Abstract
Sirt1, the closest mammalian homolog of the Sir2 yeast longevity protein, has been extensively investigated in the last few years as an avenue to understand the connection linking nutrients and energy metabolism with aging and related diseases. From this research effort the picture has emerged of an enzyme at the hub of a complex array of molecular interactions whereby nutrient-triggered signals are translated into several levels of adaptive cell responses, the failure of which underlies diseases as diverse as diabetes, neurodegeneration and cancer. Sirt1 thus connects moderate calorie intake to “healthspan,” and a decline of Sirt-centered protective circuits over time may explain the “catastrophic” nature of aging.
Keywords: Sirt1, nutrient signaling, calorie restriction, aging, metabolic disease, CR
Among the thousands of signals cells constantly exchange with each other and their microenvironment, probably only few are as important as those reporting whether nutrients are available and energy levels are high enough to maintain vital functions. Complex signaling cascades have evolved for this purpose, from the simplest organisms to primates, and the information they convey is crucial for the cell’s decision to grow and proliferate, stay quiescent and spare the few available resources, or die. Importantly, in multicellular organisms, these decisions also integrate the metabolic and nutritional status of the neighboring cells, the all organ/tissue or the whole body, as communicated by a myriad of paracrine and endocrine factors.
Research on “nutrient sensing” has recently witnessed a tremendous expansion, prompted by the increasing awareness of an intricate connection between the molecular cascades triggered by nutrient molecules and the cellular processes that underlie important diseases like diabetes, atherosclerosis, neurodegeneration and cancer, typical of advanced age. While many excellent reviews (including refs. 1 and 2) cover the general and expanding issue of nutrient signaling in disease and aging, this article will focus on the molecular interactions and pathophysiological implications of one specific nutrient sensor, Sirt1, the first discovered member of the mammalian Sirtuin family.
The Secret of Sirt1’s Success
A class 3 histone deacetylase that uses NAD+ as co-factor (Fig. 1), and is therefore exquisitely sensitive to cell metabolic status, Sirt1 owes its incredible appeal on the scientific community to a number of reasons.
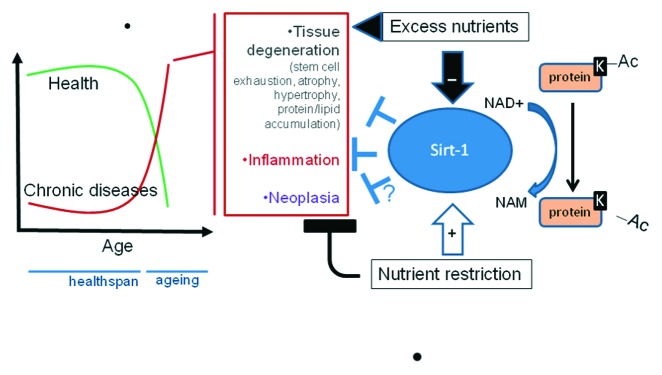
Figure 1. Role of Sirt1 in mammalian aging. Mammalian aging is the time-dependent loss of health with increased occurrence of chronic degenerative, neoplastic and inflammatory diseases. These fundamental pathologic processes are profoundly affected by nutrients and their metabolism. Current evidence points to a major role for the protein deacetylase Sirt1 as a metabolic sensor that translates nutrient availability into largely protective cell responses, thus prolonging healthspan. Molecular interactions whereby Sirt1 exerts this role are in the focus of the present article. Protein lysine deacetylation by Sirt1 involves hydrolysis of NAD+ into nicotinamide (NAM), itself an inhibitor of the enzyme activity, and yields O-acetyl-ADP-ribose (not shown) and the deacetylated substrate. The dependence on NAD links Sirt1 activity to the energy status of the cell via the cellular NAD:NADH ratio and the absolute levels of NAD.
One is the (recently questioned in ref. 3) role of its homolog Sir2, initially identified as a transcriptional repressor in yeast, in linking calorie restriction and lifespan extension in model organisms (yeast, worms and fruit flies), and the easy guess that the same could be true for Sirt1 in mammals. Calorie restriction has, for decades, represented the most reliable experimental model for extended longevity from lower eukaryotes to primates, but the molecular basis for this remarkable effect has been elusive. Activated by nutrient scarcity and able to orchestrate, as epigenetic modifiers, complex transcriptional programs aimed at stress adaptation and endurance, Sir2 and, possibly, mammalian Sirt1 were thought to represent (one of) the missing link(s) between genetic and environmental determinants of longevity.4 Of note, in spite of these early enthusiasms, the connection between Sir2/Sirt1 and longevity is still a matter of intense debate3,5-7 that has anyhow fostered an impressive amount of valuable research on molecular mechanisms and biological consequences of nutrient sensing through Sirtuin 1.
Accordingly, physiological roles of mammalian Sirt1 (and other members of the same enzyme family) appear to go well beyond chromatin remodeling and DNA repair, touching very general cell functions that range from stress responses to survival/death decision, regulation of energy metabolism, autophagy, inflammation and senescence.7,8 Many of those unanticipated activities involve interaction of Sirt1 with protein substrates different from histones, such as transcription factors or even enzymes and other protein species not directly involved in gene expression. Most important, Sirt1 is believed to interfere, through the above pleiotropic cell effects, with the most fundamental pathogenic processes (cellular degeneration, inflammation, neoplasia) that underlie aging of human beings7 (Fig. 1).
As a further element of interest, Sirt1 is a “druggable” enzyme and may mediate at least some of the long known healthy, anti-aging effects of natural compounds, like the grape-derived polyphenol Resveratrol.9 Although a direct role of Sirt1 in the “French paradox” has been often questioned over the years, with some papers dismissing the original data as in vitro artifacts, Resveratrol has become the prototype of a drug capable of activating, no matter whether indirectly10 and only in a defined range of concentrations,11 the sirtuin, thus providing a proof of principle that Sirt1 could be pharmacologically manipulated; this evidence has paved the way to an intense (and maybe exceedingly optimistic)12 search for new and more potent drugs designed to mimic the beneficial effects of calorie restriction in humans without the limitations and side effects of severe dietary regimens.13-15
Although there is still no direct proof for lifespan extension by increased dosage of Sirt1 in mammals6,7 (though there is some for another member of the sirtuin family, Sirt6),16,17 the involvement of this molecule in mammalian response to calorie restriction is rather well-supported.18-21 More generally, a large body of evidence confirms Sirt1’s unique role in translating metabolic and hormonal cues related to nutrient availability into a broad range of cell and tissue responses potentially protective against the diseases typical of later age.7,8 Since senescence of higher organisms is the result of many pathologic changes whose risk increases as the body grows old, more than a mere matter of average or maximum lifespan, the initial adventurous idea that Sirt1 delays aging may still hold, at least to some extent, true.22 Thus, more research on Sirt1, its molecular partners and targets, the cell and body functions it controls and the diseases it may help to find a cure for is warranted.
We’ll discuss current knowledge on Sirt1, following the thread of its molecular interactors, classified as Sirt1 targets (histones, transcription factors, enzymes) and Sirt1 transcriptional/post-transcriptional regulators (Fig. 2), with special attention to the emerging functional interaction between Sirt1 and the cAMP/PKA/CREB signaling cascade. In parallel, we’ll also try to outline how the above interactions relate to the proved or potential protective roles of Sirt1 in aging and related diseases.
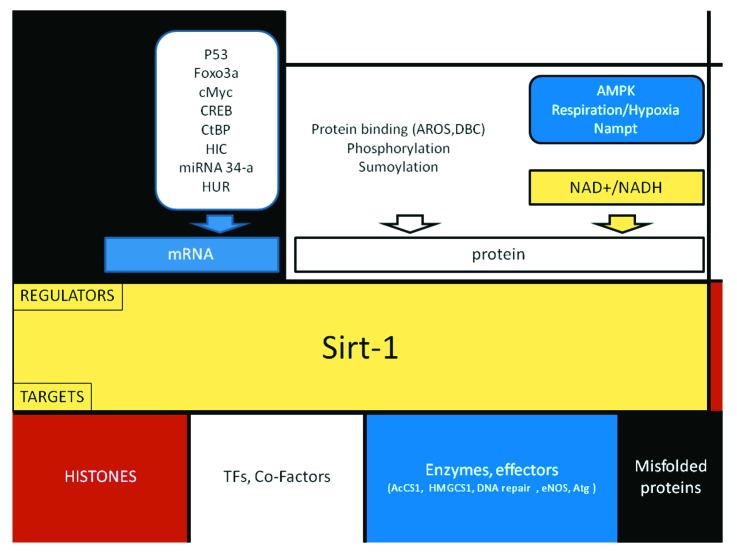
Figure 2. Mondrian’s style synopsis of the upstream regulators and molecular targets of Sirt1, discussed in detail in the article. Regulation occurs at the level of Sirt1 mRNA (transcriptional and post-transcriptional) and protein (expression level and activity). Some of the transcriptional regulators of Sirt1 are also targets of the sirtuin (not shown).
SIRT1 Targets
Sirt1 as a histone modifying enzyme
Histones were the first identified substrates of Sir2 deacetylase activity in yeast, in line with previously characterized actions of this protein in telomeric heterochromatin organization and transcriptional silencing at silent mating loci and ribosomial DNA.23 Sir2 preferentially deacetylases lysine 9 of histone H3 (H3K9Ac) and lysine 16 of histone H4 (H4K16Ac), and, interestingly, acetylation of H4K16 appears to regulate longevity in S. cerevisiae by regulating chromatin structure at telomeres.24 Mammalian Sirt1 displays very similar histone substrate preferences, and an analogous involvement of Sirt1 in heterochromatin (facultative and constitutive) maintenance was also confirmed in human cells.25 While global transcriptional silencing is largely preserved in Sirt1-null mice, suggesting that mammalian Sirt1 may act as a repressor on a smaller set of gens in mammalian cells than it does Sir2 in yeast,26 the link with telomeric DNA is interesting in view of the well-established connection between telomere erosion and cell replicative senescence.27,28 This link is further underscored by the fact that Sirt1 can also indirectly control heterochromatin formation through the deacetylation and stabilization of the histone methyl transferase Suv39H1,29 which, in turn, promotes the accumulation of trimethylated H3K9, a hallmark of constitutive pericentromeric and telomeric heterochromatin. Importantly, this activity of the sirtuin is further enhanced under some stress conditions, including calorie restriction.30 Thus, maintenance of telomeric stability may contribute to the prevention of replicative senescence by Sirt1, as observed in some cell types, like skin kertinocytes,31 while different Sirt1-dependent mechanisms likely operate in those cell contexts in which senescence is instead promoted by the sirtuin.32
Beside heterochromatin stability, histone modification by Sirt1 also regulates specific genes and cell fates in response to environmental cues. Sirt1-dependent deacetylation of H3 and H4 histones at the promoter regions of key developmental genes, for instance, promotes human embryonic stem cell (hESC) pluripotency,33 while in neuronal progenitor cells exposed to mild oxidative stress, Sirt1 docks with the co-repressor Hes-1 and deacetylates H3K9 in the promoter region of the neuron-specific gene Mash-1, thus repressing its expression and promoting cell differentiation toward the non-neuronal (glial) lineage.34 Along similar lines, under glucose restriction, Sirt1 prevents MyoD-dependent muscle cell differentiation by binding the factor and deacetylating histones on its target promoters.35 By doing so, the action of Sirt1 may preserve the muscle stem cell pool from premature exhaustion, thus explaining the prevention of age-associated sarcopenia by calorie restriction.36
As an another, indirect way of modifying histones, Sirt1 can also act on the histone acetylase p300, which is sumoylated and rapidly degraded upon lysine deacetylation by the sirtuin.37 Since p300 is a rate-limiting co-activator for a broad array of transcription factors, the interaction of Sirt1 with this important chromatin organizer is likely to underlie many of the pleiotropic effects of the deacetylase on gene expression in mammalian cells (see below).
More Sirt1 targets: Transcription factors (TFs) and transcriptional regulators.
Interactions with a growing list of transcription factors and transcriptional co-regulators support Sirt1 roles in cell responses (including death/survival, progenitor differentiation, energy metabolism, DNA repair and malignant transformation) relevant to the aging process.
The tumor suppressor p53 is acetylated and activated in response to cellular stressors and DNA damage. Sirt1 binds and deacetylates p53, thus inhibiting its transcriptional activity;38,39 accordingly, cells from Sirt1-KO mice are hypersensitive to radiation damage,40 and Sirt1 overexpression prevents p53-dependent apoptosis in response to DNA damage and stress. By inhibiting p53, Sirt1 promotes cell survival and, conceivably, delays p53-induced senescence; at the same time, it may increase the risk of malignant transformation. In support of the latter view is evidence that at least two putative tumor suppressor genes, hypermethylated in cancer (HIC1) and deleted in breast cancer (DBC1) oppose Sirt1 effects on p53, and their loss of function in malignancy leads to Sirt1-dependent p53 inactivation.41-43 Thus, functional consequences of Sirt1 action on p53 may be beneficial or potentially detrimental according to the pathophysiological context. Interestingly, p53 regulates the expression of Sirt1 by both transcriptional and micro-RNA dependent-mechanisms44-46 (see below), thus adding a further layer of complexity to the interplay between these two crucial regulators of cell fate.
The Forkhead box O (FOXO) family of transcription factors are also deacetylated and regulated by Sirt1.47,48 These factors, homologs of the C. elegans DAF-16, lie at the crossroads of multiple proliferative and metabolic signaling pathways related to longevity determination in model organisms and regulate a broad array of mammalian cell functions, including cell metabolism, cell cycle progression, susceptibility to apoptosis and DNA repair.49 Interestingly, Sirt1’s effect on FoxO activity is not univocal, but rather gene- and function-dependent, with the transcription of targets involved in cell cycle arrest and resistance to oxidative stress being increased, and genes promoting FoxO-dependent cell death instead repressed by the deacetylase.48 FoxOs are acetylated and inhibited under cellular stress, and deacetylation by Sirt1 prolongs FoxO-dependent protective responses.50 This action of FoxOs is strongly reminiscent of C. elegans DAF-16 role in dauer formation and extension of lifespan under adverse conditions, such as nutrient shortage or heat shock.51 Interestingly, the Sirt1/FoxO interplay also operates in mammalian metabolic adaptation by promoting hepatic gluconeogenesis during fasting,52 and Sirt1 is transcriptionally induced by FoxO3a in response to nutrient deprivation.44 Thus, FoxOs and Sirt1 appear to cooperate in an evolutionarily conserved fashion in orchestrating cell response to environmental and nutritional stress.
Along the same thread of evidence, linking Sirt1 to nutrient-regulated gene expression in mammalian systems, are the physical and functional interactions of the deacetylase with the lipid-responsive Peroxisome proliferator activated receptor-γ (PPARγ) and its co-factor PGC1α. PPARγ plays a dual role in white fat, where it orchestrates precursor differentiation into mature adipocytes and promotes fat accumulation in mature cells. Both these functions where found inhibited by Sirt1, which binds the co-repressors NCoR and SMRT at the promoter regions of several PPARγ targets, including PPARγ itself.53 While direct substrates of Sirt1 in this inhibitory circuit (most likely histone proteins) have remained ill-defined, it is worth note that lipolysis and a decrease in fat mass are major effects of calorie restriction and directly link to extension of lifespan from model organism to primates.54
Other important metabolic responses to fasting, including increased hepatic gluconeogenesis, mitochondrial biogenesis and increased fat oxidation and oxygen consumption are regulated by Sirt1 through its interaction with the transcriptional co-activator PGC1α. Initially discovered as a PPARγ co-factor in adaptive termogenesis, PGC1α exerts important metabolic effects by cooperating with a number of transcription factors, including, beside PPARs, also HNF4, FoxOs, the glucocorticoid receptor and the nuclear respiratory factor (NRF) 1, among the others.55 Sirt1 binds and deacetylates PGC1α specifically under nutrient deprivation, although consequences of this interaction vary in different tissues/cells: stimulation of gluconeogenesis in liver,56 increased mitochondrial oxidation of fatty acids in liver and muscle,57,58 reduced respiration in cultured pheochromocytoma cells.59 Importantly, PGC1α is deacetylated and activated in mice treated with the putative Sirt1 activator Resveratrol in parallel with a substantial increase in oxygen consumption and improvement of the dysmetabolic phenotype under high-fat diet.14 Additionally, PGC1α, like Sirt1, is crucial for brain protection from oxidative damage and age-related neurodegenerative disorders,60 and Sirt1 deletion in mouse brain reduces PGC1α induction by calorie restriction,61 further underscoring the broad relevance of the interaction between these two molecules for age-related disorders.
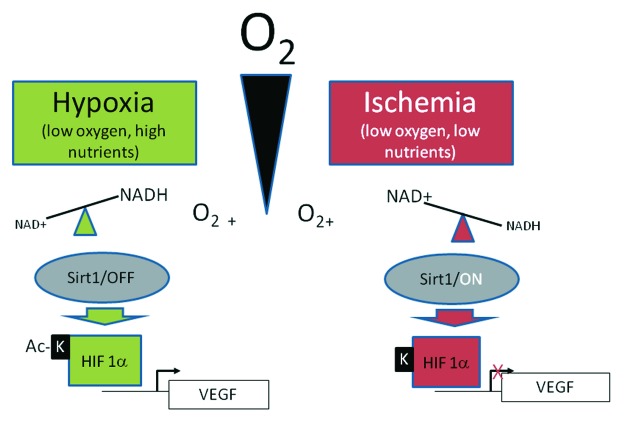
Other transcriptional regulators participate in the metabolic (and potentially anti-aging) action of Sirt1. In liver cells, Sirt1 deacetylates and activates the nuclear liver X receptor(LXR), thus inducing genes that promote cholesterol export and HDL biogenesis, and may, therefore, be protective against atherosclerosis.62 Moreover, Sirt1 modulates the hypoxic response cascade through the hypoxia induced factors (HIF) 1 and 2. Function and distribution differences among these two factors are an emerging theme in cell signaling by hypoxia63; Sirt1 appears to deacetylate both factors but with opposite effects: activation of HIF2 and inactivation of HIF1.64,65 In keeping with a recurrent finding in Sirt1 interactions with TFs (Box 1), Sirt1 is also a transcriptional target of HIFs, and is induced under hypoxia.66 On the other hand, low oxygen decreases the NAD+/NADH ratio and inhibits Sirt1 activity, which contributes to the hypoxic activation of HIF-1α. Although in part contradictory, these findings are intriguing under several aspects. Physiologically, Sirt1 inhibition may help cells discriminating between reduced blood flow, i.e., ischemia (low oxygen and low nutrients, high NAD+) and true hypoxia (low oxygen with high nutrients, low NAD+) and to tune accordingly HIF-dependent transcriptional response (Fig. 3). In a more aging-related perspective, it is noteworthy that hypoxia delays cell senescence in vitro67; moreover, HIF overexpression increases lifespan and attenuates proteotoxicity in C. elegans under specific growth conditions,68 while, in human beings, senescence-associated pathologies as diverse as cancer and diabetic proliferative retinopathy involve deregulated HIF signaling and may benefit from Sirt1 nutritional or pharmacological activation.
Box 1. Implications of Sirt1 feedback in aging.
Functional interactions of Sirt1 with transcriptional regulators are often bi-directional, with the sirtuin being transcriptionally regulated by its target. A model of positive feedback is depicted in (A), in which a transcription factor induces Sirt1 (Gene→Sirt) and is, in turn, activated by the deacetylase (TF°→TF*). For TF to prevent cell damage, or repair it, a fast kinetic of activation is critical. (B) Shows that τ, the time of TF response, is much smaller (indicating a faster response) in the feedback mode (blue squares) than in the absence of positive feedback (pink triangles); however, the feedback mode is extremely sensitive to the decrease in amount/activity of the TF (as indicated by the steep increase of τ for low TF° (i.e., c0) values) that conceivably occurs during aging.140 This behavior may in part explain the "catastrophic" nature of organ and tissue functional decline at later age. (C) Isotherm plot indicating τ ranges (z axis, color-coded) for different values of TF° (c0) and Sirt1 (S°). Note that an increase in Sirt1 (as induced by calorie restriction) effectively compensates for TF° decline. See Supplementary Information for mathematical description of the model.
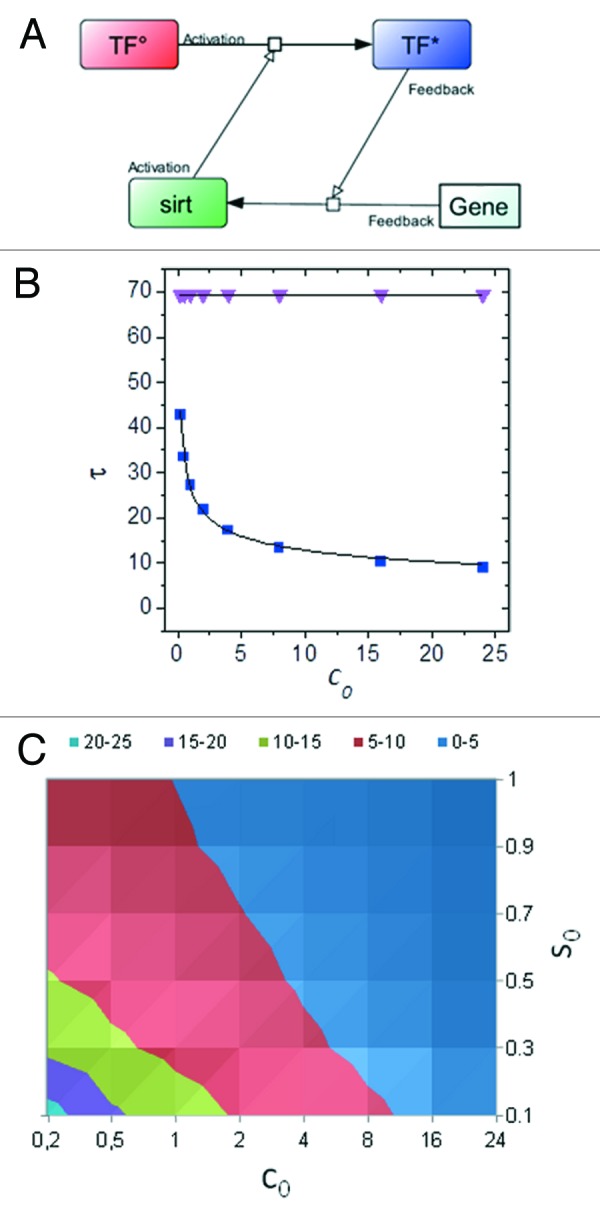
Figure 3. Sirt1 roles in the hypoxic response through HIF1α. Sirt1 may dictate distinct cell responses under different conditions of reduced oxygen availability. Two situations relevant to human pathology are depicted: hypoxia (i.e., reduced oxygen transport with normal blood flow and nutrient supply) and ischemia (decrease or block of perfusion, with reduced supply of oxygen and nutrients). In the former condition, accumulation of NADH inhibits Sirt1, thus promoting acetylation of HIF1α by p300 and transcription of HIF targets like the vascular endothelial growth factor (VEGF). Conversely, under ischemia, limited nutrient supply activates Sirt1, leading to deacetylation and Inhibition of HIF1α. This model, in part speculative, is based on reference 65 and does not include Sirt1-dependent activation of HIF2α, as described in reference 64.
While the above examples underscore the importance of Sirt1 in connecting nutritional cues and transcriptional adaptive responses in mammalian cells, Sirt1 action on STAT3 and NFkB, two master regulators of inflammatory genetic programs downstream of pathogens, cytokine and oncogene-driven signaling cascades, has huge ramifications for the emerging link among metabolism, inflammation and aging.69-71 Sirt1 deacetylates and directly inhibits STAT3 and NFκB; additionally, inhibition of the highly promiscuous co-activator p300 by Sirt1 may indirectly contribute to restraining gene expression by the two factors.37 Interestingly, STAT3 inhibition by Sirt1 also mediates important metabolic responses to fasting (enhanced liver gluconeogenesis)70 and calorie restriction (increased muscle response to insulin).71
Release of inflammatory cytokines accompanies cell replicative senescence in vitro63 as well as hyperglycemic damage to endothelial cells in vivo, and inflammation contributes to atherosclerosis and to the age-associated metabolic derangement leading to diabetes.72 Conversely, calorie restriction and the grape-derived polyphenol Resveratrol reduce markers of inflammation in obese humans,9,73 concomitant with an increased expression of Sirt1. It is also of note that Sirt1 decreases the development of immunosuppressive T lymphocytes (T-regs) by deacetylating the Treg-specific transcription factor Foxp3,74 thus enhancing specific immune response (i.e., against tumors) while inhibiting the inflammatory response. Given the strong involvement of inflammation and immune deficit in age-related diseases, these “anti-inflammatory” and immunoregulatory actions represent another important component of the complex network connecting nutrients, Sirt1 activity and mammalian healthspan.
Sirt1 meets CREB
The cAMP responsive element binding (CREB) transcription factor has been extensively studied in liver, muscle and fat as a metabolic sensor and activator of complex transcriptional programs triggered by nutrient depletion and fasting hormones (glucagon, cortisol, adrenalin), and in the brain as a key mediator of neurotrophin-triggered neuronal differentiation, survival and plasticity.75,76 Evidence involving Sirt1 as a partner in CREB-dependent gene expression has recently made a bridge across these two age-related but seemingly independent actions.
In liver cells exposed to prolonged fasting, Sirt1 limits CREB-induced gluconeogenesis to the advantage of ketogenesis, by deacetylating the CREB transcriptional co-activator (CRTC, also known as TORC, 2);77 this effect is likely relevant to the diabetic state, whereby deregulated gluconeogenesis contributes to glucose intolerance. However, in mice overexpressing Sirt1 and fed a high-fat diet, CREB deacetylation and inhibition by the sirtuin, while improving glucose homeostasis, promotes liver steatosis, dyslipidemia and accelerated atherosclerosis,78 an effect hard to reconcile with the metabolic benefits of calorie restriction; this suggests that elevated Sirt1 activity may become detrimental when uncoupled from reduced nutrient availability, as it occurs during fasting or under CR. Accordingly, forced expression of Sirt-1 at high levels in the myocardium, as opposite to low-moderate overexpression, has been reported toxic by Alcendor and Sadoshima.79 It should also be noted that Sirt1 appears to be dispensable for liver response to calorie restriction,80 and liver-specific disruption of the enzyme can either worsen57 or ameliorate80 steatosis and glucose intolerance under high fat/calorie diet, although the involvement of CREB in these effects was not tested. Instead, Sirt1 is necessary for CR-induced responses in brain, where Sirt1 activates, rather than inhibiting CREB (see below). Thus, the modality of the interplay with CREB may contribute to determining the tissue specificity of Sirt1’s roles in CR.
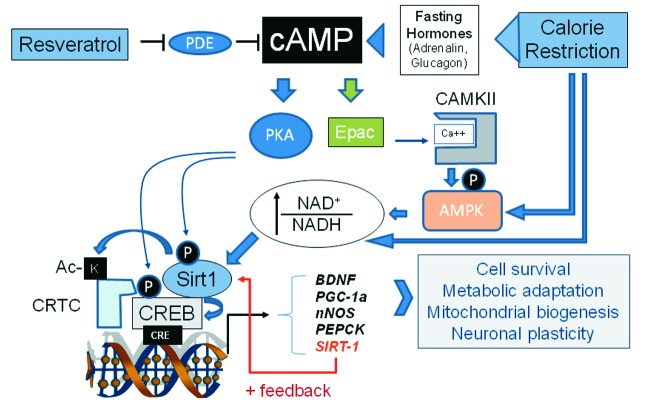
In neuronal cells, Sirt1 mediates protection from mutant Huntingtin by deacetylating TORC1, a neuron-specific CREB co-activator, and promoting the CREB-dependent expression of BDNF and other protective genes81 (Fig. 4). Moreover, CREB-dependent trans-activation of genes regulating neuronal survival, metabolism and plasticity (like PGC1α and nNOS) is induced in calorie-restricted mice in a fashion that requires Sirt1.61 In keeping, electrophysiological and cognitive brain responses to calorie restriction are similarly impaired in mice harboring brain-specific inactivation of Sirt1 or CREB.82 Finally, Sirt1 is transcriptionally induced by CREB in mouse hippocampus during calorie restriction61 and may, in turn, increase CREB expression (and function) through an miRNA-mediated mechanism.83 Thus, taken together, the above evidence suggests that the complex interplay between Sirt1 and CREB, while affecting nutrient sensing and glucose homeostasis in a complex fashion in peripheral tissues, also plays a pivotal role in the metabolic regulation of neuronal plasticity and of high-order brain functions. Knowledge on this specific aspect of neurobiology is in its infancy, but impaired cognitive function and elevated risk of Alzheimer disease are increasingly recognized complications of diabetes and obesity, while dietary restriction delays brain aging and related disorders;84 the CREB/Sirt1 interaction may hopefully provide a molecular framework to these clinical and experimental observations. Future work should also reveal whether other “metabolic” functions of CNS, like regulation of appetite and body weight85,86 and feeding-dependent synchronization of central circadian clocks,87 also involve the CREB-Sirt1 axis.
Figure 4. Nutrient sensing through the cAMP cascade and CREB/Sirt1. Fasting-induced hormones raise intracellular cAMP, leading to PKA-dependent phosphorylation and activation of both CREB and Sirt1. A parallel cascade triggered by cAMP through the adaptor Epac and CaMKII results in AMPK activation, increased NAD+/NADH ratio and metabolic activation of the sirtuin1.10 Sirt1 and CREB form a complex on cAMP responsive promoters and Sirt1 promotes CREB transcriptional activity, in part through deacetylation of the co-activator CRTC/TORC1.81 Sirt1 is also transcriptionally induced by CREB, thus creating a positive feedback loop.61 Calorie restriction and its mimic Resveratrol impinge at multiple levels on this complex circuitry. Note that some important interactions have been omitted for simplicity; for instance, in liver, Sirt1 inhibits CREB-dependent transcription of gluconeogenic enzymes by deacetylating TORC252 or directly CREB.78
Transcription-independent activities of Sirt1
DNA repair, metabolic adaptation and autophagy (a starvation-triggered catabolic process whereby bulky cellular components and damaged organelles are degraded in lysosomes)88 are phenomena relevant to aging and may underlie some of the benefits of calorie restriction. Sirt1 regulates these cell functions at different levels, including a direct interaction with their protein effectors.
Consistent with Sir2’s reported role in yeast, Sirt1 physically interacts with several components of the DNA repair machinery. These include NBS1, a target of the ATM kinase that operates in the homologous recombination route of double-strand break (DSB) repair,89 and Ku70, another DSB repair protein, but in the non-homologous end joining (NHEJ) pathway, also endowed of antiapoptotic activity by its capability of sequestering the apoptogenic factor Bax.20,90 In a similar molecular context, Tip60, an enzyme that acetylates histone H2X upon radiation DNA damage,91 and apurinic/apyrimidinic endonuclease-1 (APE1), an essential enzyme in the base excision repair pathway,20,92 may also be targets for deacetylation and regulation by Sirt1. Collectively, these observations establish an intriguing link between energy metabolism and DNA damage/repair, parallel but independent from the well-recognized one involving metabolism-generated reactive oxygen species.93
Sirt1 interaction with the cytosolic enzymes acetyl CoA synthase 194 (AcCS1) and hydroxymethylglutaryl CoA synthase 1 (HMGCS1)95 is intriguing, since these molecules, which are deacetylated and activated by Sirt1, are likely involved in metabolic adaptation to fasting. In fact, while the role of biosynthesis of acetyl CoA from acetate by AcCS1 is ill-defined in mammals, Ac-CoA may serve as precursor for ketone biosynthesis; HMGCS1, on the other hand, is the rate-limiting enzyme for the synthesis of β-hydroxybutyrate in the cytosol (another isoform of the enzyme, HMGCS2 is activated in mitochondria by Sirt3).96 Interestingly Sirt1 also deacetylates and inhibits PGAM1, a key enzyme of the glycolytic pathway.97 Combined, these actions outline a fast, transcription-independent mechanism, whereby Sirt1 may switch cell metabolism from glycolysis to ketogenesis in conditions of limited nutrients.
Along the same line of evidence lies the observation that Sirt1 promotes autophagy in nutrient-deprived cells. Activation of autophagy, which provides an extra energy source, from the degradation of damaged cell components, occurs in mammalian cells through a complex network of molecular interactions analogous to those initially described in yeast and involving an ubiquitin-like conjugation system for substrate direction to autophagosome vescicles.88 Several components of this machinery (Atg 5, Atg 7 and Atg 8) are targeted by Sirt1 in a nutrient-regulated fashion, and their hyperacetylation in Sirt1-deficient cells and tissues is accompanied by impaired autophagic activity, accumulation of damaged organelles and disrupted energy homeostasis reminiscent of the aging tissues.98 Interestingly, Resveratrol triggers Sirt1-dependent autophagy in cells from different organisms,99 and autophagy may participate in lifespan extension by calorie restriction.100
Sirt1 interaction with another key enzyme, endothelial NO synthase (eNOS), potentially links nutrient sensing with vascular physiology and its age-dependent derangement in cardiovascular diseases. Endothelial NOS mediates protective vascular relaxation by shear stress (as opposite to turbulent flow that favors endothelial damage and thrombosis), and reduced NO bioavailability contributes to vascular aging and atherosclerosis; interestingly, eNOS is upregulated, together with Sirt1, in several tissues and organs of calorie-restricted animals and is necessary for CR-induced mitochondrial biogenesis.21 Thus, the finding that Sirt1 activates eNOS has multifaceted implications. Sirt1 deacetylates two lysine residues (496 and 506) in the calmodulin-binding domain of eNOS, likely favoring enzyme interaction with its Ca2+-dependent activating partner101; importantly, deacetylation is facilitated when eNOS is phosphorylated by AMPK, another nutrient sensor stimulated by energy deprivation, as well as by endothelial shear stress.102 Of note, shear stress also induces the endothelial expression of Sirt1.102 Taken together these observation outline an intriguing flow- and nutrient-sensitive cross-talk between Sirt1, AMPK and eNOS and suggest that at least some of the protective effects of calorie restriction on vessels, and the organs they perfuse, may be related to CR capacity to mimic or amplify flow-dependent trophic signaling in endothelial cells.
Finally, a hallmark of senescent cells and tissues observed in several age-associated degenerative diseases is the accumulation of misfolded proteins and formation of degradation resistant aggregates. Sirt1 directly deacetylates the neuronal phosphorylated-tau protein, thus preventing its precipitation into Alzheimer associated fibrillary tangles.103 This action is in keeping with the protective effect exerted by transgenic Sir2.1 or the sirtuin activator Resveratrol on neuronal dysfunction phenotypes induced by mutant polyglutamines or Prion proteins (PrPs) in Caenorhabditis elegans,104,105 and, together with the indirect, protease or chaperone-mediated effects of Sirt1 on the accumulation of Abeta amyloid in AD106 and of α-synuclein in the Parkinson Lewy bodies,107 identifies in the maintenance of proteostasis another major anti-aging action of the sirtuin in mammals.
SIRT1 Regulators
Multiple levels of control govern Sirt1 action
The strong impact of Sirt1 on cell functions as diverse as metabolism, proliferation, differentiation and death raises the question of how cellular Sirt1 expression and activity are regulated. Schematically, reported levels of control include: (1) metabolic regulation by NAD+; (2) protein-protein interactions; (3) transcriptional and post-transcriptional modulation; (4) post-translational modifications (Fig. 2).
(1) NAD+ is a co-factor for Sirt1 deacetylase activity, which links Sirt1 action to metabolic conditions, like nutrient shortage or increased mitochondrial respiration, characterized by a high NAD+/NADH ratio. Conversely, reduced oxidation of NADH to NAD+ in hypoxic cells contributes to HIF acetylation and activation via inhibition of Sirt165(Fig. 2).
Besides the NAD+/NADH ratio, the total cell content of NAD is also critical for Sirt1 activity. This is mainly controlled by the NAD+ salvage synthetic pathway, whose rate-limiting enzyme, nicotinamide adenine mononucleotide phosphoribosil transferase (Nampt), has been found to regulate Sirt1 in several physiological and pathological contexts. In muscle fibers, for instance, AMPK kinase, activates Sirt1 by two distinct mechanisms: enhanced NADH oxidation in mitochondria52 and increased expression of Nampt.108 Nampt is also transcriptionally induced by the core clock proteins Clock and Bmal1, which creates a circadian fluctuation of intracellular NAD and of Sirt1 activity as a consequence109,110; in turn, Sirt1 deacetylates and inactivates Bmal1 and the feedback inhibitor Per(iod)2, thus setting the stage for a metabolically regulated molecular oscillator111 that is believed to underlie many reciprocal influences between circadian body rhythms and feeding-related cellular and organismal responses.87,112 Nampt is also induced by cMyc in tumor cells and, by activating Sirt1, further increases, in a feedforward loop, the stability and transcriptional activity of the oncoprotein.113 Finally, in a completely different pathological context, another NAD biosyntheric enzyme, Nmnat1, has been found mutated and overexpressed in Wallerian degeneration Slow (Wlds) mice, in which it prevents neuronal degeneration through both Sirt1-dependent and -independent mechanisms.114,115
(2) Systematic search for Sirt1 protein interactors has led to the discovery of both positive and negative regulators of the deacetylase. AROS (activating regulator of Sirt) was identified as a Sirt1 binding partner in a yeast two-hybrid screen and appears to act as an endogenous activator of Sirt1 that promotes p53 deacetylation and inactivation.116 Conversely, DBC1, the product of a gene frequently deleted in breast cancer, was found by two independent groups to co-precipitate with Sirt1 and to inhibit Sirt1 interaction with the tumor suppressor.42,43 Similarly, metyltransferase Set 7/9 binds Sirt1 under genotoxic stress and blocks its inhibitory action on p53.117 Accordingly, DBC or Set 7/9 inactivation, experimentally induced or naturally occurring in human cancer, results in increased Sirt/p53 binding, p53 deacetylation/inactivation and promotion of malignant growth. HIC (hypermetylated in cancer)-1, the product of another putative tumor suppressor gene, also binds Sirt1, creating a transcription inhibitory complex whereby Sirt1 represses its own expression, thus facilitating p53 activity.41 While it is not clear whether these interactions are relevant to the many other function of Sirt1 other than p53 deacetylation, they also point to a generally pro-oncogenic role of the deacetylase; this is difficult to reconcile with the tumor-suppressive action of calorie restriction and with Sirt1 participating also in tumor suppressive activities, including the deacetylation and cytoplasmic retention of β-catenin in colon cancer118; it is therefore likely that effects of Sirt1 on the neoplastic phenotype are not univocal but rather pathway-, cell- and tumor stage-specific.
(3) In keeping with its role as a nutrient sensor, the expression level of Sirt1 is regulated by nutrient availability. Calorie restriction increases Sirt1 immunoreactivity in several rodent tissues, including brain, kidney, white fat and liver.20,21,119 Initial observations revealed such increases to be inhibited by the insulin/IGF signaling cascade,20 to occur concomitantly with mitochondrial biogenesis and to depend on the expression and activity of eNOS and its gaseous product nitric oxide.21 Further mechanistic studies have demonstrated an intriguing cooperation between FoxO3a and p53, two molecules independently related to aging,120 in mediating Sirt1 transcriptional induction in nutrient-deprived rat pheochromocytoma cells.44 p53 binds the Sirt1 promoter and represses Sirt1 expression in the presence of glucose; this block is relieved under nutrient deprivation or in the absence of insulin by FoxO3a, which complexes with p53, leading to Sirt1 upregulation.44 This circuitry may also be relevant to nutritional control of Sirt1 in humans, since a genetic variant of the p53 binding site within the Sirt1 promoter impairs Sirt1 induction by calorie restriction in human muscle.121 Likewise, Sirt1 transcriptional repression by HIC-1 is alleviated in reduced nutrients through a cascade that involves a rise in NAD+ and the NAD+-dependent dissociation of HIC from the co-repressor CtBP.122 Several lines of evidence indicate that Sirt1 is also under the transcriptional control of the PKA-CREB pathway, a cascade sensitive to fasting-related metabolic hormones, including catecholamines and glucagon. CREB is activated in the brain of calorie-restricted mice and upregulates Sirt1 through multiple cAMP-responsive elements (CREs).61 Moreover, Sirt1 is induced by CREB in cultured rodent and human cells treated with growth factors, hormones or PKA agonists61,123 in a fashion that is dominantly inhibited by glucose through the activation and nuclear translocation of the carbohydrate responsive element binding protein (ChREBP).123
Other examples of transcriptional/post-transcriptional regulation of Sirt1 occur in cellular settings relevant to cancer cell biology. E2F-dependent induction of Sirt1 in response to anticancer drug etoposide prevents cell death through a protective feedback that involves E2F deacetylation and inactivation by the sirtuin.124 Likewise Sirt1 is induced by cMyc (and N-Myc) and can, in turn, deacetylate the factor, leading to either an increase or a decrease of its stability and transcriptional capacity, depending on the cell context.113,125
MiR 34a, a p53 target, can repress Sirt1 expression in human cancer cells, leading to p53 hyperacetylation/activation and, eventually, to p53-dependent cell death.45 Expression of miR34a is inhibited in normal liver tissue by the nuclear bile acid receptor farnesoid X receptor (FXR); in this setting, miR34a transcriptional repression leads to upregulation of Sirt1, which, in turn, further activates FXR by direct deacetylation, thus creating a feedforward loop; intriguingly, this circuitry appears to be dysregulated in obese mice fed a high-fat diet, a potentially hepatocarcinogenic condition.46
Finally, the ubiquitously expressed RNA-binding protein HUR has been shown to stabilizeSirt1 mRNA by binding its 3′ untranslated region in a fashion that is inhibited under oxidative stress.126 Although nutrient modulation of the interactions has not been investigated in detail, the above regulatory mechanisms, for involving Sirt1, are intrinsically sensitive to the metabolic cell status and therefore potentially important (as suggested by ref. 46) for effects of calorie restriction on malignant cell growth and response to chemotherapy.127
(4) Sirt1 can undergo sumoylation (a post-translational modification similar to ubiquitination) on lysine 734, and this modification reportedly increases Sirt1 deacetylase activity, thus promoting cell resistance to genotoxic stress128; Sirt1 is also phosphorylated on serine/threonine residues, with effects that change based on protein residues and kinases involved in the reaction. Cyclin B/CDK1, JNK1/JNK2, CK2 and DYKK1A phosphorylate Sirt1 on several N- and C-terminal residues in the context of diverse stress responses and lead to increased enzyme activity and/or stability.129-132 Conversely, mTOR and JNK1 can inhibit Sirt1 under conditions of persistent stimulation.133,134 Of note, constitutive activation of mTOR and JNK1 occurs in overnutrition and obesity and underlies insulin resistance in liver, muscle and fat,135 whereas Sirt1 promotes insulin sensitivity through multiple mechanisms;71,136,137 thus, the mTOR and JNK1-dependent inhibitory phosphorylation of Sirt1 in the context of excess nutrients may contribute to the signaling derangement that leads to impaired glucose tolerance and diabetes.
Finally, in response to β adrenergic stimulation, cAMP-activated PKA phosphorylates Sirt1 at serine 473, a highly conserved residue within the catalytic domain; this favors the NAD+-independent enzyme activation and the prompt induction of fatty acid oxidation, a response relevant for the rapid body adaptation to cold and fasting.138 Interestingly, a recently discovered, alternative pathway for Sirt1 activation by Resveratrol also involves cAMP and CaMKII/AMPK/NAD+ as intermediate transducers.10,139 This evidence, together with the effect of PKA/CREB on Sirt1 transcription61,123 and the observation that Sirt1 regulates CRE-dependent gene expression,61,77,78,81 further pinpoint the relevance of Sirt1 cross-talk with the hormone- and nutrient-sensitive cAMP signaling cascade (Fig. 4).
Conclusions
“…for even time itself is thought to be a circle.” Aristotle, Physics
One decade of intense research on Sirt1 (and other members of the sirtuin family) has provided us with an impressive amount of information on the targets, regulators, biological functions and pathophysiological implications of this molecule. While the Hamletic question of whether Sirt1 does or does not extend lifespan in mammals still remains open, what most clearly emerges from the bulk of the above studies is the role of Sirt1 as a nutrient and stress sensor and a trigger/ enhancer of protective cell responses; as such, Sirt1 perfectly qualifies as an important factor in the yet largely mysterious network of molecular interactions that underlies the effects of calorie restriction in higher organisms, and, by extension, the evolutionarily conserved connection between eating and aging.
A recurrent theme in the plethora of Sirt1 molecular interactions we have here tried to summarize is that they are often “circular,” i.e., biunivocal in a feedback or feedforward mode. Remarkable examples are those in which Sirt1 is induced by a transcription factor (i.e., Foxo3a, CREB or cMyc) and, in turn, regulates, either positively or negatively, the factor’s transcriptional activity. Alternatively, Sirt1 can modulate the expression of non-transcriptional upstream regulators such as Nampt or miR34a. Involvement of Sirt1 in the setting of the core clock circuits is another, maybe the clearest, exemplification of this emerging paradigm.
Feedbacks are quite common in biological networks; yet, it is tempting to hypothesize a connection between the circular nature of Sirt-centered adaptive mechanisms and the “non-linear,” “catastrophic” way we age, with the functional decay accelerating dramatically toward the end of the life. Along this line of speculation, it can be modeled that, if an “anti-aging” factor (like CREB or FoxO) engages in a positive feedback loop with Sirt1, protective responses to damage are triggered much faster (and are thus more effective) than in the non-feedback mode, but are also significantly more vulnerable to the factor’s decline over time140 in a fashion that is consistent with the aging catastrophe and with protection by calorie restriction (Box 1 and Supplementary Information for detailed analysis of the model).
Based on this model, “circular” in nature, as circular is the ancient philosophers’ idea of time, more and more of similar reciprocal interactions are likely to be discovered, as we keep on trying to understand what makes hungry cells stronger.
Supplementary Material
Acknowledgments
The authors wish to thank Prof. Emanuela Bartoccioni and Marco De Spirito for critically reading the manuscript. Authors’ laboratories are funded by grants from Italian Association for Cancer Research (AIRC, grant IG8634/2009 to G.P.) and by Catholic University intramural grants (linea D.1) to G.P. and G.M. S.F. is recipient of a Research Doctorate Fellowship from the Italian Ministry of University (MIUR). Costs of this publication were covered by Catholic University in the framework of a program for the support and diffusion of scientific research.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/22074
References
- 1.Guarente L, Franklin H. Franklin H. Epstein Lecture: Sirtuins, aging, and medicine. N Engl J Med. 2011;364:2235–44. doi: 10.1056/NEJMra1100831. [DOI] [PubMed] [Google Scholar]
- 2.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–6. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burnett C, Valentini S, Cabreiro F, Goss M, Somogyvári M, Piper MD, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–5. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guarente L, Picard F. Calorie restriction--the SIR2 connection. Cell. 2005;120:473–82. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 5.Cantó C, Auwerx J. Caloric restriction, SIRT1 and longevity. Trends Endocrinol Metab. 2009;20:325–31. doi: 10.1016/j.tem.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herranz D, Muñoz-Martin M, Cañamero M, Mulero F, Martinez-Pastor B, Fernandez-Capetillo O, et al. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herranz D, Serrano M. Impact of Sirt1 on mammalian aging. Aging (Albany NY) 2010;2:315–6. doi: 10.18632/aging.100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–91. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timmer S, Auwerx J, Schrauwen P. The journey of resveratrol from yeast to human. Aging (Albany NY) 2012;4:146–58. doi: 10.18632/aging.100445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park SJ, Ahmad F, Philp A, Baar K, Williams T, Luo H, et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–33. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–90. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azzi A. Can resveratrol extend your life? IUBMB Life. 2009;61:1010–1. doi: 10.1002/iub.250. [DOI] [PubMed] [Google Scholar]
- 13.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–22. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–6. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–21. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 17.Naiman S, Kanfi Y, Cohen HY. Sirtuins as regulators of mammalian aging. Aging (Albany NY) 2012 doi: 10.18632/aging.100478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bordone L, Cohen D, Robinson A, Motta MC, van Veen E, Czopik A, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–67. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 20.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–2. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 21.Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–7. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 22.Finkel T. Ageing: a toast to long life. Nature. 2003;425:132–3. doi: 10.1038/425132a. [DOI] [PubMed] [Google Scholar]
- 23.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 24.Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–7. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 26.McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, et al. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monaghan P. Telomeres and longevity. Aging (Albany NY) 2012;4:76–7. doi: 10.18632/aging.100437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol. 2011;192:547–56. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–4. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 30.Bosch-Presegué L, Raurell-Vila H, Marazuela-Duque A, Kane-Goldsmith N, Valle A, Oliver J, et al. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol Cell. 2011;42:210–23. doi: 10.1016/j.molcel.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 31.Vorovich E, Ratovitski EA. Dual regulation of TERT activity through transcription and splicing by DeltaNP63alpha. Aging (Albany NY) 2009;1:58–67. doi: 10.18632/aging.100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chua KF, Mostoslavsky R, Lombard DB, Pang WW, Saito S, Franco S, et al. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress. Cell Metab. 2005;2:67–76. doi: 10.1016/j.cmet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Calvanese V, Fraga MF. SirT1 brings stemness closer to cancer and aging. Aging (Albany NY) 2011;3:162–7. doi: 10.18632/aging.100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prozorovski T, Schulze-Topphoff U, Glumm R, Baumgart J, Schröter F, Ninnemann O, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10:385–94. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 35.Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/S1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 36.McKiernan SH, Colman RJ, Aiken E, Evans TD, Beasley TM, Aiken JM, et al. Cellular adaptation contributes to calorie restriction-induced preservation of skeletal muscle in aged rhesus monkeys. Exp Gerontol. 2012;47:229–36. doi: 10.1016/j.exger.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, et al. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280:10264–76. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 38.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–59. doi: 10.1016/S0092-8674(01)00527-X. [DOI] [PubMed] [Google Scholar]
- 39.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–48. doi: 10.1016/S0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 40.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–9. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–48. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 42.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–6. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 43.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–90. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–8. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 45.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–6. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J, Kemper JK. Controlling SIRT1 expression by microRNAs in health and metabolic disease. Aging (Albany NY) 2010;2:527–34. doi: 10.18632/aging.100184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–63. doi: 10.1016/S0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 48.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–5. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 49.Burgering BM. A brief introduction to FOXOlogy. Oncogene. 2008;27:2258–62. doi: 10.1038/onc.2008.29. [DOI] [PubMed] [Google Scholar]
- 50.van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279:28873–9. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 51.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–62. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 52.Cantó C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–60. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–6. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Picard F, Guarente L. Molecular links between aging and adipose tissue. Int J Obes (Lond) 2005;29(Suppl 1):S36–9. doi: 10.1038/sj.ijo.0802912. [DOI] [PubMed] [Google Scholar]
- 55.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–8. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 57.Purushotham A, Schug TT, Li X. SIRT1 performs a balancing act on the tight-rope toward longevity. Aging (Albany NY) 2009;1:669–73. doi: 10.18632/aging.100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–23. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1alpha. J Biol Chem. 2005;280:16456–60. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 60.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jäger S, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 61.Fusco S, Ripoli C, Podda MV, Ranieri SC, Leone L, Toietta G, et al. A role for neuronal cAMP responsive-element binding (CREB)-1 in brain responses to calorie restriction. Proc Natl Acad Sci USA. 2012;109:621–6. doi: 10.1073/pnas.1109237109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 63.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, et al. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–93. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- 65.Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38:864–78. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 66.Chen R, Dioum EM, Hogg RT, Gerard RD, Garcia JA. Hypoxia increases sirtuin 1 expression in a hypoxia-inducible factor-dependent manner. J Biol Chem. 2011;286:13869–78. doi: 10.1074/jbc.M110.175414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–7. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mehta R, Steinkraus KA, Sutphin GL, Ramos FJ, Shamieh LS, Huh A, et al. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science. 2009;324:1196–8. doi: 10.1126/science.1173507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, et al. Modulation of NFκB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–80. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nie Y, Erion DM, Yuan Z, Dietrich M, Shulman GI, Horvath TL, et al. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol. 2009;11:492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schenk S, McCurdy CE, Philp A, Chen MZ, Holliday MJ, Bandyopadhyay GK, et al. Sirt1 enhances skeletal muscle insulin sensitivity in mice during caloric restriction. J Clin Invest. 2011;121:4281–8. doi: 10.1172/JCI58554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–7. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 73.Timmers S, Konings E, Bilet L, Houtkooper RH, van de Weijer T, Goossens GH, et al. Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab. 2011;14:612–22. doi: 10.1016/j.cmet.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Loosdregt J, Vercoulen Y, Guichelaar T, Gent YY, Beekman JM, van Beekum O, et al. van LJ Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood. 2010;115:965–74. doi: 10.1182/blood-2009-02-207118. [DOI] [PubMed] [Google Scholar]
- 75.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–51. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finkbeiner S. CREB couples neurotrophin signals to survival messages. Neuron. 2000;25:11–4. doi: 10.1016/S0896-6273(00)80866-1. [DOI] [PubMed] [Google Scholar]
- 77.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–73. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qiang L, Lin HV, Kim-Muller JY, Welch CL, Gu W, Accili D. Proatherogenic abnormalities of lipid metabolism in SirT1 transgenic mice are mediated through Creb deacetylation. Cell Metab. 2011;14:758–67. doi: 10.1016/j.cmet.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, et al. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–21. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 80.Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–7. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jeong H, Cohen DE, Cui L, Supinski A, Savas JN, Mazzulli JR, et al. Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat Med. 2012;18:159–65. doi: 10.1038/nm.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cohen DE, Supinski AM, Bonkowski MS, Donmez G, Guarente LP. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23:2812–7. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao J, Wang WY, Mao YW, Gräff J, Guan JS, Pan L, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–9. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schwartz MW, Porte D., Jr. Diabetes, obesity, and the brain. Science. 2005;307:375–9. doi: 10.1126/science.1104344. [DOI] [PubMed] [Google Scholar]
- 85.Coppari R, Ramadori G, Elmquist JK. The role of transcriptional regulators in central control of appetite and body weight. Nat Clin Pract Endocrinol Metab. 2009;5:160–6. doi: 10.1038/ncpendmet1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramadori G, Coppari R. Does hypothalamic SIRT1 regulate aging? Aging (Albany NY) 2011;3:325–8. doi: 10.18632/aging.100311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–37. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 88.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 89.Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149–62. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, et al. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 91.Yamagata K, Kitabayashi I. Sirt1 physically interacts with Tip60 and negatively regulates Tip60-mediated acetylation of H2AX. Biochem Biophys Res Commun. 2009;390:1355–60. doi: 10.1016/j.bbrc.2009.10.156. [DOI] [PubMed] [Google Scholar]
- 92.Yamamori T, DeRicco J, Naqvi A, Hoffman TA, Mattagajasingh I, Kasuno K, et al. SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res. 2010;38:832–45. doi: 10.1093/nar/gkp1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nemoto S, Finkel T. Ageing and the mystery at Arles. Nature. 2004;429:149–52. doi: 10.1038/429149a. [DOI] [PubMed] [Google Scholar]
- 94.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. 2006;103:10230–5. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hirschey MD, Shimazu T, Capra JA, Pollard KS, Verdin E. SIRT1 and SIRT3 deacetylate homologous substrates: AceCS1,2 and HMGCS1,2. Aging (Albany NY) 2011;3:635–42. doi: 10.18632/aging.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–61. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hallows WC, Yu W, Denu JM. Regulation of glycolytic enzyme phosphoglycerate mutase-1 by Sirt1 protein-mediated deacetylation. J Biol Chem. 2012;287:3850–8. doi: 10.1074/jbc.M111.317404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105:3374–9. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morselli E, Galluzzi L, Kepp O, Criollo A, Maiuri MC, Tavernarakis N, et al. Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging (Albany NY) 2009;1:961–70. doi: 10.18632/aging.100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martins I, Galluzzi L, Kroemer G. Hormesis, cell death and aging. Aging (Albany NY) 2011;3:821–8. doi: 10.18632/aging.100380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mattagajasingh I, Kim CS, Naqvi A, Yamamori T, Hoffman TA, Jung SB, et al. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc Natl Acad Sci USA. 2007;104:14855–60. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen Z, Peng IC, Cui X, Li YS, Chien S, Shyy JY. Shear stress, SIRT1, and vascular homeostasis. Proc Natl Acad Sci USA. 2010;107:10268–73. doi: 10.1073/pnas.1003833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Min SW, Cho SH, Zhou Y, Schroeder S, Haroutunian V, Seeley WW, et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron. 2010;67:953–66. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bizat N, Peyrin JM, Haïk S, Cochois V, Beaudry P, Laplanche JL, et al. Neuron dysfunction is induced by prion protein with an insertional mutation via a Fyn kinase and reversed by sirtuin activation in Caenorhabditis elegans. J Neurosci. 2010;30:5394–403. doi: 10.1523/JNEUROSCI.5831-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, Catoire H, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37:349–50. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 106.Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–32. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 107.Donmez G, Arun A, Chung CY, McLean PJ, Lindquist S, Guarente L. SIRT1 protects against α-synuclein aggregation by activating molecular chaperones. J Neurosci. 2012;32:124–32. doi: 10.1523/JNEUROSCI.3442-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 108.Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–73. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–7. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–4. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Belden WJ, Dunlap JC. SIRT1 is a circadian deacetylase for core clock components. Cell. 2008;134:212–4. doi: 10.1016/j.cell.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Froy O, Miskin R. The interrelations among feeding, circadian rhythms and ageing. Prog Neurobiol. 2007;82:142–50. doi: 10.1016/j.pneurobio.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 113.Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Lüscher B, et al. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci USA. 2012;109:E187–96. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–3. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 115.Wang J, Zhai Q, Chen Y, Lin E, Gu W, McBurney MW, et al. A local mechanism mediates NAD-dependent protection of axon degeneration. J Cell Biol. 2005;170:349–55. doi: 10.1083/jcb.200504028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–90. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 117.Liu X, Wang D, Zhao Y, Tu B, Zheng Z, Wang L, et al. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1) Proc Natl Acad Sci USA. 2011;108:1925–30. doi: 10.1073/pnas.1019619108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kanfi Y, Peshti V, Gozlan YM, Rathaus M, Gil R, Cohen HY. Regulation of SIRT1 protein levels by nutrient availability. FEBS Lett. 2008;582:2417–23. doi: 10.1016/j.febslet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 120.Vigneron A, Vousden KH. p53, ROS and senescence in the control of aging. Aging (Albany NY) 2010;2:471–4. doi: 10.18632/aging.100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Naqvi A, Hoffman TA, DeRicco J, Kumar A, Kim CS, Jung SB, et al. A single-nucleotide variation in a p53-binding site affects nutrient-sensitive human SIRT1 expression. Hum Mol Genet. 2010;19:4123–33. doi: 10.1093/hmg/ddq331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang Q, Wang SY, Fleuriel C, Leprince D, Rocheleau JV, Piston DW, et al. Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc Natl Acad Sci USA. 2007;104:829–33. doi: 10.1073/pnas.0610590104. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 123.Noriega LG, Feige JN, Canto C, Yamamoto H, Yu J, Herman MA, et al. CREB and ChREBP oppositely regulate SIRT1 expression in response to energy availability. EMBO Rep. 2011;12:1069–76. doi: 10.1038/embor.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–31. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 125.Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;185:203–11. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Abdelmohsen K, Pullmann R, Jr., Lal A, Kim HH, Galban S, Yang X, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–57. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Raffaghello L, Lee C, Safdie FM, Wei M, Madia F, Bianchi G, et al. Starvation-dependent differential stress resistance protects normal but not cancer cells against high-dose chemotherapy. Proc Natl Acad Sci USA. 2008;105:8215–20. doi: 10.1073/pnas.0708100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang Y, Fu W, Chen J, Olashaw N, Zhang X, Nicosia SV, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9:1253–62. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Guo X, Williams JG, Schug TT, Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem. 2010;285:13223–32. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kang H, Jung JW, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PLoS ONE. 2009;4:e6611. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, et al. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS ONE. 2009;4:e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, et al. Phosphorylation regulates SIRT1 function. PLoS ONE. 2008;3:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Back JH, Rezvani HR, Zhu Y, Guyonnet-Duperat V, Athar M, Ratner D, et al. Cancer cell survival following DNA damage-mediated premature senescence is regulated by mammalian target of rapamycin (mTOR)-dependent Inhibition of sirtuin 1. J Biol Chem. 2011;286:19100–8. doi: 10.1074/jbc.M111.240598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gao Z, Zhang J, Kheterpal I, Kennedy N, Davis RJ, Ye J. Sirtuin 1 (SIRT1) protein degradation in response to persistent c-Jun N-terminal kinase 1 (JNK1) activation contributes to hepatic steatosis in obesity. J Biol Chem. 2011;286:22227–34. doi: 10.1074/jbc.M111.228874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tanti JF, Jager J. Cellular mechanisms of insulin resistance: role of stress-regulated serine kinases and insulin receptor substrates (IRS) serine phosphorylation. Curr Opin Pharmacol. 2009;9:753–62. doi: 10.1016/j.coph.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 136.Sun C, Zhang F, Ge X, Yan T, Chen X, Shi X, et al. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. 2007;6:307–19. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 137.Li Y, Xu W, McBurney MW, Longo VD. SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 2008;8:38–48. doi: 10.1016/j.cmet.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gerhart-Hines Z, Dominy JE, Jr., Blättler SM, Jedrychowski MP, Banks AS, Lim JH, et al. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+) Mol Cell. 2011;44:851–63. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chung JH. Using PDE inhibitors to harness the benefits of calorie restriction: lessons from resveratrol. Aging (Albany NY) 2012;4:144–5. doi: 10.18632/aging.100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kauffman AL, Ashraf JM, Corces-Zimmerman MR, Landis JN, Murphy CT. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biol. 2010;8:e1000372. doi: 10.1371/journal.pbio.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.