Proteins of the RecA family carry out the central reaction in homologous recombination by forming stretches of hybrid DNA that connect two identical or closely related DNA duplexes. During the mitotic cell cycle, formation of hybrid DNA can serve to align sequences for accurate repair of DNA double-strand breaks or damaged replication forks. During meiosis, recombination serves to create new combinations of alleles and to create connections between homologous chromosomes as required for reductional chromosome segregation. In all eukaryotic organisms, Rad51, a relative of the E. coli RecA protein, is the sole strand exchange protein responsible for mitotic recombinational repair of DNA. Rad51 is also critical for meiotic recombination, and thus rad51 mutants are sterile. Many eukaryotic organisms, including fungi, plants and vertebrates, encode a second RecA-like strand exchange protein, Dmc1, which is meiosis-specific. A key area of research in the recombination field is to determine the functional relationship between Rad51 and Dmc1 during normal meiosis. Earlier studies used mutant strains to show that Rad51 and Dmc1 each can act alone to promote meiotic recombination, but also provided evidence that they often cooperate during a single recombination event.1-3 Biochemical studies showed that both proteins have RecA-like homology search and strand exchange activity.4
Why then do meiotic recombination events in many organisms involve two strand exchange proteins? New observations in budding yeast make it clear that Dmc1, not Rad51, catalyzes homology search and strand exchange for most, if not all, meiotic recombination events.5 A separation of function mutant showed that although Rad51’s ability to form filaments on DNA is required for normal meiotic recombination, its strand exchange activity is fully dispensable. Furthermore, biochemical experiments showed that Rad51 stimulates Dmc1’s strand exchange activity by more than 20-fold. The hypothesis that Rad51 is a Dmc1 accessory factor was prompted by discovery of Hed1, a protein that inhibits Rad51’s activity.6 Hed1 prevents Rad51 from completing recombination in a dmc1 mutant; a dmc1 single mutant does not form meiotic recombination products, but a dmc1 hed1 double mutant does. Thus, meiotic induction of Hed1 expression converts Rad51 from a recombination enzyme to a recombination regulatory factor (Fig. 1).
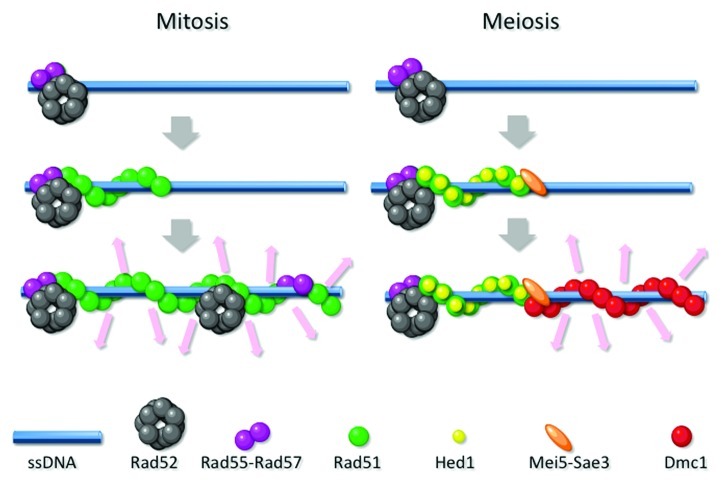
Figure 1. Model for assembly of Rad51 and Dmc1 filaments. The left cartoon depicts the major pathway for Rad51 filament formation in mitotic cells; Rad52 and Rad55-Rad57 stimulate initiation of a Rad51 filament on ssDNA and contribute to filament stability. The homology search and strand exchange activity of the mitotic Rad51 filament is represented by the pink arrows. The right cartoon shows a speculative model for the major pathway of meiotic recombination in budding yeast. An elongating Rad51 filament is kept inactive by binding of the inhibitory protein Hed1. Binding of Mei5-Sae3 to the elongating Rad51 filament forms a mediator complex capable of stimulating initiation of Dmc1 assembly. Because Hed1 inhibition is specific for Rad51, the Dmc1 filament carries out homology search and strand exchange in this case. Note that no attempt was made to draw the various proteins to scale.
The molecular mechanism through which Rad51 controls Dmc1’s activity remains to be fully characterized. However, key mechanistic insight is provided by the observation that Rad51-mediated stimulation of Dmc1 depends on a previously characterized Dmc1 co-factor called Mei5-Sae3. Mei5-Sae3 stimulates Dmc1’s strand exchange activity by enhancing its ability to form nucleoprotein filaments on ssDNA.7 This type of stimulatory activity is referred to as “mediator” activity. Mediator proteins act by simulating filament initiation, especially on tracts of single-strand DNA (ssDNA) bound by ssDNA binding proteins such as the eukaryotic RPA protein. Mediators also promote filament stability. Discovery of a mediator function for Rad51 is novel, but Rad51 is not the first RecA-related protein found to have mediator function. Rad51 paralogs Rad55 and Rad57 form a heterodimeric mediator complex.4,8 Vertebrates have five Rad51 paralogs that combine in at least two distinct complexes to regulate Rad51.4,8 A number of proteins with no structural similarity to Rad51 also have mediator activity, including Rad52 and, in vertebrates, the breast cancer suppressor protein BRCA2.4,8 Much remains to be done to determine how the numerous mediators interact with one another and what regulatory functions are provided by individual pathways of mediated Rad51 and Dmc1 assembly. The finding that Rad55-Rad57 is activated by the budding yeast ATM/ATR DNA damage-dependent kinase pathway is just one example of the important role mediators play in regulation of recombination.9
Having found that the role of promoting homology search and strand exchange is transferred from Rad51 to Dmc1 when cells exit the mitotic cell cycle and enter meiosis, the next critical question becomes: what is accomplished by this change of roles? Meiotic recombination is subject to unique regulatory processes that promote high-fidelity chromosome segregation at the first meiotic division. These regulatory pathways direct recombination to occur between homologous chromatids (as opposed to sister chromatids) and also distribute reciprocal crossover recombination events, such that all homologous chromosome pairs are connected by at least one crossover. A mechanistic understanding of these modes of regulation is just beginning to emerge. Unpublished collaborative work from our lab and that of Neil Hunter indicates that meiosis-specific mechanisms for regulation of recombination in budding yeast require that Dmc1 play the role of strand exchange protein; regulation fails when Rad51 is allowed to take Dmc1’s role. The switch is Rad51’s role is likely occur in other organisms that encode Dmc1, including humans. On the other hand, Dmc1 and its accessory proteins have been lost in a number of evolutionary lineages, and these lineages appear to have evolved mechanisms that allow Rad51 to retain the leading role in homology search and strand exchange during meiosis. Understanding differences in strand exchange protein interaction between species is an important new focus of the recombination field.
Discovery of a second role for Rad51 in recombination is not entirely surprising given that RecA has long been known to be a multifunctional protein. One of RecA’s non-strand exchange functions is its co-protease activity. RecA-stimulated protein cleavage is responsible for the transcriptional response to DNA damage as well as for post-transcriptional regulation of error prone polymerase-mediated DNA damage tolerance.10 Given the diverse functions of RecA, it could well be that key functions of the eukaryotic RecA-like proteins remain to be discovered.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/22396
References
- 1.Bishop DK. Cell. 1994;79:1081–92. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 2.Schwacha A, et al. Cell. 1997;90:1123–35. doi: 10.1016/S0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 3.Shinohara A, et al. Genes Cells. 1997;2:615–29. doi: 10.1046/j.1365-2443.1997.1480347.x. [DOI] [PubMed] [Google Scholar]
- 4.San Filippo J, et al. Annu Rev Biochem. 2008;77:229–57. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 5.Cloud V, et al. Science. 2012;337:1222–5. doi: 10.1126/science.1219379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsubouchi H, et al. Genes Dev. 2006;20:1766–75. doi: 10.1101/gad.1422506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrari SR, et al. J Biol Chem. 2009;284:11766–70. doi: 10.1074/jbc.C900023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gasior SL, et al. Proc Natl Acad Sci USA. 2001;98:8411–8. doi: 10.1073/pnas.121046198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herzberg K, et al. Mol Cell Biol. 2006;26:8396–409. doi: 10.1128/MCB.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinagawa H. EXS. 1996;77:221–35. doi: 10.1007/978-3-0348-9088-5_14. [DOI] [PubMed] [Google Scholar]