Abstract
In studies on parentage assignment with both parents unknown, the exclusion power of a marker set is generally computed under the hypothesis that the potential families tested are independent and unrelated samples. This tends to produce overly optimistic exclusion power estimates. In this work, we have developed a new formula that gives almost unbiased results at the population level.
Findings
Parentage assignment using genomic markers, usually microsatellites, is now widely used for research on population ecology and evolution [1], as well as in selective breeding, particularly for aquatic species. Indeed, maintaining pedigrees for these species is a challenge because of the very small size of individuals at hatching, which prevents physical tagging [2]. When developing a marker set for parentage assignment, it is important to be able to predict the assignment efficiency from a priori data. Exclusion probabilities are easily calculated from allele frequencies and are commonly used to quantify the efficiency of individual markers for parentage assignment. The most frequently used exclusion probability is the probability to exclude a random parent pair that is unrelated to the individual tested (named Q3 in [3], here Q3i for each locus i). Since a single locus is generally not sufficient to exclude all potential parent pairs, several loci have to be combined to reach an appropriate combined exclusion probability Q3, which is calculated as the product of the individual non-exclusion probabilities of all L loci:
| (1) |
Then, the combined exclusion probability is raised to the power of the total number of potential parental pairs to be excluded. With N possible parent pairs (including the correct one), this number is N-1, and the probability to have all parent pairs excluded except the correct one is the theoretical probability of having a unique assignment [4,5]:
| (2) |
However, experience shows that the predicted assignment rates using this formula are often too optimistic, especially in factorial designs, i.e. when the mating structure is unknown and thus all possible mother-father combinations must be taken into consideration [4,6]. It is then necessary to make two assumptions when applying formulae (1) and (2), i.e. (i) exclusion of the N-1 incorrect parent pairs represents N-1 independent tests and (ii) all excluded parents are unrelated to the offspring, which justifies the use of probability Q3. However, in practice, these assumptions are never met. While the lack of independence between tests does not prevent formula (2) to yield good approximations [5], the second problem is generally overlooked.
The most commonly encountered situation is when offspring are collected from a population that has a number of potential parents. The mating structure may be known (in some farmed populations) or not (in the wild or in farmed populations where parents are allowed to mate “naturally”). The practical aim of such studies is to identify the true parent pair of every genotyped offspring that derives from the sampled parents, which means excluding all parent pairs except the true one. Except in very specific cases where only single pair matings occur according to a perfectly known mating structure, the sole use of Q3 is disqualified because some potential half-sib families will have to be excluded. This is especially true when no mating structure is assumed (all mother-father combinations are considered possible, as in Figure 1) and thus the half-sib families cannot be considered to be unrelated to the correct family under consideration. The general approach is to exclude all mother-father combinations other than the true one, without taking a mating structure into account since, in most cases, the aim is to establish or check the mating structure.
Figure 1.
Types of family relationships to be excluded for an offspring. Types of family relationships to be excluded for an offspring with Nm potential mothers and Nf potential fathers; black = true family of an offspring; grey = Nf -1 families that share the same mother and Nm -1 families that share the same father, that have to be excluded; white = (Nm -1)(Nf -1) pairs of parents that are unrelated to the true parents and that also have to be excluded.
Another exclusion probability Q1 was initially proposed by Jamieson [7] to calculate the probability to exclude one parent when the other parent is known, which is relevant to the exclusion of parents from half-sib families sharing one parent with the correct family. Probabilities Q1i and Q3i can be calculated for each locus with the following formulae [3]:
| (3) |
| (4) |
with and pj the frequency of the jth allele of locus i in the population. Combined probabilities over all loci, Q1 and Q3 can be calculated with formula (1).
Then, the combined probability of having a unique assignment among parent pairs that share one parent with the true parental pair is:
| (5) |
while the probability of having a unique assignment among unrelated parent pairs is:
| (6) |
Since the probability of having a unique assignment requires having both unique assignments within related pairs and within unrelated pairs, the global probability of having a unique assignment (also named exclusion power) is:
| (7) |
It is then clear that the probability of having a unique assignment decreases exponentially as the number of potential parents increases, as already underlined by Wang [[5]]. However, the rate of decrease depends on whether term Q1 or term Q3 in formula (7) is most influential. Dodds et al. [3] have already shown that Q3i is always greater than Q1i for a given locus regardless of the allelic frequencies [3].
In the work reported here, we studied the relative importance of Q1 and Q3 using idealized loci, with three, five or eight equally frequent alleles. Individual Q1i values were 0.370 for a locus with three alleles, 0.595 for a locus with five alleles and 0.743 for a locus with eight alleles, while the values for Q3i were 0.519 for a locus with three alleles, 0.772 with five alleles and 0.898 with eight alleles. In most cases, these values reflect microsatellites with low, moderate or high variability.
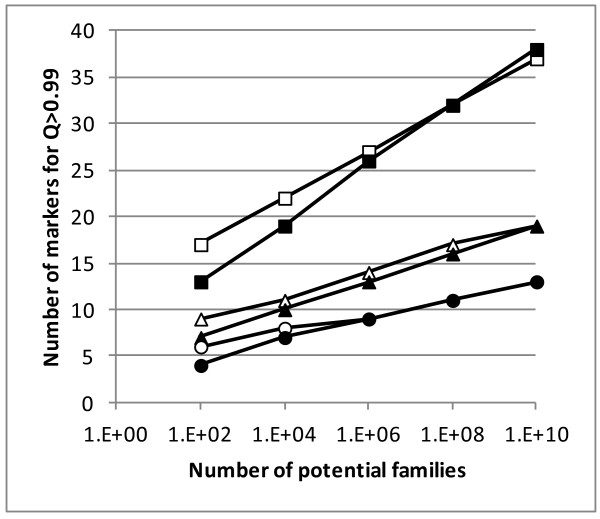
As shown in Figure 2, in general a larger number of loci were needed for the Q1 term to exceed 0.99 compared to the Q3 term, except for very high numbers of potential families (≥ 106 for loci with eight alleles, ≥ 1010 for loci with five alleles and ≥ 108 for loci with three alleles). The only case in which the Q3 term required more loci than the Q1 term to reach 0.99 was with tri-allelic (low variability) loci and more than 1010 potential families. Thus in most cases, and especially when the number of potential families is moderate and the variability of the markers is low or intermediate, Pu will be governed by the Q1 term, contrary to the general view [4].
Figure 2.
Number of markers to achieve exclusion power greater than 0.99 for both terms in formula (7). Number of markers to achieve exclusion power greater than 0.99 for both terms in formula (7); white symbols for the Q1 term; black symbols for the Q3 term; squares = loci with three equally frequent alleles; triangles = loci with five equally frequent alleles; circles = loci with eight equally frequent alleles; the situations simulated included N potential fathers and N potential mothers and, thus, N2 families.
One important thing to note is that formula (7) does not assume a mating structure. This is because no mother-father combination is excluded a priori on the basis of pre-existing knowledge about mating structure and, thus, exclusion is performed on the basis of a full factorial design (Figure 1), which is the general case when no mating structure is assumed. It may be possible to consider fewer combinations when the mating structure is known and thus, modify the exponents of Q1 and Q3 in formula (7), but this approach is not recommended since it limits the generality of the estimated assignment power.
When comparing our results with those previously reported in the literature [4,6], we found that, except for marker sets with a very low assignment power, formula (7) gives much more accurate results than formula (2) (Table 1). When assignment power is low, formula (7) tends to underestimate it, making it a conservative estimate. Other problems (linkage between markers, genotyping errors, inbreeding, use of relatives as parents, sampling errors, etc.) may further decrease the assignment power of a marker set but the systematic gap between the assignment power computed with formula (2) and the theoretical one (up to now approached only by simulation) is the main cause of overestimation of the power of marker sets for parentage assignment [6]. Since formula (7) is easily computed based on allele frequencies in a spreadsheet, we recommend its use to design marker sets with an appropriate exclusion power.
Table 1.
Comparison of predicted and simulated exclusion power Pu of idealized and real marker sets
| |
Exclusion power Pu |
|||||
|---|---|---|---|---|---|---|
| Type of markers | Size of factorial design (Nf x Nm) | Alleles/ locusa | Number of loci | Predicted (Eq.2)* | Simulated | Predicted (Eq.7)** |
| Idealized markers (equally frequent alleles) |
10x10 |
5 |
3 |
0.3064 |
0.2123 |
0.1104 |
| |
10x10 |
5 |
6 |
0.9861 |
0.9163 |
0.9131 |
| |
10x10 |
5 |
9 |
0.9998 |
0.9947 |
0.9946 |
| |
10x10 |
5 |
12 |
1.0000 |
0.9997 |
0.9996 |
| |
10x10 |
10 |
3 |
0.9690 |
0.8448 |
0.8334 |
| |
10x10 |
10 |
6 |
1.0000 |
0.9987 |
0.9986 |
| |
20x20 |
5 |
3 |
0.0085 |
0.0321 |
0.0001 |
| |
20x20 |
5 |
6 |
0.9453 |
0.8143 |
0.8037 |
| |
20x20 |
5 |
9 |
0.9993 |
0.9884 |
0.9884 |
| |
20x20 |
5 |
12 |
1.0000 |
0.9993 |
0.9993 |
| |
20x20 |
10 |
3 |
0.8810 |
0.6717 |
0.6409 |
| |
20x20 |
10 |
6 |
1.0000 |
0.9972 |
0.9971 |
| Real microsatellites |
76x13 |
20.1 |
8 |
1.0000 |
0.9994 |
0.9993 |
| |
75x26 |
21.7 |
6 |
0.9999 |
0.9934 |
0.9934 |
| |
41x8 |
19.3 |
6 |
0.9999 |
0.9928 |
0.9920 |
| |
20x2 |
16.3 |
4 |
0.9986 |
0.9465 |
0.9421 |
| |
147x8 |
7.5 |
8 |
0.9911 |
0.8604 |
0.8636 |
| |
96x8 |
7.6 |
8 |
0.9975 |
0.9473 |
0.9422 |
| |
24x10 |
7.8 |
8 |
0.9990 |
0.9712 |
0.9782 |
| 100x101 | 7.5 | 12 | 0.9968 | 0.9708 | 0.9696 | |
Predicted and simulated values from Villanueva et al. [4] for idealized marker sets and from Vandeputte et al. [6] for real marker sets; simulated values were obtained for 800 offspring per cross in [4] and 1000 offspring per cross in 100 independent parent samples in [6]; both used *formula (2) to calculate predicted values, which were compared to the values obtained with **formula (7) described here; afor real loci, average number of alleles per locus.
Competing interests
The author declares no competing interests.
Authors’ contributions
MV identified the question, established the formula, tested it on real data and wrote the article.
Author information
MV works in fish quantitative genetics at the INRA-Ifremer research group on sustainable fish breeding. One of the main tools used for fish quantitative genetics studies is parentage assignment with microsatellite markers, which he contributes to optimize.
References
- Blouin MS. DNA-based methods for pedigree reconstruction and kinship analysis in natural populations. Trends Ecol Evol. 2003;18:503–511. doi: 10.1016/S0169-5347(03)00225-8. [DOI] [Google Scholar]
- Liu ZJ, Cordes JF. DNA marker technologies and their applications in aquaculture genetics. Aquaculture. 2004;238:1–37. doi: 10.1016/j.aquaculture.2004.05.027. [DOI] [Google Scholar]
- Dodds KG, Tate ML, McEwan JC, Crawford AM. Exclusion probabilities for pedigree testing farm animals. Theor Appl Genet. 1996;92:966–975. doi: 10.1007/BF00224036. [DOI] [PubMed] [Google Scholar]
- Villanueva B, Verspoor E, Visscher PM. Parental assignment in fish using microsatellite genetic markers with finite numbers of parents and offspring. Anim Genet. 2002;33:33–41. doi: 10.1046/j.1365-2052.2002.00804.x. [DOI] [PubMed] [Google Scholar]
- Wang J. Parentage and sibship exclusions: higher statistical power with more family members. Heredity. 2007;99:205–217. doi: 10.1038/sj.hdy.6800984. [DOI] [PubMed] [Google Scholar]
- Vandeputte M, Rossignol MN, Pincent C. From theory to practice: empirical evaluation of the assignment power of marker sets for pedigree analysis in fish breeding. Aquaculture. 2011;314:80–86. doi: 10.1016/j.aquaculture.2011.01.043. [DOI] [Google Scholar]
- Jamieson A. The genetics of transferrins in cattle. Heredity. 1965;20:419–441. doi: 10.1038/hdy.1965.54. [DOI] [PubMed] [Google Scholar]