Abstract
To clarify the role of stem cells in hepatocarcinogenesis, the expression of epithelial cell adhesion molecule (EpCAM) and proliferating cell nuclear antigen (PCNA) was investigated in mouse hepatic tumors and embryonic cell lineages. Ten ICR mice were treated with diethylnitrosamine (DEN) at 14 days of age and sacrificed at 36 weeks subsequent to DEN treatment to obtain the hepatic tumors. Mouse embryonic stem cells, hepatic progenitor cells and hepatocyte-like cells, representing 0, 22 and 40 days of differentiation, respectively, were treated in vitro with DEN at four doses (0, 1, 5 and 15 mM; G1, G2, G3 and G4, respectively) for 24 h and RNA was isolated. A total of 71 hepatic tumors were obtained from the DEN-treated mice. EpCAM expression was increased mainly in hepatic tumor cells, although it was also detected in the surrounding visually normal cells. Double staining showed that EpCAM and PCNA were co-expressed in numerous tumor cells. In vitro, EpCAM expression was significantly different for G4 at day 0 (P<0.01) and for G2, G3 and G4 at day 40 (P<0.01) compared with the control (G1) at the corresponding time-point. PCNA expression was significantly different for G3 and G4 at day 0 (P<0.01), for G2, G3 and G4 at day 22 (P<0.01) and for G2 at day 40 (P<0.01) compared with G1 at the corresponding time-point. In summary, the expression of EpCAM and PCNA was increased in DEN-induced tumors and the expression of EpCAM and PCNA was altered by DEN treatment in cultured cells. This suggests that EpCAM expression may be modulated in the progeny of adult liver stem cells during their differentiation toward hepatocytes and may be increased during DEN-induced hepatocarcinogenesis.
Keywords: mouse, hepatocarcinogenesis, epithelial cell adhesion molecule, proliferating cell nuclear antigen, in vivo, in vitro
Introduction
Carcinogenesis is considered to be a multi-stage process involving the initiation, promotion and progression of tumor cells, which may arise as a result of DNA damage, mutations, clonal expansion of preneoplastic cells and dysregulation of oncogenes and/or tumor suppressor genes.
Embryonic stem cells (ESCs) have self-renewal potential and may be differentiated into lineage-specific cell types, including hepatocytes (1). In the liver, stem cells are located in the ductal plates in fetuses and neonates and in the canals of Hering in infants and adults (2). As stem cells have the potential to survive, DNA damage induced by carcinogens may remain as cell proliferation occurs. Furthermore, stem cells with a loss of DNA repair function may be susceptible to malignant transformation, either directly or through the emergence of cancer-prone stem cells (3). Cell proliferation at the time of carcinogen exposure may be pivotal for the fixation of genotoxic injury as a heritable form (4).
It has been proposed that hepatocellular and ductal carcinomas originate from liver stem cells and that enzyme-altered foci and nodular changes are adaptive non-oncogenic responses to the toxic effects of carcinogens (5). Liver tumors appear to be hierarchically organized and sustained by a distinct subpopulation of cancer stem cells (6). This suggests that stem cells have a significant role in hepatocarcinogenesis.
As a putative stem cell marker, the epithelial cell adhesion molecule (EpCAM) is a membrane glycoprotein highly expressed on the majority of cancer cells (7), although it is also expressed on the majority of normal epithelial cells. In humans, EpCAM-positive hepatocytes have been found to be rare in the early stages of liver disease. However, they became increasingly prominent during the later stages and are consistently arrayed around the periphery of cords of keratin 19-positive hepatobiliary cells in the ductular reaction (8). Human EpCAM-positive hepatocellular carcinomas (HCCs) exhibit a distinct molecular signature with the features of hepatic progenitor cells (HPCs), including the presence of known stem cell/progenitor cell markers, including cytokeratin 19 and c-Kit, whereas EpCAM-negative HCCs exhibit a gene expression profile with the features of mature hepatocytes (9). Thus, EpCAM expression may be elevated during human liver tumor progression.
EpCAM expression in mouse liver carcinogenesis has not yet been reported. In the present study, we investigated the expression of EpCAM in diethylnitrosamine (DEN)-induced hepatic tumors. DEN is widely used in mouse liver cancer models (10) and has been used in several mouse strains (11–14). Based on the finding that young animals treated with DEN exhibited a higher incidence of liver tumors than older animals (15), we also assessed the effects of DEN on the expression of EpCAM and proliferating cell nuclear antigen (PCNA) in hepatic cells. The assessment was carried out at various developmental stages, from ESCs to HPCs and hepatocyte-like cells (HCs).
Materials and methods
Animals and treatment
ICR mice (Koatec Inc., Pyungtaek, Korea) were housed in a room maintained on a 12 h light/dark cycle and at a constant temperature and humidity. The mice were allowed free access to a pellet chow diet (Koatec Inc.) during the experiment. Male mice were bred with females, yielding the F1 generation. Male F1 mice at 14 days of age were injected intraperitoneally with DEN (10 mg/kg body weight; Sigma, St. Louis, MO, USA) dissolved in 0.9% saline. Ten mice treated with DEN were sacrificed 36 weeks later and hepatic masses were sampled for histopathological examination.
Histopathological examination of hepatic tumors
The hepatic masses (n=71) were fixed in 10% neutral phosphate-buffered formalin, embedded in paraffin, sectioned to a thickness of 4 μm and stained with hematoxylin and eosin. Tumor characteristics were classified based on histopathological and cytological criteria.
Immunohistochemical analysis of EpCAM in hepatic tumors
The avidin-biotin complex method was used to stain EpCAM in 4-μm sections of liver tissues. The sections were dewaxed in xylene, hydrated using a graded ethanol series and boiled in sodium citrate buffer (pH 6.0) in an autoclave for 20 min. Then, they were sequentially treated with 0.3% hydrogen peroxide, blocking buffer containing skimmed milk and the anti-EpCAM antibody (ab32392; Abcam, Cambridge, MA, USA; diluted 1:400). The sections were washed with TBS-T and subjected to the ABC-peroxidase procedure (ABC kit; Vector Laboratories, Burlingame, CA, USA). As a negative control, skimmed milk was used instead of the primary antibody.
The immune complexes were visualized using the chromogen 3,3′-diaminobenzidine tetrahydrochloride (DAB). The sections were counterstained with hematoxylin to facilitate their examination under a light microscope.
Double staining of EpCAM and PCNA in hepatic tumors
The sections were dewaxed in xylene, hydrated using a graded ethanol series and boiled in a sodium citrate buffer (pH 6.0) in an autoclave for 20 min. Then they were sequentially treated with 0.3% hydrogen peroxide, blocking buffer containing horse serum and anti-PCNA antibody (M879; Dako, Carpinteria, CA, USA; diluted 1:500) for 1 h. The sections were washed with TBS and incubated with HRP-polymer (MRT621; Biocare Medical, Concord, CA, USA). The immune complexes were visualized using DAB.
For concomitant labeling of EpCAM, the sections were washed with TBS, blocked with goat serum, incubated with anti-EpCAM antibody (ab32392, Abcam) for 1 h and then incubated with AP-polymer (RMR625; Biocare Medical) for 30 min. The sections were washed with TBS, treated with Vulcan Fast Red (FR805H; Biocare Medical), washed again with TBS, counterstained with hematoxylin and viewed under a light microscope.
Culture of mouse ESCs and differentiation of hepatic lineage cells
Mouse NVRQS-11F ESCs were cultured on mitomycin C-treated mouse embryonic fibroblasts (feeder cells) grown on 0.1% gelatin-coated dishes in Dulbecco’s modified Eagle’s medium (Millipore, Billerica, MA, USA), supplemented with 15% fetal bovine serum (Hyclone, Rockville, MD, USA), 2 mM L-glutamine (Millipore), 0.1% non-essential amino acids (Invitrogen, Carlsbad, CA, USA), 1% penicillin-streptomycin (Millipore) and 10 ng/ml mouse leukemia inhibitory factor (Millipore).
To differentiate the ESCs into hepatic lineage cells, we used the culture conditions for differentiating ESCs into HPCs and HCs reported by Zhou et al(16).
DEN treatment of ESCs, HPCs and HCs
To determine a non-cytotoxic concentration of DEN, ESCs were treated with DEN at concentrations of 0–90 mM for 24 h. Cell viability was estimated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and the fluorescence intensity was analyzed using an ArrayScan VTI HCS Reader (Thermo Scientific, Rockford, IL, USA). Subsequently, ESCs (day 0), HPCs (day 22 of differentiation) and HCs (day 40 of differentiation) were treated with four concentrations of DEN (0, 1, 5 and 15 mM; G1, G2, G3, G4, respectively) for 24 h.
EpCAM and PCNA mRNA expression in ESC, HPCs and HCs
RNA was isolated from cultured cells using an easy-spin Total RNA Extraction kit (Intron Biotechnology, Scottsdale, AZ, USA), dissolved in DEPC-treated distilled water and stored at −80°C until use. RNA concentrations were measured using a UV/Vis Spectrophotometer (DU730; Beckman Coulter, Miami, FL, USA). The quality of the isolated RNA was assessed using an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA) and an Agilent RNA 6000 Nano kit (Agilent).
EpCAM and PCNA mRNA expression was determined by relative quantitative real-time PCR in 96-well optical plates using an ABI 7500 Real Time PCR System (Applied Biosystems, Foster City, CA, USA). Assay-on-Demand TaqMan probes (Applied Biosystems) were used to measure EpCAM and PCNA mRNA (Table I). A master mix was created containing the following: 6.25 μl water, 1.25 μl forward primer (9 μM) and reverse primer (9 μM), 2.5 μl probe mixture (2.5 μM) and 12.5 μl TaqMan PCR 2X master mixture (Applied Biosystems). Reverse transcribed total RNA (40 ng in 5 μl) was added as the PCR template.
Table I.
Probe sequences.
| Assay ID | Probe sequence | Gene symbol | Amplicon size (bp) |
|---|---|---|---|
| Mm00493214_m1 | TTGAAAAAGATGTGAAGGGGGAGTC | EpCAM | 95 |
| Mm00448100_g1 | CAACTTGGAATCCCAGAACAGGAGT | PCNA | 117 |
The following PCR conditions were used: initial activation of uracyl-N-glycosylase at 50°C for 2 min; activation of AmpliTaq Gold at 95°C for 10 min; and 45 cycles of denaturation at 95°C for 15 sec and annealing/extension at 60°C for 1 min. During PCR, the amplified products were continuously monitored by measuring the fluorescence emission. All PCR assays were performed in triplicate.
The expression levels of the target genes were normalized to mouse GAPDH mRNA and were presented as relative expression. The expression of the genes was normalized to GAPDH, using the comparative Ct method. The cycle number at which the fluorescence signal of the target product was detectable (threshold cycle, Ct) was normalized against the Ct of GAPDH, to give ΔCt. The expression of the genes relative to a reference was calculated as 2−ΔΔCt, where ΔΔCt referred to the difference between the ΔCt values of the test group and the reference.
Statistical analysis
Data were analyzed using the Student’s t-test with JMP software (SAS Institute, Cary, NC, USA). For all comparisons, P<0.05 was considered to indicate a statistically significant difference.
Results
Histopathological examination of hepatic masses
Histo-pathological examination of the 71 hepatic masses showed that 48 samples were adenomas and 23 were adenocarcinomas. The tumor tissues exhibited nuclear pleomorphism and alteration of cellular structure, with or without fatty liver and inflammatory cell infiltration.
Immunohistochemical examination of EpCAM
EpCAM was immunohistochemically detected mainly in hepatic tumor cells, with some expression in the surrounding visually normal cells, and exhibited a cytoplasmic staining pattern (Fig. 1). EpCAM expression was similar in benign and malignant tumors.
Figure 1.
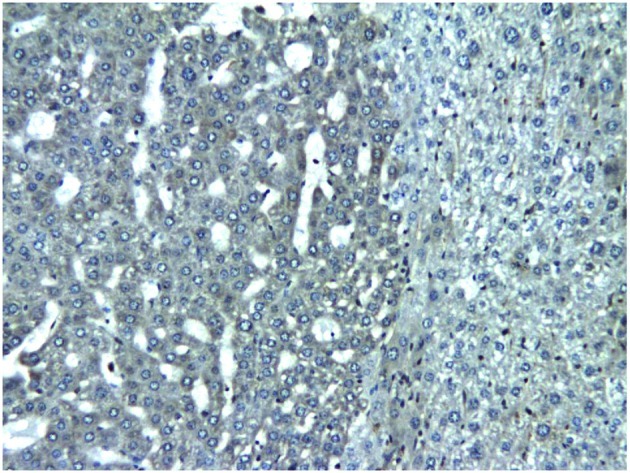
EpCAM expression in a DEN-induced hepatocellular tumor. Magnification ×200. Note the high expression levels of EpCAM and its cytoplasmic localization in the liver tissue of the DEN-induced tumor. EpCAM, epithelial cell adhesion molecule; DEN, diethylnitrosamine.
Double staining of EpCAM and PCNA
Double staining showed that EpCAM and PCNA were co-expressed in numerous tumor cells, particularly in dysplastic ductal tumor cells (Fig. 2). PCNA showed nuclear staining and EpCAM exhibited cytoplasmic staining.
Figure 2.

Co-expression of PCNA and EpCAM in a DEN-induced tumor. Magnification ×200. Note the nuclear staining of PCNA (brown) and the cytoplasmic staining of EpCAM (red) in the tumor and the co-expression in dysplastic ductal tumor cells. PCNA, proliferating cell nuclear antigen; EpCAM, epithelial cell adhesion molecule; DEN, diethylnitrosamine.
EpCAM and PCNA mRNA expression in ESCs, HPCs and HCs
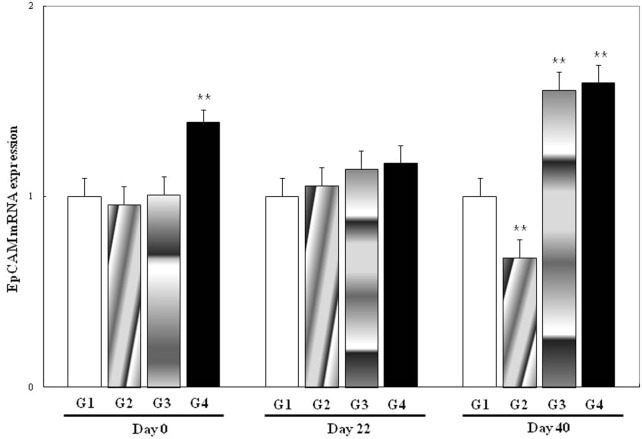
The expression of EpCAM mRNA was significantly different for G4 at day 0 (P<0.01) and for G2, G3 and G4 at day 40 (P<0.01) compared with the control (G1) at the corresponding time-points (Fig. 3). There were no differences for G2 or G3 at day 0, or for G2, G3 or G4 at day 22.
Figure 3.
Expression of epithelial cell adhesion molecule (EpCAM) mRNA in mouse embryonic stem cells (ESCs), hepatic progenitor cells (HPCs) and hepatocyte-like cells (HCs). ESCs (day 0), HPCs (day 22) and HCs (day 40) were treated with DEN at four doses (0, 1, 5 and 15 mM; G1, G2, G3 and G4, respectively) for 24 h and the expression of EpCAM mRNA was determined by PCR. **Significantly different from G1 (P<0.01). DEN, diethylnitrosamine.
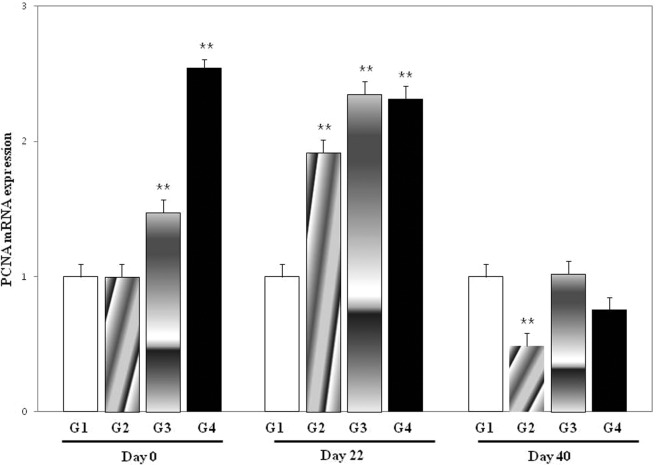
PCNA mRNA expression was significantly different for G3 and G4 at day 0 (P<0.01), for G2, G3 and G4 at day 22 (P<0.01) and for G2 at day 40 (P<0.01) compared with G1 at the corresponding time-points (Fig. 4). There were no differences for G2 at day 0 and for G3 or G4 at day 40.
Figure 4.
Expression of proliferating cell nuclear antigen (PCNA) mRNA in mouse embryonic stem cells (ESCs), hepatic progenitor cells (HPCs) and hepatocyte-like cells (HCs). ESCs (day 0), HPCs (day 22) and HCs (day 40) were treated with DEN at four doses (0, 1, 5 and 15 mM; G1, G2, G3 and G4, respectively) for 24 h and the expression of PCNA mRNA was determined by PCR. **Significantly different from G1 (P<0.01). DEN, diethylnitrosamine.
Discussion
In the present study, EpCAM expression was increased in DEN-induced tumors and was associated with PCNA. Although EpCAM expression was also detected in surrounding visually normal cells, its expression was stronger in hepatic tumor cells. It has been reported that EpCAM-positive HCCs express stem cell and progenitor cell markers (9). Double staining revealed that EpCAM and PCNA were co-expressed in numerous tumor cells, suggesting that EpCAM-positive tumor cells may have the potential to proliferate.
Previously, EpCAM was identified as an additional marker of cancer-initiating cells (7) and it was detected in the cytoplasm of hepatic stem cells and in the plasma membranes of hepatoblasts (2). EpCAM-positive cells in the rat liver were shown to be bipotential adult hepatic epithelial progenitors (17). These findings indicate that EpCAM may be involved in the early stages of liver carcinogenesis.
To investigate the role of EpCAM in cancer initiation, we examined its expression in hepatic cells at various stages of differentiation. Hepatic differentiation of mouse ESCs may be induced in a stepwise manner by adding several specific growth factors following embryoid body formation (18) and functional hepatocytes may be generated using chemically defined culture conditions (19). In the present study, mouse NVRQS-11F ESCs were efficiently differentiated into hepatic lineage cells, HPCs and HCs. This multi-step generation of HCs from ESCs resembles in vivo hepatogenesis and ESC-derived hepatogenesis may be useful as a novel integrative model for hepatocarcinogenesis or for the hepatic toxicity evaluation of a number of chemicals.
DEN treatment enhanced EpCAM expression in ESCs, although EpCAM expression may be lost as the progeny of adult liver stem cells differentiate toward HPCs. However, DEN treatment induced the upregulation of EpCAM in HCs. This indicates that carcinogen treatment altered EpCAM expression in cells at each stage of differentiation.
Notably, DEN treatment increased PCNA expression at days 0 and 22, but not at day 40. This may be significant, since any proliferative cell in the liver may be susceptible to neoplastic transformation at the time of carcinogen exposure. As there was a time lag between increased EpCAM expression and cell proliferation, additional stem cell proteins may be involved. Further studies are also warranted to investigate the role of other stem cell markers in the context of hepatocarcinogenesis.
It has been hypothesized that stem cells and/or progenitor cells are transformed into cancer stem cells (20) by a process involving the dysregulation of stem cell self-proliferation (21). Considering that the induction of hepatic tumors by DEN treatment in animals was dependent on age at carcinogen treatment (22), there may be more hepatic stem and/or progenitor cells in young animals. Generally, compared with older animals, young animals are more sensitive to chemical-induced carcinogenesis (23). Hepatic tumors have been generated in young animals upon the administration of only a single injection of DEN (22) and ∼1% of cells treated with a tumor promoter developed into altered hepatic foci (24).
In the present study, low-dose DEN treatment decreased cell proliferation at day 40. Although it is not known why cell proliferation was not observed at day 40 following middle- and high-dose DEN treatment, it may be that the differentiation stage of hepatic cells is a significant factor in cellular proliferation caused by carcinogen treatment. Further studies are required to investigate the effects of various hepatocarcinogens on hepatic cells at various stages of differentiation.
In summary, EpCAM and PCNA expression was increased in DEN-induced tumors and the expression of EpCAM and PCNA in ESCs, HPCs and HCs was modulated by DEN treatment. This study contributes to cancer research by clarifying EpCAM expression in hepatic tumors and during the early stages of hepatocarcinogenesis.
Acknowledgments
The authors would like to thank Ms. He Jin Jung, Sung Chan Kim, Ji Ae Seo and Eun-Joo Park for their technical assistance. This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2010-0003829).
References
- 1.Rambhatla L, Chiu CP, Kundu P, Peng Y, Carpenter MK. Generation of hepatocyte-like cells from human embryonic stem cells. Cell Transplant. 2003;12:1–11. doi: 10.3727/000000003783985179. [DOI] [PubMed] [Google Scholar]
- 2.Turner R, Lozoya O, Wang Y, Cardinale V, Gaudio E, Alpini G, Mendel G, Wauthier E, Barbier C, Alvaro D, Reid LM. Human hepatic stem cell and maturational liver lineage biology. Hepatology. 2011;53:1035–1045. doi: 10.1002/hep.24157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenyon J, Gerson SL. The role of DNA damage repair in aging of adult stem cells. Nucleic Acids Res. 2007;35:7557–7565. doi: 10.1093/nar/gkm1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alison MR. Liver stem cells: implications for hepatocarcinogenesis. Stem Cell Rev. 2005;1:253–260. doi: 10.1385/SCR:1:3:253. [DOI] [PubMed] [Google Scholar]
- 5.Sell S, Dunsford HA. Evidence for the stem cell origin of hepatocellular carcinoma and cholangiocarcinoma. Am J Pathol. 1989;134:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 6.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 7.van der Gun BT, Melchers LJ, Ruiters MH, de Leij LF, McLaughlin PM, Rots MG. EpCAM in carcinogenesis: the good, the bad or the ugly. Carcinogenesis. 2010;31:1913–1921. doi: 10.1093/carcin/bgq187. [DOI] [PubMed] [Google Scholar]
- 8.Yoon SM, Gerasimidou D, Kuwahara R, Hytiroglou P, Yoo JE, Park YN, Theise ND. Epithelial cell adhesion molecule (EpCAM) marks hepatocytes newly derived from stem/progenitor cells in humans. Hepatology. 2011;53:964–973. doi: 10.1002/hep.24122. [DOI] [PubMed] [Google Scholar]
- 9.Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX, Tang ZY, Wang XW. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 10.Fausto N, Campbell JS. Mouse models of hepatocellular carcinoma. Semin Liver Dis. 2010;30:87–98. doi: 10.1055/s-0030-1247135. [DOI] [PubMed] [Google Scholar]
- 11.Diwan BA, Rice JM, Ward JM, Ohshima M, Lynch PH. Inhibition by phenobarbital and lack of effect of amobarbital on the development of liver tumors induced by N-nitrosodiethylamine in juvenile B6C3F1 mice. Cancer Lett. 1984;23:223–234. doi: 10.1016/0304-3835(84)90157-5. [DOI] [PubMed] [Google Scholar]
- 12.Jang JJ, Weghorst CM, Henneman JR, Devor DE, Ward JM. Progressive atypia in spontaneous and N-nitrosodiethylamine-induced hepatocellular adenomas of C3H/HeNCr mice. Carcinogenesis. 1992;13:1541–1547. doi: 10.1093/carcin/13.9.1541. [DOI] [PubMed] [Google Scholar]
- 13.Tamano S, Merlino GT, Ward JM. Rapid development of hepatic tumors in transforming growth factor alpha transgenic mice associated with increased cell proliferation in precancerous hepatocellular lesions initiated by N-nitrosodiethylamine and promoted by phenobarbital. Carcinogenesis. 1994;15:1791–1798. doi: 10.1093/carcin/15.9.1791. [DOI] [PubMed] [Google Scholar]
- 14.Jensen MR, Factor VM, Fantozzi A, Helin K, Huh CG, Thorgeirsson SS. Reduced hepatic tumor incidence in cyclin G1-deficient mice. Hepatology. 2003;37:862–870. doi: 10.1053/jhep.2003.50137. [DOI] [PubMed] [Google Scholar]
- 15.Vesselinovitch SD, Koka M, Mihailovich N, Rao KV. Carcinogenicity of diethylnitrosamine in newborn, infant, and adult mice. J Cancer Res Clin Oncol. 1984;108:60–65. doi: 10.1007/BF00390974. [DOI] [PubMed] [Google Scholar]
- 16.Zhou QJ, Xiang LX, Shao JZ, Hu RZ, Lu YL, Yao H, Dai LC. In vitro differentiation of hepatic progenitor cells from mouse embryonic stem cells induced by sodium butyrate. J Cell Biochem. 2007;100:29–42. doi: 10.1002/jcb.20970. [DOI] [PubMed] [Google Scholar]
- 17.Yovchev MI, Grozdanov PN, Zhou H, Racherla H, Guha C, Dabeva MD. Identification of adult hepatic progenitor cells capable of repopulating injured rat liver. Hepatology. 2008;47:636–647. doi: 10.1002/hep.22047. [DOI] [PubMed] [Google Scholar]
- 18.Hamazaki T, Iiboshi Y, Oka M, Papst PJ, Meacham AM, Zon LI, Terada N. Hepatic maturation in differentiating embryonic stem cells in vitro. FEBS Lett. 2001;497:15–19. doi: 10.1016/s0014-5793(01)02423-1. [DOI] [PubMed] [Google Scholar]
- 19.Touboul T, Hannan NR, Corbineau S, Martinez A, Martinet C, Branchereau S, Mainot S, Strick-Marchand H, Pedersen R, Di Santo J, Weber A, Vallier L. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–1765. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- 20.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea - a paradigm shift. Cancer Res. 66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. discussion 1895–1886, 2006. [DOI] [PubMed] [Google Scholar]
- 21.Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–7282. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- 22.Kang JS, Wanibuchi H, Morimura K, Gonzalez FJ, Fukushima S. Role of CYP2E1 in diethylnitrosamine-induced hepatocarcinogenesis in vivo. Cancer Res. 2007;67:11141–11146. doi: 10.1158/0008-5472.CAN-07-1369. [DOI] [PubMed] [Google Scholar]
- 23.Gray R, Peto R, Brantom P, Grasso P. Chronic nitrosamine ingestion in 1040 rodents: the effect of the choice of nitrosamine, the species studied, and the age of starting exposure. Cancer Res. 1991;51:6470–6491. [PubMed] [Google Scholar]
- 24.Pitot HC, Dragan YP, Teeguarden J, Hsia S, Campbell H. Quantitation of multistage carcinogenesis in rat liver. Toxicol Pathol. 1996;24:119–128. doi: 10.1177/019262339602400116. [DOI] [PubMed] [Google Scholar]