Abstract
Following genotoxic stress, cells activate a complex, kinase-based signaling network to arrest the cell cycle and initiate DNA repair or apoptosis. The tumor suppressor p53 lies at the heart of this DNA damage response. p53 mediates the transactivation of both cell cycle-regulating and pro-apoptotic clusters of target genes. However, it remains incompletely understood which signaling molecules dictate the choice between these two opposing p53-dependent cellular outcomes. Over recent years, numerous regulatory mechanisms impacting on the cellular outcome of p53 signaling have been described. However, no single dominant mechanism has thus far been identified to regulate the cellular choice between p53-driven apoptosis or senescence. The transcriptional regulator AATF has recently emerged as a novel factor impacting on the cellular outcome of the p53 response. Upon genotoxic stress, cytoplasmic pools of MRLC-bound AATF are phosphorylated through the p38MAPK/MK2 checkpoint kinase complex. This AATF phosphorylation results in the disruption of cytoplasmic MRLC3:AATF complexes followed by rapid nuclear localization of AATF. Once in the nucleus, AATF binds to the PUMA, BAX and BAK promoters to repress the DNA damage-induced expression of these pro-apoptotic p53 target genes. Depletion of AATF in tumor cells results in a dramatically enhanced response to DNA-damaging chemotherapeutics, both in vitro and in vivo. Furthermore, focal copy number gains at the AATF locus in neuroblastoma correlate with adverse prognosis and reduced overall survival in this typically p53-proficient malignancy. These data identify the p38/MK2/AATF signaling pathway as a critical repressor of p53-driven apoptosis in tumor cells and implicate this signaling cascade as a novel target for chemotherapy-sensitizing therapeutic efforts.
Keywords: AATF, DNA damage, MK2, apoptosis, checkpoint, kinase signaling, p53
The Tumor Suppressor p53 Serves as a Critical Signaling Hub to Determine Cell Fate in Response to Genotoxic Stress
In response to genotoxic stress, cells activate a complex signaling network to protect genomic stability through mounting a transient or permanent cell cycle arrest, activation of the DNA repair machinery or the induction of apoptotic cell death, if the extent of DNA damage is beyond repair capacity.1-3 This signaling network is collectively referred to as the DNA damage response (DDR). The DDR has traditionally been divided into two major kinase branches operating through the upstream kinases ATM and ATR, together with their respective effector kinases Chk2 and Chk1.4,5 In recent years, numerous reports have pointed to an important role for p38MAPK and its downstream substrate mitogen-activated protein kinase-activated protein kinase-2 (MAPKAP-kinase-2/MK2) as a third checkpoint effector kinase complex operating downstream of ATM and ATR and parallel to Chk1.3,6-10 p38MAPK and MK2 are components of a general stress kinase pathway that is activated in response to a variety of stimuli, including inflammatory signals, oxidative stress, heat shock, hyperosmolar stress and DNA damage.11 Specifically, in response to genotoxic stress, the p38MAPK/MK2 signaling complex appears to be recruited into the DDR network through the canonical DDR kinases ATM and ATR to maintain prolonged cell cycle checkpoints.8,9
One of the major downstream targets of the DDR network is the tumor suppressor p53. Upon DNA damage-dependent phosphorylation by ATM, ATR, DNA-PKcs, Chk1, Chk2, MK2 and others, p53 becomes stabilized to ultimately act as a transcription factor.12,13 This phosphorylation at N-terminal sites close to the MDM2-binding region is thought to reduce ubiquitin-dependent degradation, allowing tetramerization and accumulation in the nucleus, where p53 signaling contributes to two distinct cellular responses13,14: p53 promotes apoptosis in response to genotoxic stress through the transactivation of a set of target genes, such as PUMA, NOXA, BAX, BAK and DR5.15–18 On the other hand, p53-mediated transcriptional activation of CDKN1A, GADD45A or RPRM promotes the induction of a stable cell cycle checkpoint arrest, which serves a protective function by allowing time for the repair of genotoxic lesions.19-21 In addition to nuclear accumulation, p53 has also been shown to be recruited to mitochondria in response to genotoxic stress.22,23 In a recent study, Trinh et al. could show that Tid1 directly interacted with p53 through its DnaJ domain. Furthermore, RNAi-mediated Tid1 depletion led to a failure of p53 accumulation at mitochondria and resistance to apoptosis under hypoxic or genotoxic stresses.22 At first glance, both cell cycle-regulating and apoptotic cellular outcomes of p53 signaling appear to have evolved to repress the uncontrolled proliferation of incipient cancer cells carrying severely damaged genomic material. However, these diverse p53-mediated cellular responses pose a difficult challenge for the treatment of p53-proficient tumors in the clinical setting. While p53 is clearly a potent therapeutic target due to its ability to induce the apoptotic demise of tumor cells, p53-mediated cell cycle arrest might counteract these beneficial effects by allowing the tumor cells time to repair genotoxic lesions set by DNA damaging therapeutics. Which molecular cues dictate the cellular decision between a protective p53-mediated cell cycle arrest and p53-driven apoptosis remains a matter of active debate to date.12 It also remains somewhat unclear to what extent known p53-target genes contribute to apoptosis and senescence. For example, recent data from Kuribayashi et al. suggest that the intrinsic p53-dependent apoptotic pathway mediated through Puma may rely on extrinsic signals relayed through DR5 to promote cell death in a cell- and tissue-dependent manner following DNA damage.15 The cell type in which p53 activation occurs clearly is a major determinant of the functional outcome of p53 signaling. Such a cell type-specific effect of DNA damage-driven p53 activation is apparent in thymocytes, which typically undergo apoptosis, while fibroblasts rather initiate cellular senescence when exposed to genotoxic stress.24,25 These different effects may reflect the biological function, where thymocytes are rapidly turned over, while fibroblasts function as scaffolds in connective tissues, requiring physical integrity. However, no single dominant mechanism has thus far been identified to regulate the cellular choice between p53-driven apoptosis or senescence. Instead, numerous regulatory mechanisms involving selective DNA binding of p53 to its target gene promoters, selective transactivation of p53-bound target genes, differential stability of p53-dependent transcripts and differential responses depending on p53 posttranslational modifications or cellular signaling network state have been reported to mediate cell fate decisions in response to p53 activation.26-36 For instance, ASPP1 and -2, apoptosis-stimulating proteins of p53, bind to conserved contact residues in p53 via their C termini and selectively promote p53-driven apoptosis by directing the binding of p53 to the promoters of the pro-apoptotic p53 target genes BAX and PIG3 but not those of CDKN1A or MDM2.26 The hematopoietic zinc-finger (HZF) is a known p53 co-factor that has been suggested to be a regulator of the functional outcome of p53 signaling. HZF is transactivated by p53, and its gene product forms a complex with the p53 DNA-binding domain. It has recently been shown that HZF promotes binding of p53 to the promoters of the cell cycle-regulatory target genes CDKN1A and 14-3-3σ, while it prevents p53 binding to the promoters of the pro-apoptotic target genes BAX, PERP and NOXA.27 Thus, HZF promotes p53-driven cell cycle arrest over apoptosis, which is underscored by the observation that murine embryonic fibroblasts (MEFs) derived from HZF−/− mice show impaired cell cycle arrest and enhanced apoptosis in response to DNA damage.27 Furthermore, posttranslational modifications of p53 itself have been suggested to impact on the functional outcome of p53 signaling. Phosphorylation of p53 on Ser-46 through HIPK2 has been reported to be critical for the p53-mediated transactivation of the pro-apoptotic p53 target genes p53AIP1, PIG3, NOXA, BAX, PUMA and KILLER/DR5.28-32
Hill et al. recently showed that the NHEJ kinase DNA-PKcs is recruited to the CDKN1A promoter, where it forms a protein complex with p53 under pro-apoptotic conditions.37 Intriguingly, DNA-PKcs-associated p53 displays post-translational modifications that are distinct from those under pro-arrest conditions, ultimately ablating CDKN1A transcription and promoting apoptosis. DNA-PKcs inhibition prevented DNA-PKcs binding to p53 on the CDKN1A promoter, restoring CDKN1A transcription, and significantly reduced apoptosis.37 These data demonstrate that DNA-PKcs negatively regulates CDKN1A expression by directly interacting with the CDKN1A transcription machinery via p53, directing the p53 response toward apoptosis. Although the above-mentioned reports suggest that binding selectivity of p53 is the primary determinant that regulates the cellular decision between pro- or anti-apoptotic outcome of p53 signaling, others have reported that the pattern of p53 occupancy does not differ significantly between different p53-activating stimuli, leading to distinct biological outcomes.33,34 These observations suggest that target gene regulation is not necessarily achieved through selectivity of p53 binding. In fact, human genes are typically targeted by numerous transcription factors. For example, Myc represses CDKN1A transcription via binding to and repression of Miz1, a zinc-finger transcription factor that binds to the CDKN1A promoter, thus preventing p53-driven CDKN1A expression.35 Intriguingly, Myc does not significantly repress p53-driven PUMA, BAX or PIG3 expression.35 Hence, Myc selectively shifts the p53 response toward an apoptotic outcome. These data are in keeping with a recent report from Carr-Wilkinson et al. showing that MYCN-amplified neuroblastoma cell lines carrying wild type TP53 fail to undergo ionizing radiation-induced G1 arrest and instead undergo p53-dependent apoptosis.38 A similar apoptosis-promoting role has recently been reported for ATM.39,40 When Jiang and colleagues depleted p53-proficient MEFs of ATM, they found these cells to be exquisitely resistant to DNA damage-induced apoptosis when compared with their ATM-proficient counterparts.39,40 This failure to properly induce apoptosis was the result of an impaired transactivation of the p53 target genes PUMA and NOXA. Intriguingly, DNA damage-induced p53 phosphorylation on Ser-23 (corresponding to human Ser-20), a residue known to be phosphorylated by ATM, ATR, DNA-PKcs and others, appeared to be largely unaffected in ATM-depleted MEFs, compared with their ATM-proficient controls. In contrast, no difference in the DNA damage-dependent induction of the cell cycle-regulating p53 target genes CDKN1A and GADD45A was observed when ATM-proficient and ATM-depleted MEFs were analyzed.39,40 These data indicate that ATM is critical for the induction of p53-driven apoptosis. However, ATM-mediated phosphorylation of p53 appears to be dispensable for this process. There appear to be additional pathways that impact on the functional outcome of p53 signaling. Blagosklonny and coworkers recently showed that nutlin-3a (a small molecule that activates p53 without causing DNA damage) induces quiescence in certain cell lines.41 Further investigation into the molecular mechanisms revealed that nutlin-3a caused quiescence by actively suppressing the senescence program.42,43 Interestingly, in these cells nutlin-3a inhibited mTOR signaling, which is known to be involved in the senescence program. The group went on to show that shRNA-mediated knockdown of TSC2, a negative regulator of mTOR, partially converted quiescence into senescence in these nutlin-arrested cells. In keeping with these findings, nutlin-3a failed to inhibit mTOR in melanoma cell lines and MEFs, which both readily undergo senescence in response to p53 activation. Furthermore, the mTOR inhibitor rapamycin converted nutlin-3a-induced senescence into quiescence in these senescence-prone cells. It could further be shown that a cell cycle arrest in the presence of hyperactive mTOR leads to cellular senescence.44 While cell cycle withdrawal at high levels of p53 is associated with mTOR repression, leading to the induction of reversible quiescence instead of senescence. Super-induction of p53 by either nutlin-3a or high concentrations of the anthracycline drug doxorubicin prevented the induction of senescence that had been observed with low doses of doxorubicin, converting it into quiescence. This observation might rationalize that in order to cause senescence, DNA damaging drugs should be used at low concentrations, which arrest cell cycle but do not induce p53 at levels sufficient to suppress mTOR.44 Thus, the activation status of the mTOR pathway appears to, at least partially, determine the choice between senescence and quiescence in p53-arrested cells.
The MK2 Substrate AATF Acts as a Phosphorylation-Dependent Molecular Modulator to Repress p53-Driven Apoptosis
Apoptosis-antagonizing transcription factor (AATF) is a conserved RNA Pol II-binding protein involved in transcriptional regulation of gene expression that has been shown to be a substrate of the canonical DNA damage response kinases ATM/ATR and Chk1 and -2 45. This phosphorylation appears to be critical for the DNA damage-induced stabilization of AATF.45 Much of our current understanding of the role of AATF within the DDR stems from genetic gain- and loss-of-function experiments. For example, inducible AATF overexpression in NIH3T3 cells has been shown to promote cell cycle progression, mainly through an accelerated S-phase entry.46 Exogenously expressed AATF has been shown to compete with HDAC1 for binding to Rb/E2F complexes. Replacement of HDAC1 with AATF relieved the transcriptional repression of E2F target genes, ultimately promoting enhanced S-phase entry.46 In contrast, overexpression of AATF in NIH3T3-, HEK293- and p53-proficient HCT116 cells resulted in an enhanced G2/M arrest in response to doxorubicin exposure.45 Höpker and colleagues recently reported data corroborating these observations, showing an enhanced stability of DNA damage-induced cell cycle checkpoints upon overexpression of exogenous AATF.47 These observations suggest that the effects of AATF overexpression depend at least partially on the activation status of the DDR network with AATF promoting cell cycle progression in the absence of DNA damage and enforcing a cell cycle arrest in response to genotoxic stress. These contrasting effects of AATF signaling correlate with distinct promoter binding of AATF in the different scenarios. In cycling, non-damaged cells, AATF has been shown to bind to the promoters of the E2F target genes CCNA1, DHFR and TK1.46 This promoter usage changes upon the infliction of genotoxic stress, where AATF has been shown to dissociate from the CCNA1, DHFR and TK1 promoters.45 However, AATF promoter occupancy on pro-apoptotic p53 target genes has not been evaluated in this study.
AATF depletion has previously been shown to enhance the cytotoxic effects of doxorubicin.45 Furthermore, when analyzing the relative survival of TP53+/+ and TP53−/− HCT116 cells, Bruno et al. documented an apoptosis-repressing effect of AATF overexpression that was specific to TP53+/+ cells.45 However, the molecular details of this phenotypic observation remained largely enigmatic. Recently, published data hint at the regulation of apoptotic processes as a critical mechanism for AATF-mediated chemotherapy resistance.48 Using gain- and loss-of-function genetics, Fanciulli and colleagues showed that AATF overexpression results in the induction of antiapoptotic XIAP, even in the absence of genotoxic stress.48 Furthermore, AATF depletion prevented the doxorubicin-induced expression of XIAP.48 However, the mechanisms regulating AATF-mediated transcriptional control remained largely unclear.
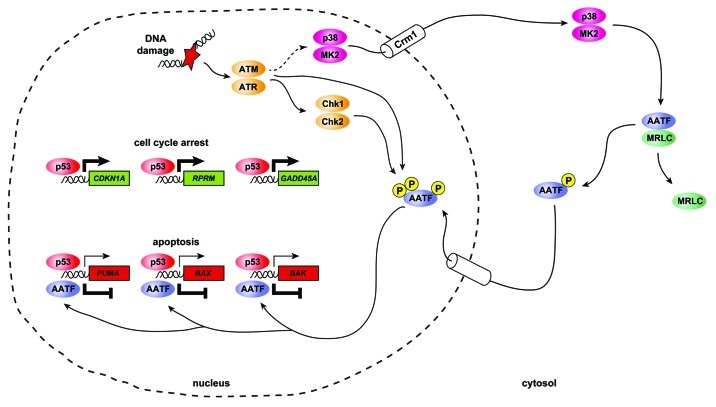
In a very recent paper, Höpker et al. shed some light on the molecular nuts and bolts of AATF-mediated repression of p53-dependent apoptosis.47 Using a library vs. library phospho-proteomics screening approach, they identified a cytoplasmic protein complex consisting of myosin regulatory light chain (MRLC) and AATF. Further experiments revealed that this protein complex was disrupted upon DNA damage-induced phosphorylation of AATF on Thr-366, and that this dissociation from MRLC led to the subsequent nuclear accumulation of AATF. Intriguingly, the peptide sequence spanning Thr-366 was an ideal match for the substrate motif selected by the DNA damage checkpoint effector kinases Chk1, Chk2 and MK2.7,49 Further investigation aimed at the identification of the DNA damage-responsive kinase responsible for AATF phosphorylation showed that treatment of cells with hyperosmotic solutions resulted in a similar disruption of the MRLC:AATF complex than that observed after genotoxic treatment with UV irradiation or adriamycin. Osmotic stress is known to be a strong stimulus for MK2 activation, with little or no effect on Chk1 or Chk2.47,50 Furthermore, MK2 was capable of directly phosphorylating AATF in vitro. Lastly, MK2/3 double-knockout MEFs failed to show nuclear enrichment of AATF following genotoxic stress, strongly suggesting that MK2 is the checkpoint kinase responsible for AATF phosphorylation in response to DNA damage. In further experiments, Höpker and colleagues were able to confirm the data previously reported by Bruno et al., showing that AATF depletion results in significantly increased apoptosis of p53-proficient cells following DNA-damaging chemotherapy both in vitro and in vivo. Using chromatin immunoprecipitation experiments, it could be shown that AATF specifically engages the promoter regions of the pro-apoptotic p53 target genes PUMA, BAX and BAK in response to UV-induced genotoxic stress. In marked contrast, no increase in AATF binding to the promoter regions of the cell cycle-regulating p53 target genes CDKN1A, RPRM and GADD45A could be documented in UV-exposed cells. When these experiments were repeated in MK2/3 double-knockout MEFs, the authors failed to see any UV-induced AATF binding to the PUMA, BAX and BAK promoters, strongly suggesting that MK2 is necessary to allow AATF engagement of at least a subset of its target promoters.47 Binding of AATF to the promoter regions of PUMA, BAX and BAK correlated with significantly reduced expression of the protein products of these genes, both in vitro and in vivo. Thus, AATF emerges as a phosphorylation-dependent molecular regulator of the p53 response acting to repress p53-driven apoptosis in response to genotoxic damage. Intriguingly, a primarily nuclear signal emanating from damaged genomic material appears to be relayed into the cytoplasm through MK2 in order to mobilize AATF, which, in turn, acts as a potent modifier of the p53 response in the nucleus (Fig. 1).
Figure 1. AATF acts as a phosphorylation-dependent molecular switch to dictate the functional outcome of p53 activation in response to genotoxic stress. Depicted is a simplified schematic overview of the regulatory imposed on the p53 response through the p38MAPK/MK2/AATF signal transduction cascade. Following genotoxic stress, the canonical DNA damage response kinases ATM and ATR are activated. Through yet uncharacterized mechanisms, ATM and ATR mediate activation of p38MAPK ultimately leading to MK2 activation. Active p38MAPK/MK2 complexes subsequently translocate from the nucleus into the cytoplasm in a Crm1-dependent process. Once in the cytosolic subcellular compartment, MK2 phosphorylates AATF on Thr-366, leading to a disruption of AATF:MRLC complexes and subsequent nuclear translocation of AATF. In addition to MK2-mediated phosphorylation on Thr-366, AATF is also directly phosphorylated by the canonical DDR kinases ATM/ATR, Chk1 and Chk2. These phosphorylation events likely occur in the nucleus. Nuclear AATF specifically engages the promoter regions of the pro-apoptotic p53 target genes PUMA, BAX and BAK leading to transcriptional repression of these genes. In contrast, AATF does not appear to bind to the promoters of the cell cycle-regulating p53 target genes CDKN1A, RPRM or GADD45A. Thus, the overall outcome of nuclear AATF activity is a repression of p53-driven apoptosis and a promotion of p53-dependent cell cycle checkpoints.
Based on the above-mentioned observations, one might predict that p53-proficient human tumors display an enrichment of nuclear AATF, as this might serve as a critical barrier against p53-driven apoptosis. In contrast, p53-defective tumors might not show such a behavior, as nuclear AATF localization is likely not to be selected for in the absence of functional p53. To directly test this hypothesis in a relevant human tumor entity, Höpker and colleagues went on to examine a large cohort of endometrial cancer specimens. The group could demonstrate that nuclear AATF enrichment indeed appears to be selected for in p53-proficient tumors, while p53-defective lesions showed significantly less nuclear AATF staining. Furthermore, using genomic profiling of a large cohort of neuroblastoma samples, the authors were able to identify a substantial number of cases in which focal copy number gains at the AATF locus could be detected. Importantly, the cancer genes most frequently altered in adult neoplastic disease, such as TP53, CDKN2A or RAS, are rarely aberrant in neuroblastoma. Specifically, inactivating TP53 mutations are extremely rare in primary neuroblastomas.51 The samples examined by Höpker et al. were obtained at diagnosis prior to any cytotoxic treatment and are thus highly likely to contain wild type TP53. AATF amplification, as determined by array-CGH, was shown to correlate with increased AATF mRNA expression. Furthermore, copy number gains at the AATF locus and increased AATF mRNA abundance were associated with significantly reduced event-free and overall survival in these neuroblastoma patients. Collectively, these data strongly suggest that AATF is genomically altered in human tumors, and that increased AATF expression levels are associated with poor prognosis and reduced survival in (p53-proficient) neuroblastomas.
Concluding Remarks and Future Challenges
The molecular regulation of p53-governed cell fate decisions is highly complex and only partially understood.12,13,33,52 Over recent years, AATF has emerged as an additional regulator of the p53 response.45,47 AATF has been shown to repress p53-driven apoptosis in response to genotoxic stress.45,47 This AATF-dependent repression of the apoptotic arm of the p53 response requires the MK2-dependent mobilization of cytoplasmic MRLC-bound AATF pools.47 Once liberated, AATF translocates to the nucleus, where it specifically engages the promoter regions of pro-apoptotic p53 target genes to repress the expression of their respective gene products, thus selectively repressing p53-dependent apoptosis.47 However, many questions regarding this novel p53-regulating pathway remain unanswered today. For instance, it is not clear how exactly the reduced expression of Puma, Bax and Bak observed upon AATF binding to their promoter regions is brought about. As AATF does not contain any DNA-binding domains, it is intriguing to speculate that AATF acts as a co-repressor, requiring the presence of additional DNA-binding proteins to exert its postulated repressive function on the PUMA, BAX and BAK promoters. Although AATF-mediated transcriptional repression is a very attractive hypothesis given the above-mentioned data, it remains formally unclear whether the reduced abundance of Puma, Bax and Bak protein is the result of reduced transcription, for example brought about through the recruitment of polycomb group proteins, posttranscriptional or even posttranslational mechanisms. Another interesting, yet unanswered, question pertains to a potential competition between AATF and p53 on the promoters of pro-apoptotic p53 target genes. Does AATF repel p53 from these genomic regions to prevent p53-mediated transactivation of PUMA, BAX and BAK? One aspect worth further study in this regard is the determination of the mode of AATF binding to its target promoters. Does AATF recognize a distinct sequence motif? Is it strictly dependent on one or more different DNA-binding proteins to help its recruitment to the DNA?
It was shown that AATF recruitment into the nuclear compartment critically hinges on MK2 activity. However, previous studies demonstrated that phosphorylation of AATF by the canonical checkpoint kinases ATM/ATR, Chk1 and -2 was important for AATF protein stabilization and involved in the control of cell cycle progression.45 Thus, the relative contribution of these phosphorylation events to the apoptosis-repressing role of AATF remains somewhat unclear and should be elucidated in future experiments. It is tempting to speculate that cytoplasmic MK2-phosphorylation that triggers the translocation of AATF to the nucleus is a prerequisite for subsequent nuclear hyperphosphorylation of AATF by the nuclear kinases ATM/ATR, Chk1 and Chk2. Lastly, the strong correlation between focal copy number gains at the AATF locus with reduced survival in neuroblastoma patients, together with the prominent chemotherapy-sensitizing effects of AATF-depletion in vivo, recommend targeting the p38MAPK/MK2/AATF pathway as a novel therapeutic strategy for the treatment of p53-proficient malignancies.
Acknowledgments
We apologize to our colleagues for the omission of many seminal contributions to the field, and their references, owing to space constraints. This work was supported by the Deutsche Forschungsgemeinschaft (SFB-829 and SFB-832 to T.B., RE2246/1–1, RE2246/2–1, SFB-829 and SFB-832 to H.C.R.), the Volkswagenstifung (Lichtenberg Program to H.C.R.), the Ministry of Science, NRW (313–005–0910–0102 to H.C.R.), the BMBF (0313921 to T.B.) and the Köln Fortune Program (K.H.). We thank the members of our laboratories for helpful discussions.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21997
References
- 1.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–45. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Opin Cell Biol. 2009;21:245–55. doi: 10.1016/j.ceb.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–9. doi: 10.1016/S1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 5.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–68. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 6.Bulavin DV, Higashimoto Y, Popoff IJ, Gaarde WA, Basrur V, Potapova O, et al. Initiation of a G2/M checkpoint after ultraviolet radiation requires p38 kinase. Nature. 2001;411:102–7. doi: 10.1038/35075107. [DOI] [PubMed] [Google Scholar]
- 7.Manke IA, Nguyen A, Lim D, Stewart MQ, Elia AE, Yaffe MB. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol Cell. 2005;17:37–48. doi: 10.1016/j.molcel.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 8.Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11:175–89. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinhardt HC, Cannell IG, Morandell S, Yaffe MB. Is post-transcriptional stabilization, splicing and translation of selective mRNAs a key to the DNA damage response? Cell Cycle. 2011;10:23–7. doi: 10.4161/cc.10.1.14351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raman M, Earnest S, Zhang K, Zhao Y, Cobb MH. TAO kinases mediate activation of p38 in response to DNA damage. EMBO J. 2007;26:2005–14. doi: 10.1038/sj.emboj.7601668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81:807–69. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 12.Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 2012;28:128–36. doi: 10.1016/j.tig.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–23. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 14.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 15.Kuribayashi K, Finnberg N, Jeffers JR, Zambetti GP, El-Deiry WS. The relative contribution of pro-apoptotic p53-target genes in the triggering of apoptosis following DNA damage in vitro and in vivo. Cell Cycle. 2011;10:2380–9. doi: 10.4161/cc.10.14.16588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–94. doi: 10.1016/S1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 17.Selvakumaran M, Lin HK, Miyashita T, Wang HG, Krajewski S, Reed JC, et al. Immediate early up-regulation of bax expression by p53 but not TGF beta 1: a paradigm for distinct apoptotic pathways. Oncogene. 1994;9:1791–8. [PubMed] [Google Scholar]
- 18.Farrow SN, White JH, Martinou I, Raven T, Pun KT, Grinham CJ, et al. Cloning of a bcl-2 homologue by interaction with adenovirus E1B 19K. Nature. 1995;374:731–3. doi: 10.1038/374731a0. [DOI] [PubMed] [Google Scholar]
- 19.Ohki R, Nemoto J, Murasawa H, Oda E, Inazawa J, Tanaka N, et al. Reprimo, a new candidate mediator of the p53-mediated cell cycle arrest at the G2 phase. J Biol Chem. 2000;275:22627–30. doi: 10.1074/jbc.C000235200. [DOI] [PubMed] [Google Scholar]
- 20.Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–97. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 21.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-P. [DOI] [PubMed] [Google Scholar]
- 22.Trinh DL, Elwi AN, Kim SW. Direct interaction between p53 and Tid1 proteins affects p53 mitochondrial localization and apoptosis. Oncotarget. 2010;1:396–404. doi: 10.18632/oncotarget.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Brate A, Giannakakou P. The importance of p53 location: nuclear or cytoplasmic zip code? Drug Resist Updat. 2003;6:313–22. doi: 10.1016/j.drup.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Clarke AR, Purdie CA, Harrison DJ, Morris RG, Bird CC, Hooper ML, et al. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–52. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 25.Di Leonardo A, Linke SP, Clarkin K, Wahl GM. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–51. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 26.Samuels-Lev Y, O’Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, et al. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8:781–94. doi: 10.1016/S1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 27.Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, et al. Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell. 2007;130:624–37. doi: 10.1016/j.cell.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 2002;4:11–9. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- 29.Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–62. doi: 10.1016/S0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 30.Pistritto G, Puca R, Nardinocchi L, Sacchi A, D’Orazi G. HIPK2-induced p53Ser46 phosphorylation activates the KILLER/DR5-mediated caspase-8 extrinsic apoptotic pathway. Cell Death Differ. 2007;14:1837–9. doi: 10.1038/sj.cdd.4402186. [DOI] [PubMed] [Google Scholar]
- 31.Di Stefano V, Rinaldo C, Sacchi A, Soddu S, D’Orazi G. Homeodomain-interacting protein kinase-2 activity and p53 phosphorylation are critical events for cisplatin-mediated apoptosis. Exp Cell Res. 2004;293:311–20. doi: 10.1016/j.yexcr.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 32.Puca R, Nardinocchi L, Givol D, D’Orazi G. Regulation of p53 activity by HIPK2: molecular mechanisms and therapeutical implications in human cancer cells. Oncogene. 2010;29:4378–87. doi: 10.1038/onc.2010.183. [DOI] [PubMed] [Google Scholar]
- 33.Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–19. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 34.Espinosa JM. Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene. 2008;27:4013–23. doi: 10.1038/onc.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seoane J, Le HV, Massagué J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–34. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen D, Zajac-Kaye M, Rubinstein L, Voeller D, Tomaszewski JE, Kummar S, et al. Poly(ADP-ribose) polymerase inhibition enhances p53-dependent and -independent DNA damage responses induced by DNA damaging agent. Cell Cycle. 2011;10:4074–82. doi: 10.4161/cc.10.23.18170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill R, Madureira PA, Waisman DM, Lee PW. DNA-PKCS binding to p53 on the p21WAF1/CIP1 promoter blocks transcription resulting in cell death. Oncotarget. 2011;2:1094–108. doi: 10.18632/oncotarget.378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Carr-Wilkinson J, Griffiths R, Elston R, Gamble LD, Goranov B, Redfern CP, et al. Outcome of the p53-mediated DNA damage response in neuroblastoma is determined by morphological subtype and MYCN expression. Cell Cycle. 2011;10:3778–87. doi: 10.4161/cc.10.21.17973. [DOI] [PubMed] [Google Scholar]
- 39.Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, et al. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895–909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinhardt HC, Jiang H, Hemann MT, Yaffe MB. Exploiting synthetic lethal interactions for targeted cancer therapy. Cell Cycle. 2009;8:3112–9. doi: 10.4161/cc.8.19.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korotchkina LG, Demidenko ZN, Gudkov AV, Blagosklonny MV. Cellular quiescence caused by the Mdm2 inhibitor nutlin-3A. Cell Cycle. 2009;8:3777–81. doi: 10.4161/cc.8.22.10121. [DOI] [PubMed] [Google Scholar]
- 42.Santoro R, Blandino G. p53: The pivot between cell cycle arrest and senescence. Cell Cycle. 2010;9:4262–3. doi: 10.4161/cc.9.21.13853. [DOI] [PubMed] [Google Scholar]
- 43.Korotchkina LG, Leontieva OV, Bukreeva EI, Demidenko ZN, Gudkov AV, Blagosklonny MV. The choice between p53-induced senescence and quiescence is determined in part by the mTOR pathway. Aging (Albany NY) 2010;2:344–52. doi: 10.18632/aging.100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leontieva OV, Gudkov AV, Blagosklonny MV. Weak p53 permits senescence during cell cycle arrest. Cell Cycle. 2010;9:4323–7. doi: 10.4161/cc.9.21.13584. [DOI] [PubMed] [Google Scholar]
- 45.Bruno T, De Nicola F, Iezzi S, Lecis D, D’Angelo C, Di Padova M, et al. Che-1 phosphorylation by ATM/ATR and Chk2 kinases activates p53 transcription and the G2/M checkpoint. Cancer Cell. 2006;10:473–86. doi: 10.1016/j.ccr.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 46.Bruno T, De Angelis R, De Nicola F, Barbato C, Di Padova M, Corbi N, et al. Che-1 affects cell growth by interfering with the recruitment of HDAC1 by Rb. Cancer Cell. 2002;2:387–99. doi: 10.1016/S1535-6108(02)00182-4. [DOI] [PubMed] [Google Scholar]
- 47.Höpker K, Hagmann H, Khurshid S, Chen S, Hasskamp P, Seeger-Nukpezah T, et al. AATF/Che-1 acts as a phosphorylation-dependent molecular modulator to repress p53-driven apoptosis. EMBO J. 2012 doi: 10.1038/emboj.2012.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruno T, Iezzi S, De Nicola F, Di Padova M, Desantis A, Scarsella M, et al. Che-1 activates XIAP expression in response to DNA damage. Cell Death Differ. 2008;15:515–20. doi: 10.1038/sj.cdd.4402284. [DOI] [PubMed] [Google Scholar]
- 49.O’Neill T, Giarratani L, Chen P, Iyer L, Lee CH, Bobiak M, et al. Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J Biol Chem. 2002;277:16102–15. doi: 10.1074/jbc.M111705200. [DOI] [PubMed] [Google Scholar]
- 50.Engel K, Kotlyarov A, Gaestel M. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J. 1998;17:3363–71. doi: 10.1093/emboj/17.12.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 52.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]