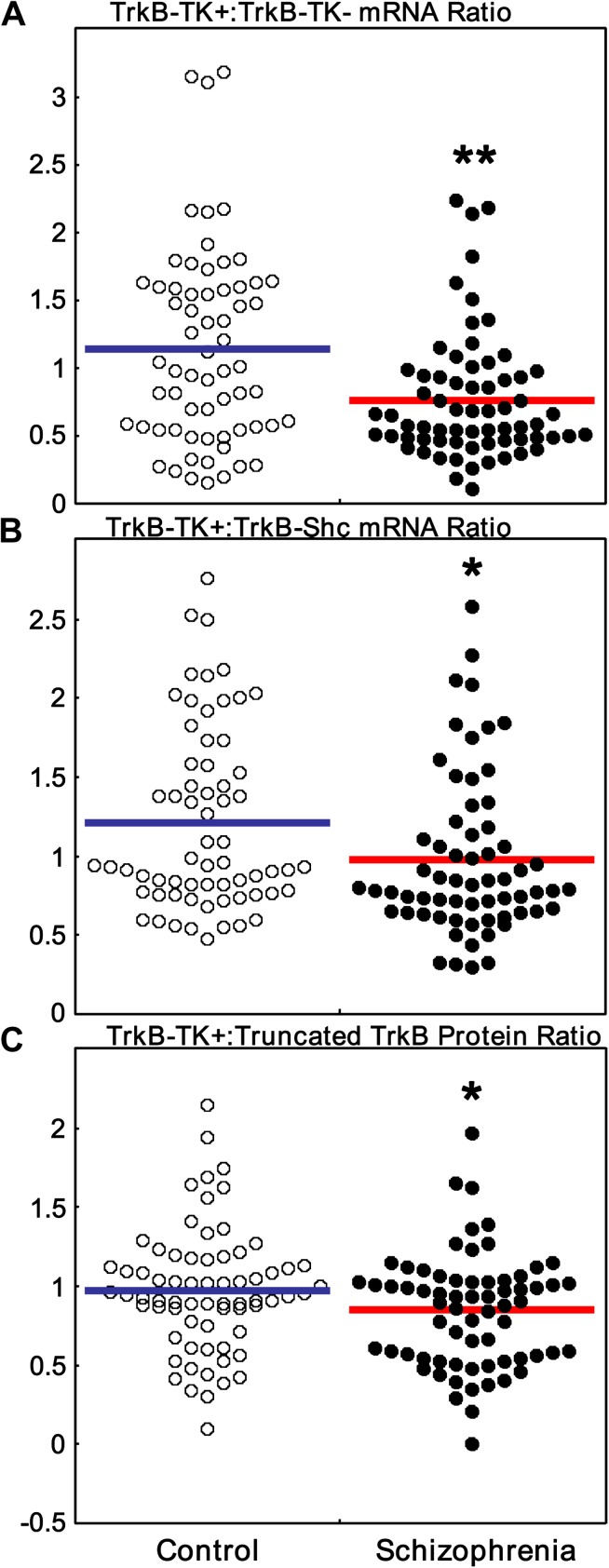
We next determined whether the TrkB-TK+:truncated TrkB receptor ratios were altered in schizophrenia. In the DLPFC of people with schizophrenia, we observed a highly significant reduction in the relative ratios of TrkB-TK+:TrkB-TK− mRNA (ANOVA diagnosis main effect: F = 8.21, df = 1, 127, P = .005) (figure 4A) and TrkB-TK+:TrkB-Shc mRNA (ANOVA diagnosis main effect: F = 5.90, df = 1, 124, P = .017) (figure 4B). Since reductions in the ratio of full-length to truncated TrkB mRNA expression were observed in schizophrenia, we tested whether the ratio of TrkB-TK+:truncated TrkB protein was also decreased in people with schizophrenia and confirmed that this reduction extended to the protein level (unpaired t test: t = −2.01, df = 134, P = .02, one-tailed) (figure 4C).
Fig. 4.
Full-length TrkB (TrkB-TK+):truncated TrkB (TrkB-TK−) mRNA and protein expression ratios. (A) The mRNA ratio between (A) TrkB-TK+ and TrkB-TK− or (B) TrkB-TK+ and sarc homology containing TrkB (TrkB-Shc) were calculated by dividing normalized TrkB-TK+ with normalized TrkB-TK− or TrkB-Shc. (C) The protein ratios between TrkB-TK+ and truncated TrkB were calculated by dividing normalized TrkB-TK+ with normalized truncated TrkB. Data are expressed as quantity mean/geomean or TrkB density/β-actin density of individual subjects. **P < .01, *P < .05.
The Effect of Gender on TrkB Expression
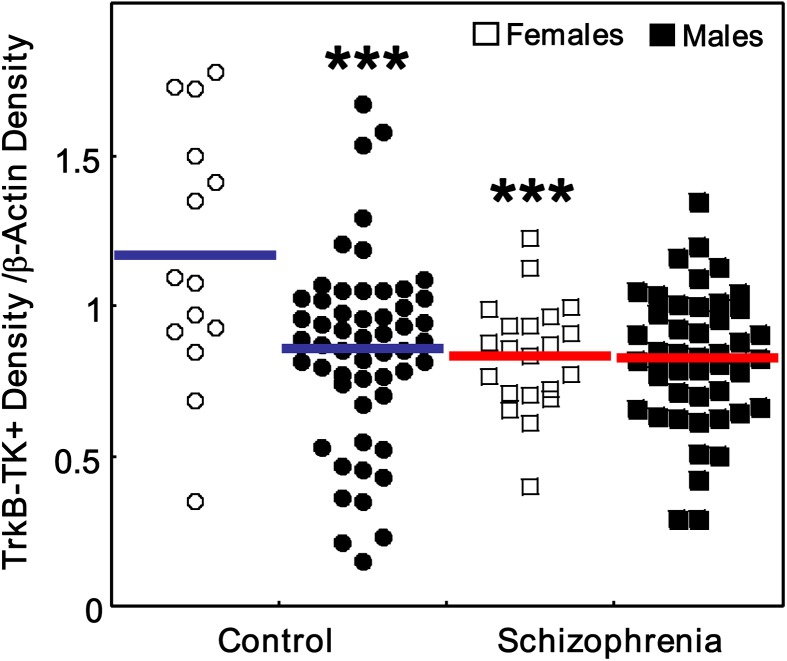
No significant main effect of gender or diagnosis × gender interaction was found for TrkB-TK+ mRNA expression by 2-way ANOVA (all F < 0.34, df = 1, 130, all P > .57). Strikingly, when we examined the expression levels of TrkB-TK+ protein, we found highly significant main effects of gender (F = 8.16, df = 1, 132, P = .005) and diagnosis × gender interaction (F = 7.23, df = 1, 132, P = .008) (figure 5). We found that control females expressed higher levels of TrkB-TK+ protein than control males (post hoc LSD, P = .0003) and that females with schizophrenia showed a significant reduction in TrkB-TK+ protein compared with control females (post hoc LSD, P = .0009).
Fig. 5.
Gender-specific changes in TrkB isoform expression in schizophrenia. Full-length TrkB (TrkB-TK+) protein expression was grouped based on gender: females (white) and males (black). Protein expression of TrkB-TK+ was normalized by the corresponding internal controls; hence, expression levels are on the same scale. Data is expressed as TrkB-TK+ density/β-actin density of individual subjects. ***P < .001 compared with control females.
No significant main effect of gender (F = 0.77, df = 1, 129, P = .38) or diagnosis × gender interaction effect (F = 0.001, df = 1, 129, P = .98) was observed for TrkB-TK− mRNA expression. For TrkB-Shc mRNA, no significant main effect of gender or diagnosis × gender interaction effect (both F < .91, df = 1, 132, P > 0.34) was observed. We did not detect a significant main effect of gender (F = 1.87, df = 1, 133, P = .16) or a significant diagnosis × gender interaction effect on truncated TrkB protein (F = 0.44, df = 1, 133, P = .51). No main effect of gender or diagnosis × gender interaction were observed for any of the ratio measures (all gender: F < 0.55, all P > .37).
Antipsychotics May Down-Regulate TrkB-TK+ mRNA
We next examined the relationship between clinical variables and TrkB mRNA expression in people with schizophrenia. TrkB-TK+ mRNA expression showed no significant correlation with age of onset or duration of illness (trend level) but was negatively correlated with lifetime antipsychotic use (r = −.50, P = .00003) (see online supplementary material for table S1). Considering that lifetime antipsychotic use is influenced by age, we conducted partial correlations and found a similar significant correlation between TrkB-TK+ mRNA and lifetime antipsychotic use (r = −.37, P = 0.003). TrkB-TK− and TrkB-Shc mRNA levels did not correlate with age of onset, duration of illness, or lifetime antipsychotic use (see online supplementary material for table S1).
We then determined whether the class of antipsychotics used by people with schizophrenia had differential effects on TrkB transcript expression. Considering that most cases were on both typical and atypical antipsychotics during their lifetime, we analyzed only those who were just on typical or atypical antipsychotics. In the schizophrenia group, we found that individuals on typical antipsychotics had significantly lower TrkB-TK+ mRNA levels (24% lower; ANOVA main effect: F = 14.09, df = 1, 47, P = .0005) compared with those individuals on atypical antipsychotics. TrkB-TK− and TrkB-Shc mRNA levels were comparable between groups (all P > .10).
When we examined the effect of clinical variables on TrkB protein, we found no significant correlations with expression of the TrkB-TK+ protein (see online supplementary material for table S1). Interestingly, we observed a significant positive correlation between truncated TrkB protein expression and lifetime antipsychotic use (r = .24, P = .05). However, when we controlled for the effect of age by partial correlation, the significance of antipsychotics on truncated TrkB protein expression was lost (r = .15, P = .23). Expression of truncated TrkB protein did not correlate with age of onset or duration of illness (see online supplementary material for table S1).
We found no significant difference in TrkB protein (both full-length and truncated) in people with schizophrenia on typical or atypical antipsychotics (all P > .40).
Relationship Between TrkB mRNA and GAD67 mRNA
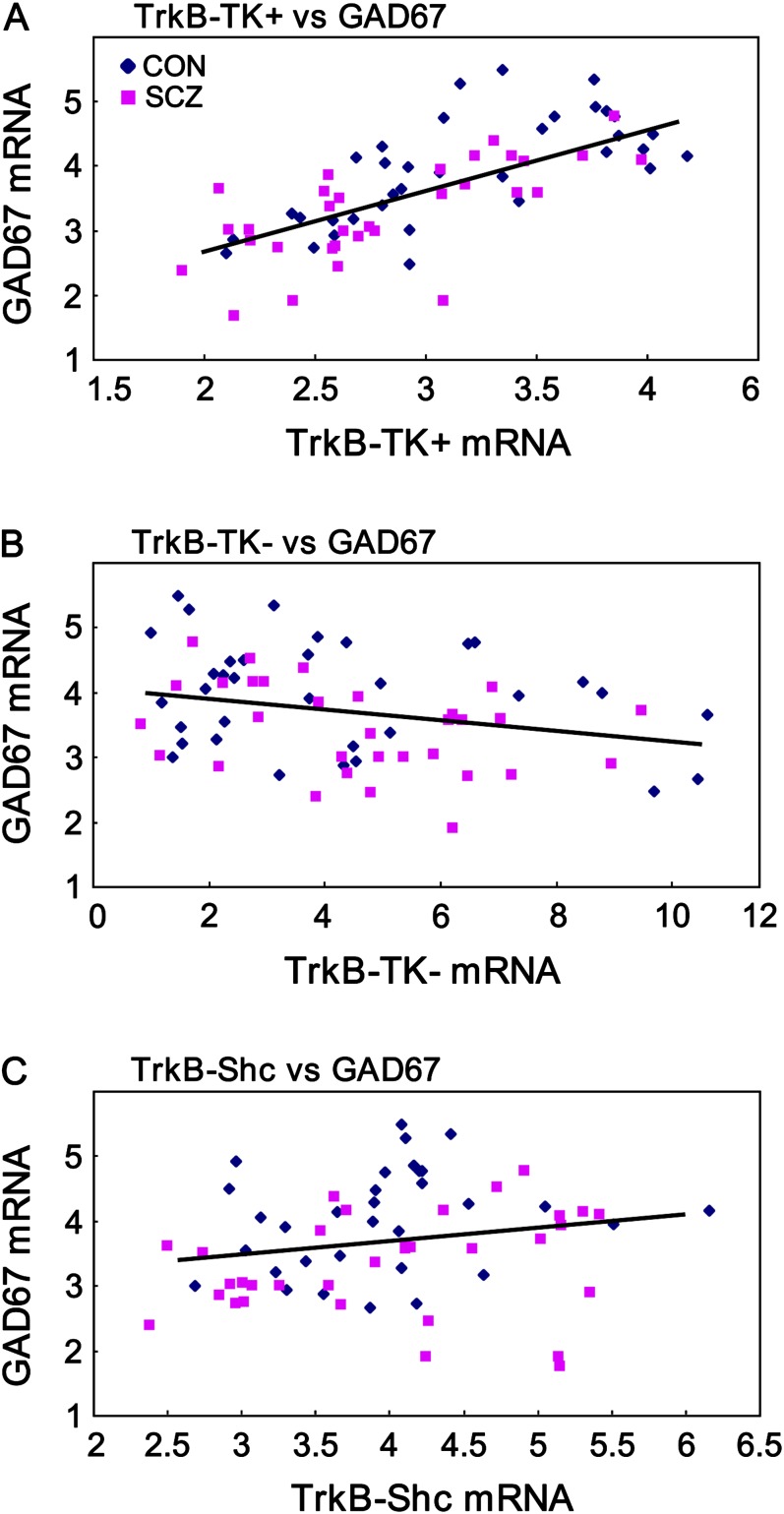
Previously, a reduction in TrkB-TK+ mRNA expression was found to correlate with a reduction in GAD67 mRNA expression.3 However, a relationship between either of the truncated TrkB receptors with GAD67 mRNA has yet to be demonstrated. Using the TRC cohort, we found a highly significant positive correlation between TrkB-TK+ and GAD67 mRNA levels (r = .69, P < .000001) (figure 6A). Conversely, a moderately strong negative correlation between TrkB-TK− and GAD67 mRNA was observed (r = −.32, P = .008) (figure 6B). TrkB-Shc and GAD67 mRNA expression were not significantly correlated (r = .19, P = .13) (figure 6C).
Fig. 6.
Correlations between TrkB and GAD67 mRNA. GAD67 mRNA expression levels were positively correlated with (A) full-length TrkB (TrkB-TK+) (P < .000001) and (B) negatively correlated with truncated TrkB (TrkB-TK−) (P = .008) but not correlated with (C) sarc homology containing TrkB (TrkB-Shc) (P = .13). Diamonds represent control and squares represent schizophrenic cases.
Relationship Between BDNF and TrkB Expression
In our previous study, we found a significant decrease in BDNF transcript II-IX and trends toward reduction in BDNF transcript VI-IX in the DLPFC of people with schizophrenia.4 Considering that we find changes in TrkB transcripts in this current study, we determined whether changes in TrkB were correlated with BDNF. We found significant positive correlations between TrkB-TK+ mRNA levels and BDNF transcripts II-IX (r = .56, P = .000003) and IV-IX (r = .42, P = .0003); and significant negative correlations between TrkB-TK− mRNA and BDNF transcripts IV-IX (r = −.45, P = .0001) and VI-IX (r = −.33, P = .01). Moreover, a significant negative correlation was also found between expression of the truncated TrkB proteins and BDNF transcript II-IX (r = −.31, P = .02).
Discussion
The major finding we report is the novel and highly significant increase in both TrkB-TK− and TrkB-Shc mRNA levels in the DLPFC of people with schizophrenia. The increased expression of truncated TrkBs in schizophrenia was the most robust change found in this study using a large cohort (n = 144). Our finding of large increases in truncated TrkB mRNA levels, along with more limited increases in truncated TrkB proteins, suggests that additional to elevated steady-state levels of truncated TrkB protein, there is a possibility of greater TrkB-TK− and/or TrkB-Shc protein turnover and thus, a higher demand on mRNA synthesis. However, further studies examining the regulation of truncated TrkB mRNA are needed to determine if alterations in these mRNAs are primary in nature or are changed in response to alterations in TrkB signaling or usage in schizophrenia. Regardless of the molecular mechanism involved, our findings suggest that adequate stimulation of neurotrophin signaling in the cortex of people with schizophrenia will require not only augmentation of BDNF and TrkB-TK+ signaling but also simultaneous blocking of the increased truncated TrkBs.
Another novel finding we report is the reduction in the TrkB-TK+:truncated TrkB ratios on the mRNA and protein levels. The fact that reductions in the TrkB-TK+:truncated TrkB (both mRNA and protein) ratios were significant in both males and females with schizophrenia suggests that a reduced ratio of full-length to truncated TrkB may be a generalizable measure of altered neurotrophin response in schizophrenia. Indeed, increased TrkB-TK− and TrkB-Shc in neurons may act as dominant-negative receptors by forming inactive heterodimers with TrkB-TK+ (tyrosine kinase signaling requires 2 functional tyrosine kinase units) or compete with neuronal TrkB-TK+ homodimers for BDNF binding.
Finally, while we replicate previous observations of reduced TrkB-TK+ mRNA levels in the DLPFC of people with schizophrenia,3,11 the overall reduction we found was small and not statistically significant. However, we did find significant reductions in TrkB-TK+ mRNA in those people with schizophrenia on typical antipsychotics and the difference between our study and others may be because earlier cohorts had a greater proportion of patients on typical antipsychotics as compared with our study. Despite a modest reduction in TrkB-TK+ mRNA, a significant decrease was observed in TrkB-TK+ protein expression, consistent with a previous report that found a reduction in TrkB-TK+ protein in the DLPFC of people with schizophrenia.33 However, when we examined gender-specific levels of TrkB-TK+ protein, we found that only females showed a significant reduction in schizophrenia, whereas males showed no change (please refer to the online supplementary material for Discussion). The age range of our cohort spanned young adulthood to aged (18–78 y) and consistent with previous findings.3,11 we found that TrkB-TK+ mRNA continued to decrease gradually throughout adult life in the DLPFC.
Implications of Increased Truncated TrkB Isoforms
The neuronal response to BDNF is proposed to depend principally upon the relative levels of TrkB-TK+ and TrkB-TK−.21,24 While little has been reported on TrkB-Shc, this truncated isoform is unable to be phosphorylated by TrkB-TK+,13 suggesting that it shares a similar biological function to TrkB-TK−. However, dissimilar to TrkB-TK−, which can be expressed in multiple tissues and by both neurons (pyramidal and interneurons) and glia (astrocytes),16–18 TrkB-Shc is brain-specific and neuron-specific.13,25. This would suggest that in neuronal membranes, direct physical interactions between TrkB-TK+ and TrkB-Shc are likely to occur and that increased TrkB-Shc may be anatomically positioned to compromise neuronal second messenger activation by BDNF. Previously, we and others have found that cortical BDNF mRNA and protein are reduced in schizophrenia.2–4 When we correlated TrkB transcript levels with BDNF alternative transcript expression,4 we found that expression of BDNF transcripts II-, IV-, and VI-IX were significantly correlated with TrkB-TK+ and TrkB-TK− mRNA levels. Importantly, BDNF transcript II-IX expression, the transcript we found most affected in the DLPFC in schizophrenia,4 was significantly and negatively correlated with expression of the truncated TrkB protein. This finding suggests that changes in this ligand-receptor pair may be related or reflective of a shared underlying pathology and supports our hypothesis that alterations in BDNF, TrkB-TK+, and/or truncated TrkB receptor expression may work in combination to alter neuronal neurotrophin signaling in the DLPFC of people with schizophrenia.
Interestingly, the diagnostic changes we observed in the ratios of TrkB-TK+ and the truncated TrkB receptors were very dramatic. Since neural activity up-regulates BDNF and TrkB mRNA and that BDNF mRNA is decreased in schizophrenia, we expected to find a large decrease in overall TrkB levels. However, we only found subtle reductions in TrkB-TK+ mRNA despite the fact that some BDNF transcripts did positively correlate with TrkB-TK+ as would be expected if they were co-regulated. Instead, we found an increase in mRNA encoding truncated forms of TrkB and a negative correlation with BDNF mRNA. We speculate that the subtle decrease in TrkB-TK+ mRNA and dramatic increases in truncated TrkB receptors (TrkB-TK−/TrkB-Shc) may be due to alterations in TrkB splicing and increased stability of the truncated TrkB transcripts. Increases in the stability of truncated TrkB mRNAs coupled with increased translation may result in the different direction and magnitude of change observed between TrkB-TK+ and the truncated TrkB receptors. This is plausible as other transcripts associated with the schizophrenia neuropathology, such as BDNF, have demonstrated altered alternative transcript expression and posttranscriptional regulation in schizophrenia.4,34
Putative Cause/Consequence of Altered Cortical TrkB
Alterations in TrkB ratios are associated with cortical maldevelopment. Deficits in neurotrophin signaling have been proposed as an upstream event in the neuropathology of schizophrenia. In particular, deficits in neurotrophin signaling may be a principle factor leading to deficiencies in cortical inhibitory interneurons in schizophrenia.3,35–37 Considering that BDNF is primarily synthesized by excitatory neurons, expression of TrkB-TK+, particularly when expressed by the inhibitory interneurons, may be critical for enabling them to gain sufficient target-derived neurotrophic support. Indeed, reductions in cortical TrkB-TK+ and BDNF expression have been found to correlate with reductions in GAD67 mRNA, a key pathological hallmark of schizophrenia.3,38,39 Previously, we found a significant reduction in GAD67 mRNA levels in the DLPFC of people with schizophrenia in our cohort40 and in this current study, we found a highly significant positive correlation between TrkB-TK+ and GAD67 mRNA levels. Interestingly, we also found a significant negative correlation between TrkB-TK−, but not TrkB-Shc, and GAD67 mRNA. This would suggest that increased expression of TrkB-TK− in neurons and glia may be deleterious for interneuron health and viability as increased expression of TrkB-TK− would serve to limit the amount of BDNF secreted by pyramidal neurons for interneuron TrkB-TK+ binding. Together, these findings support the notion that adequate balance of TrkB isoforms may be crucial for proper cortical interneuron development, differentiation, and/or maintenance.
Increases in TrkB-TK− expression in glia may negatively impact interneuron function in several ways. An increase in glia-specific expression of TrkB-TK− may compete with interneurons for BDNF binding and reduce overall neurotrophic support to interneurons that do not produce BDNF. Alternatively, TrkB-TK− has been shown to be important in mediating calcium release from astrocytes.41 An increase in TrkB-TK− expression in astrocytes may dysregulate calcium signaling and negatively impact interneuron health and viability. Interestingly, expression of the calcium binding protein, parvalbumin within a specific set of interneurons is consistently decreased in the prefrontal cortex in schizophrenia.37,42,43 Furthermore, astrocytic TrkB-TK− is coupled to the Rho signaling pathway and TrkB-TK− is critical in regulating astrocyte morphology in response to BDNF44 and changes in astrocyte morphology can impact communication at synapses where astrocytic processes are involved in neurotransmitter removal.
Potential Limitations
We found that TrkB-TK+ mRNA expression was correlated negatively with lifetime antipsychotic usage. However, when we grouped individuals with schizophrenia into the class of antipsychotics prescribed, we found that those on typical antipsychotics had lower levels of TrkB-TK+ mRNA compared with those on atypical antipsychotics. These findings would suggest that the trending reduction in TrkB-TK+ mRNA found in this study or the reduction in TrkB-TK+ mRNA found previously3,11 may have occurred as a consequence of antipsychotic medication. However, we previously found no significant correlation between TrkB-TK+ mRNA and lifetime antipsychotic exposure,11 and previous studies have also found no change in TrkB-TK+ mRNA levels in the prefrontal cortex of monkeys after long-term exposure to haloperidol3 or in the frontal cortex of rodents following chronic exposure to antipsychotics.45,46 Thus, antipsychotic usage may be a potential confound in this study, and further work delineating the effects of specific classes of antipsychotics on TrkB mRNA and protein expression in schizophrenia is required.
Conclusions
Our findings show for the first time that increases in truncated TrkB receptor isoforms may contribute to deficits in BDNF/TrkB signaling and neuronal plasticity in schizophrenia. However, the compromised signaling potential of BDNF/TrkB, be it from reduced BDNF, reduced TrkB-TK+, or increased truncated TrkB receptors, would be expected to converge on the same consequence of reduced neurotrophic support for cortical neurons leading to impairments in inhibitory transmission and cortical plasticity in people with schizophrenia. Further work aimed at understanding how dysregulation of TrkB occurs in schizophrenia would be needed; however, our results suggest that there may be multiple points of dysregulation in this gene and gene product in schizophrenia. In particular, our study highlights the need to engineer treatments that can stimulate TrkB-TK+ function while attenuating the effects of truncated TrkBs if we are to restore proper BDNF signaling in the brains of people with schizophrenia.
Funding
Schizophrenia Research Institute utilizing infrastructure funding from New South Wales (NSW) Health; Macquarie Group Foundation; Neuroscience Research Australia; University of NSW. National Health and Medical Research Council Postdoctoral Training Fellowship (568884 to J.W.).
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Acknowledgments
We thank Shan Yuan Tsai, Duncan Sinclair, and Heng Giap Woon for technical assistance. Tissues were from the Australian Brain Donor Programs NSW TRC, which is supported by The University of Sydney, National Health and Medical Research Council of Australia, Schizophrenia Research Institute, National Institute of Alcohol Abuse and Alcoholism, National Institutes of Health, and the NSW Department of Health. There are no conflicts of interest.
References
- 1.Weickert TW, Goldberg TE. First- and second-generation antipsychotic medication and cognitive processing in schizophrenia. Curr Psychiatry Rep. 2005;7:304–310. doi: 10.1007/s11920-005-0085-5. [DOI] [PubMed] [Google Scholar]
- 2.Weickert CS, Hyde TM, Lipska BK, Herman MM, Weinberger DR, Kleinman JE. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8:592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- 3.Hashimoto T, Bergen SE, Nguyen QL, et al. Relationship of brain-derived neurotrophic factor and its receptor TrkB to altered inhibitory prefrontal circuitry in schizophrenia. J Neurosci. 2005;25:372–383. doi: 10.1523/JNEUROSCI.4035-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong J, Hyde TM, Cassano HL, Deep-Soboslay A, Kleinman JE, Weickert CS. Promoter specific alterations of BDNF mRNA in schizophrenia. Neuroscience. 2010;169:1071–1084. doi: 10.1016/j.neuroscience.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poo MM. Neurotrophins as synaptic modulators. Nat Rev Neurosci. 2001;2:24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- 7.Thoenen H. Neurotrophins and neuronal plasticity. Science. 1995;270:593–598. doi: 10.1126/science.270.5236.593. [DOI] [PubMed] [Google Scholar]
- 8.McAllister AK, Katz LC, Lo DC. Neurotrophins and synaptic plasticity. Annu Rev Neurosci. 1999;22:295–318. doi: 10.1146/annurev.neuro.22.1.295. [DOI] [PubMed] [Google Scholar]
- 9.Klein R, Nanduri V, Jing SA, et al. The trkB tyrosine protein kinase is a receptor for brain-derived neurotrophic factor and neurotrophin-3. Cell. 1991;66:395–403. doi: 10.1016/0092-8674(91)90628-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu B, Zang K, Ruff NL, et al. Cortical degeneration in the absence of neurotrophin signaling: dendritic retraction and neuronal loss after removal of the receptor TrkB. Neuron. 2000;26:233–245. doi: 10.1016/s0896-6273(00)81153-8. [DOI] [PubMed] [Google Scholar]
- 11.Weickert CS, Ligons DL, Romanczyk T, et al. Reductions in neurotrophin receptor mRNAs in the prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2005;10:637–650. doi: 10.1038/sj.mp.4001678. [DOI] [PubMed] [Google Scholar]
- 12.Luberg K, Wong J, Weickert CS, Timmusk T. Human TrkB gene: novel alternative transcripts, protein isoforms and expression pattern in the prefrontal cerebral cortex during postnatal development. J Neurochem. 2010;113:952–964. doi: 10.1111/j.1471-4159.2010.06662.x. [DOI] [PubMed] [Google Scholar]
- 13.Stoilov P, Castren E, Stamm S. Analysis of the human TrkB gene genomic organization reveals novel TrkB isoforms, unusual gene length, and splicing mechanism. Biochem Biophys Res Commun. 2002;290:1054–1065. doi: 10.1006/bbrc.2001.6301. [DOI] [PubMed] [Google Scholar]
- 14.Klein R, Parada LF, Coulier F, Barbacid M. trkB, a novel tyrosine protein kinase receptor expressed during mouse neural development. EMBO J. 1989;8:3701–3709. doi: 10.1002/j.1460-2075.1989.tb08545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein R, Conway D, Parada LF, Barbacid M. The trkB tyrosine protein kinase gene codes for a second neurogenic receptor that lacks the catalytic kinase domain. Cell. 1990;61:647–656. doi: 10.1016/0092-8674(90)90476-u. [DOI] [PubMed] [Google Scholar]
- 16.Ohira K, Shimizu K, Yamashita A, Hayashi M. Differential expression of the truncated TrkB receptor, T1, in the primary motor and prefrontal cortices of the adult macaque monkey. Neurosci Lett. 2005;385:105–109. doi: 10.1016/j.neulet.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 17.Ohira K, Hayashi M. Expression of TrkB subtypes in the adult monkey cerebellar cortex. J Chem Neuroanat. 2003;25:175–183. doi: 10.1016/s0891-0618(02)00096-0. [DOI] [PubMed] [Google Scholar]
- 18.Frisen J, Verge VM, Fried K, et al. Characterization of glial trkB receptors: differential response to injury in the central and peripheral nervous systems. Proc Natl Acad Sci U S A. 1993;90:4971–4975. doi: 10.1073/pnas.90.11.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biffo S, Offenhauser N, Carter BD, Barde YA. Selective binding and internalisation by truncated receptors restrict the availability of BDNF during development. Development. 1995;121:2461–2470. doi: 10.1242/dev.121.8.2461. [DOI] [PubMed] [Google Scholar]
- 20.Snapyan M, Lemasson M, Brill MS, et al. Vasculature guides migrating neuronal precursors in the adult mammalian forebrain via brain-derived neurotrophic factor signaling. J Neurosci. 2009;29:4172–4188. doi: 10.1523/JNEUROSCI.4956-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eide FF, Vining ER, Eide BL, Zang K, Wang XY, Reichardt LF. Naturally occurring truncated trkB receptors have dominant inhibitory effects on brain-derived neurotrophic factor signaling. J Neurosci. 1996;16:3123–3129. doi: 10.1523/JNEUROSCI.16-10-03123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ninkina N, Adu J, Fischer A, Pinon LG, Buchman VL, Davies AM. Expression and function of TrkB variants in developing sensory neurons. Embo J. 1996;15:6385–6393. [PMC free article] [PubMed] [Google Scholar]
- 23.Armanini MP, McMahon SB, Sutherland J, Shelton DL, Phillips HS. Truncated and catalytic isoforms of trkB are co-expressed in neurons of rat and mouse CNS. Eur J Neurosci. 1995;7:1403–1409. doi: 10.1111/j.1460-9568.1995.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 24.Arevalo JC, Wu SH. Neurotrophin signaling: many exciting surprises! Cell Mol Life Sci. 2006;63:1523–1537. doi: 10.1007/s00018-006-6010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romanczyk TB, Weickert CS, Webster MJ, Herman MM, Akil M, Kleinman JE. Alterations in trkB mRNA in the human prefrontal cortex throughout the lifespan. Eur J Neurosci. 2002;15:269–280. doi: 10.1046/j.0953-816x.2001.01858.x. [DOI] [PubMed] [Google Scholar]
- 26.Weickert CS, Sheedy D, Rothmond DA, et al. Selection of reference gene expression in a schizophrenia brain cohort. Aust N Z J Psychiatry. 2010;44:59–70. doi: 10.3109/00048670903393662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webster MJ. Tissue preparation and banking. Prog Brain Res. 2006;158:3–14. doi: 10.1016/S0079-6123(06)58001-X. [DOI] [PubMed] [Google Scholar]
- 28.Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The stanley foundation brain collection and neuropathology consortium. Schizophr Res. 2000;44:151–155. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
- 29.Sarris M, Garrick TM, Sheedy D, Harper CG. Banking for the future: an Australian experience in brain banking. Pathology. 2002;34:225–229. doi: 10.1080/00313020220131260. [DOI] [PubMed] [Google Scholar]
- 30.Kozlovsky N, Shanon-Weickert C, Tomaskovic-Crook E, Kleinman JE, Belmaker RH, Agam G. Reduced GSK-3beta mRNA levels in postmortem dorsolateral prefrontal cortex of schizophrenic patients. J Neural Transm. 2004;111:1583–1592. doi: 10.1007/s00702-004-0166-3. [DOI] [PubMed] [Google Scholar]
- 31.Wong J, Webster MJ, Cassano H, Weickert CS. Changes in alternative brain-derived neurotrophic factor transcript expression in the developing human prefrontal cortex. Eur J Neurosci. 2009;29:1311–1322. doi: 10.1111/j.1460-9568.2009.06669.x. [DOI] [PubMed] [Google Scholar]
- 32.Lipska BK, Deep-Soboslay A, Weickert CS, et al. Critical factors in gene expression in postmortem human brain: focus on studies in schizophrenia. Biol Psychiatry. 2006;60:650–658. doi: 10.1016/j.biopsych.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi M, Shirakawa O, Toyooka K, et al. Abnormal expression of brain-derived neurotrophic factor and its receptor in the corticolimbic system of schizophrenic patients. Mol Psychiatry. 2000;5:293–300. doi: 10.1038/sj.mp.4000718. [DOI] [PubMed] [Google Scholar]
- 34.Mellios N, Huang HS, Baker SP, Galdzicka M, Ginns E, Akbarian S. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry. 2009;65:1006–1014. doi: 10.1016/j.biopsych.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 35.Morris HM, Hashimoto T, Lewis DA. Alterations in somatostatin mRNA expression in the dorsolateral prefrontal cortex of subjects with schizophrenia or schizoaffective disorder. Cereb Cortex. 2008;18:1575–1587. doi: 10.1093/cercor/bhm186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woo NH, Lu B. Regulation of cortical interneurons by neurotrophins: from development to cognitive disorders. Neuroscientist. 2006;12:43–56. doi: 10.1177/1073858405284360. [DOI] [PubMed] [Google Scholar]
- 37.Fung SJ, Webster MJ, Sivagnanasundaram S, Duncan C, Weickert CS. Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am J Psychiatry. 2009;167:1479–1488. doi: 10.1176/appi.ajp.2010.09060784. [DOI] [PubMed] [Google Scholar]
- 38.Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- 39.Ray M, Weickert CS, Wyatt E, Webster M. Decreased BDNF, trkB-TK+ and GAD67 mRNA expression in the hippocampus of individuals with schizophrenia and mood disorders. J Psychiatr Res. 2011;36:195–203. doi: 10.1503/jpn.100048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duncan CE, Webster MJ, Rothmond DA, Bahn S, Elashoff M, Shannon Weickert C. Prefrontal GABA(A) receptor alpha-subunit expression in normal postnatal human development and schizophrenia. J Psychiatr Res. 2010;44:673–681. doi: 10.1016/j.jpsychires.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 41.Lidow MS. Calcium signaling dysfunction in schizophrenia: a unifying approach. Brain Res Brain Res Rev. 2003;43:70–84. doi: 10.1016/s0165-0173(03)00203-0. [DOI] [PubMed] [Google Scholar]
- 42.Hashimoto T, Volk DW, Eggan SM, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23:6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reynolds GP, Zhang ZJ, Beasley CL. Neurochemical correlates of cortical GABAergic deficits in schizophrenia: selective losses of calcium binding protein immunoreactivity. Brain Res Bull. 2001;55:579–584. doi: 10.1016/s0361-9230(01)00526-3. [DOI] [PubMed] [Google Scholar]
- 44.Ohira K, Kumanogoh H, Sahara Y, et al. A truncated tropomyosin-related kinase B receptor, T1, regulates glial cell morphology via Rho GDP dissociation inhibitor 1. J Neurosci. 2005;25:1343–1353. doi: 10.1523/JNEUROSCI.4436-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nibuya M, Morinobu S, Duman RS. Regulation of BDNF and trkB mRNA in rat brain by chronic electroconvulsive seizure and antidepressant drug treatments. J Neurosci. 1995;15:7539–7547. doi: 10.1523/JNEUROSCI.15-11-07539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linden AM, Vaisanen J, Lakso M, Nawa H, Wong G, Castren E. Expression of neurotrophins BDNF and NT-3, and their receptors in rat brain after administration of antipsychotic and psychotrophic agents. J Mol Neurosci. 2000;14:27–37. doi: 10.1385/JMN:14:1-2:027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.