Abstract
Purpose
Poor physical fitness and obesity are risk factors for all cause morbidity and mortality. We aimed to clarify whether common genetic variants of key energy intake determinants in leptin (LEP), leptin receptor (LEPR), and fat mass and obesity-associated (FTO) are associated with aerobic and neuromuscular performance, and whether aerobic fitness can alter the effect of these genotypes on body composition.
Methods
846 healthy Finnish males of Caucasian origin were genotyped for FTO (rs8050136), LEP (rs7799039) and LEPR (rs8179183 and rs1137101) single nucleotide polymorphisms (SNPs), and studied for associations with maximal oxygen consumption, body fat percent, serum leptin levels, waist circumference and maximal force of leg extensor muscles.
Results
Genotype AA of the FTO SNP rs8050136 associated with higher BMI and greater waist circumference compared to the genotype CC. In general linear model, no significant interaction for FTO genotype-relative VO2max (mL·kg−1·min−1) or FTO genotype-absolute VO2max (L·min−1) on BMI or waist circumference was found. Main effects of aerobic performance on body composition traits were significant (p<0.001). Logistic regression modelling found no significant interaction between aerobic fitness and FTO genotype. LEP SNP rs7799039, LEPR SNPs rs8179183 and rs1137101 did not associate with any of the measured variables, and no significant interactions of LEP or LEPR genotype with aerobic fitness were observed. In addition, none of the studied SNPs associated with aerobic or neuromuscular performance.
Conclusions
Aerobic fitness may not modify the effect of FTO variation on body composition traits. However, relative aerobic capacity associates with lower BMI and waist circumference regardless of the FTO genotype. FTO, LEP and LEPR genotypes unlikely associate with physical performance.
Introduction
Obesity and low physical fitness frequently associate with each other, and are individual risk factors for many pathological conditions and cardiovascular mortality [1], [2]. Body composition and physical performance are outcomes of cumulative effects of common and rare variants in a large number of genes with environmental and gene-gene interactions [3]. The effects of behavioural factors on human performance, including physical activity and dietary habits have been widely studied, yet knowledge on genetic background is sparse. The heritability estimates for obesity range from 40% to 70% [3], from 20 to 40% for aerobic performance [4], [5], and approximately 60% for muscle force [6]. Single nucleotide polymorphisms (SNPs) represent the most common type of genetic variation in the human genome [7]. However, it is not known whether genetic variants in genes encoding fat and obesity-associated (FTO), leptin (LEP), and leptin receptor (LEPR) associate with physical performance, and whether fitness level modifies the risk for obesity associated with these gene variants.
Genome-wide association studies have identified the FTO gene as the first susceptibility locus for common obesity [8], [9]. Minor allele of this variant is associated with increased risk for obesity and elevated body weight [8], [9]. In a recent meta-analysis, physical activity level was shown to modify the relationship between the FTO risk variant and body weight and risk of obesity [10]. However, effects of FTO genotype and its interaction with aerobic fitness on body composition have not been reported so far. Despite the fact that FTO is under extensive research, the role and function of the FTO gene product remains incompletely understood.
Leptin is a peptide hormone secreted mainly from white adipose tissue. It regulates hunger, body temperature and energy metabolism, and has been used as a biomarker of energy deficiency [11], [12]. Recent studies have shown that obese individuals are in fact leptin resistant [12], and that physical exercise may restore leptin sensitivity [13]. Associations of leptin receptor gene (LEPR) SNP Gln223Arg (rs1137101) and leptin (LEP) promoter region SNP -2548G/A (rs7799039) with BMI and other body composition-related traits have been reported [14]–[18]. We previously demonstrated that the rs7799039 associates with changes in body composition in response to physical training [19]. Furthermore, another LEPR SNP Lys656Asn (rs8179183) has been reported to associate with substrate oxidation and basal metabolic rate [20], [21]. However, the interaction effects of these SNPs with aerobic performance on body composition are not known.
The present study investigated the association of selected SNPs in FTO, LEP and LEPR with body composition, neuromuscular and cardiorespiratory performance and plasma leptin levels in 846 healthy male subjects. In addition, genotype-aerobic fitness interactions on body composition traits were studied. Our hypothesis was that these common variants are associated with human performance, body composition and health-related risk factors. Furthermore, we hypothesized that aerobic fitness modifies the relationship between the genetic variants and body composition. The information this study provides may be used to identify those individuals having higher health risks and tendency for poor physical fitness, and for better understanding of the effect of genetic factors on human physical performance.
Materials and Methods
Study Subjects
The subjects of Caucasian origin were 846 healthy male Finnish volunteers with mean age (SD) of 25±5 years. All subjects were informed on the purpose of the study, gave a written informed consent and underwent medical examination prior to the tests. This study was approved by the ethics committee of the University of Jyväskylä and the hospital district of Central Finland. Anthropometric data and blood samples were collected after an overnight fast. The subjects consumed light breakfast 1 to 2 hours before the exercise tests.
Physical Performance and Body Composition
Height, weight and waist circumference were recorded, and body mass index (BMI, kg·m−2) was calculated. Body fat and lean mass percentage were recorded by using the eight-polar bioimpedance method with multifrequency current (InBody 720; Biospace Company, Seoul, Korea). Bioimpedance was recorded after an overnight fast and with at least one day off from any intensive physical activity.
Maximal isometric force production of the leg extensor muscles was measured with a dynamometer (Department of Biology of Physical Activity, University of Jyväskylä, Finland). The test was performed in sitting position with 107-degree knee angle. The subjects were supervised to generate maximal force as fast as possible and maintain this force for 3 seconds. The data were analysed with a 16-bit AD converter (CED power 1401, Cambridge Electronic Design ltd, England) and Signal (2.16) software.
Aerobic fitness was assessed by maximal bicycle ergometer test (Ergoline 800 S, Ergoselect 100 K, Ergoselect 200 K, Bitz, Germany) as previously described [22]. The initial workload was 50 W with 25 W increase on 2-minute intervals until exhaustion. Heart rate was monitored throughout the test (Polar T-31; Polar Vantage, Kempele, Finland). The analyzed variables were maximum heart rate, maximal workload and maximal oxygen consumption (mL·kg−1·min−1) estimated by software (Milfit4/Fitware, Finland) [(11.016 · maximum work load) x (1·body weight−1) +7.0]. The test was terminated when the subject could not maintain the required cycling speed (60–90 rpm). Physical activity was assessed with international physical activity questionnaire (IPAQ) [23].
Blood Samples and Genotyping
The fasting blood samples were collected and analysed immediately with a hemacytometer (Sysmex Co., Kobe, Japan). Plasma was separated from the whole blood and stored at -80C° until analysis. Plasma leptin concentrations were assayed by commercial ELISA according to the manufacturer’s instructions (Quantikine, R&D Systems, Minneapolis, MN, USA). Assay specifications were as follows: sensitivity limit 7.8 pg·mL−1, and maximum intra- and interassay CV% 3.3% and 5.4%, respectively.
The SNP genotyping was performed using allele-specific PCR assays. Briefly, genomic DNA was first isolated from the blood mononuclear cells using QIAamp DNA Blood kit (Qiagen, Hilden, Germany). Next, 50 nanograms of the DNA was amplified with Brilliant QPCR Master Mix (Stratagene, La Jolla, CA, USA) and allele-specific SNP assays on a Mx3000P Real-time PCR System (Stratagene). For FTO (rs8050136) and LEPR SNP Lys656Asn (rs8179183), the commercially available TaqMan SNP genotyping assays were used (Applied Biosystems, Foster City, CA, USA), and for LEP (rs7799039) and LEPR SNP Gln223Arg (rs1137101), the following molecular beacons SNP genotyping assays were used: LEP SNP forward primer 5′-CCTGTAATTTTCCCATGAGAAC-3′ and reverse primer 5′-TGCAACATCTCAGCACTTAG-3′, and the molecular beacons 5′-FAM/HEX-CGTGCCCGACAGGGTTGC(G/A)CTGATCGGCACG -BHQ1; LEPR SNP forward primer 5′-TCAACGACACTCTCCTTATG-3′ and reverse primer 5′-TTATGGGCTGAACTGACAT-3′, and the molecular beacons 5′-FAM/HEX- CGGACGTGGAGTAATTTTCC(A/G)GTCACCTCCGTCCG -BHQ1-3′.
Statistics
Calculations were performed with SPSS software (Illinois, Chicago, USA) by using one-way ANCOVA or non-parametric tests, when appropriate. Age, smoking and earlier physical activity were set as covariates. Smoking (smoker or non-smoker) and physical activity (vigorous physical activity more than 3 times per week) were set as dichotomous variables, whereas age VO2max, BMI, waist circumference were set as continuous variables.
Genotype-VO2max interaction on BMI, waist circumference and fat percent were analyzed by using general linear model. The general linear model included main effects terms for earlier physical activity, smoking, age, genotype, VO2max and genotype-VO2max interaction. Physical activity and smoking were set as dichotomous variables, and VO2max, age and body composition traits were set as continuous variables. Genotype was set as discrete variable in every analysis.
Additionally, the FTO genotype and FTO genotype-VO2max interaction related odds for overweight (BMI over 25 kg·m−2) and abdominal obesity (waist circumference over 90 cm) were analyzed by logistic regression. Recessive model was applied in analysis (AA genotype vs. CA and CC genotype). Age, smoking and physical activity were set as covariates. The subjects were divided into quartiles according to VO2max. The 75% percentile limits for VO2max were 3.7 L·min−1 and 46.9 mL·kg−1·min−1. The subjects in the highest 25% quartile were defined as high aerobic fitness group, and compared with the 75% percentile of subjects (VO2max <3.7 L·min−1 or <46.9 mL·kg−1·min−1).
Data are presented as mean ± standard deviation unless otherwise stated. Statistical significance was set at p<0.0125 to account for multiple testing. Bonferroni correction was applied to post-hoc comparisons. Statistical power and sample sizes were estimated with SISA web calculator [24]. A 90% level was chosen for power calculations, and the number of subjects needed to demonstrate 1 S.D. difference in continuous parameters was estimated.
Results
All SNPs conformed to Hardy-Weinberg’s equilibrium. None of the SNPs were associated with earlier physical activity estimated with IPAQ.
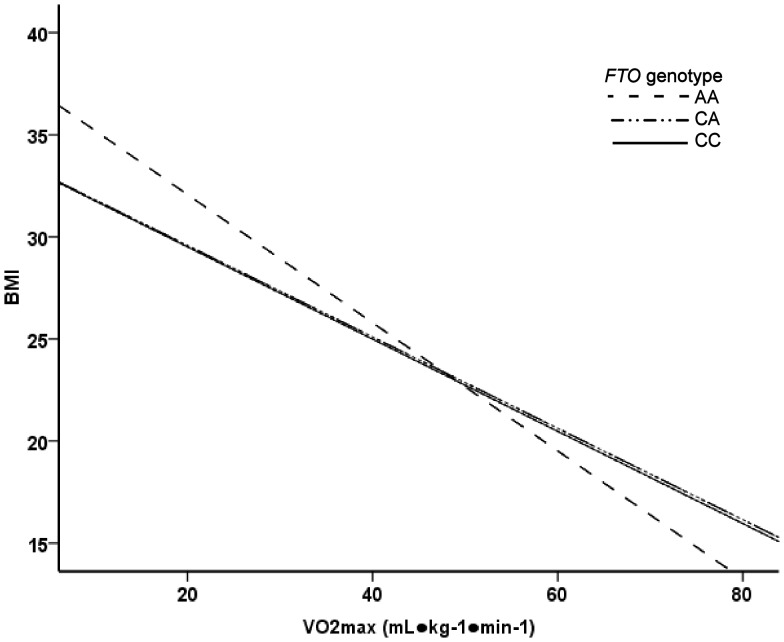
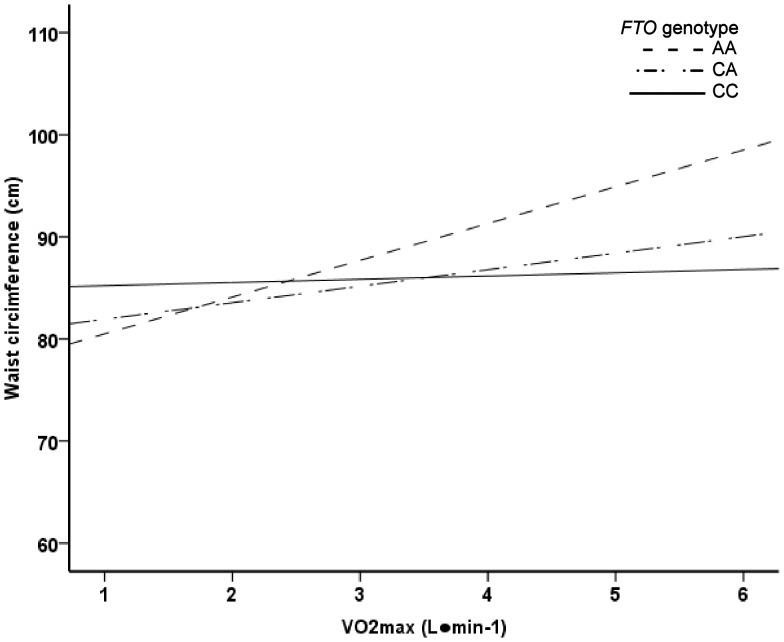
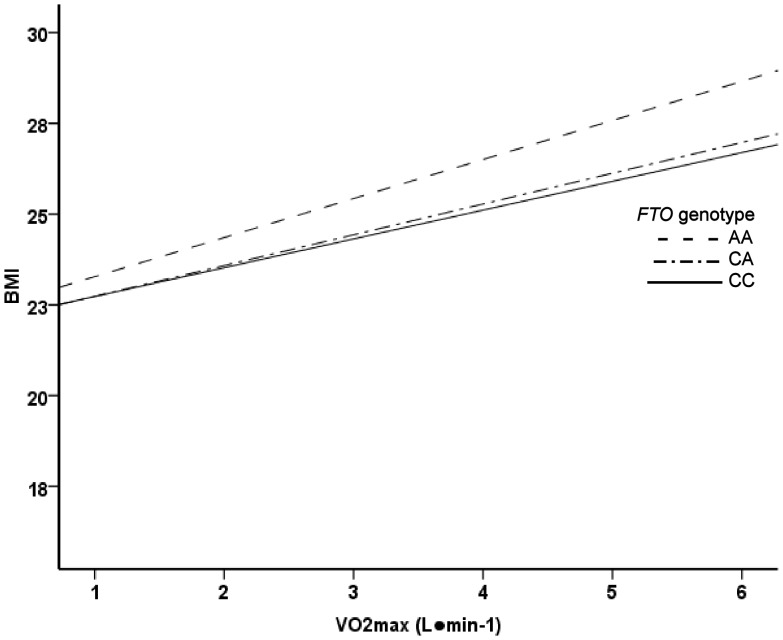
In ANCOVA analysis, the FTO SNP rs8050136 associated with BMI and waist circumference (Table 1). Genotype AA carriers had significantly higher BMI and greater waist circumference compared to the genotype CC carriers. In general linear model, the main effects of relative and absolute VO2max on BMI and waist circumference were significant (p<0.001). However, no significant FTO genotype-relative VO2max (mL·kg−1·min−1) interaction on BMI (p = 0.081) or waist circumference (p = 0.093) was found. In addition, the test result was insignificant when FTO genotype-absolute VO2max (L·min−1) interaction on BMI (p = 0.937) or waist circumference (p = 0.262) were analyzed (Figures 1–4). In logistic regression, genotype AA increased odds for waist circumference over 90 cm. However, the interaction of genotype AA with high VO2max was insignificant (Table 2). No association was found with aerobic fitness (VO2max expressed as L·min−1 or mL·kg−1·min−1) (Table 1).
Table 1. Showing the results of ANCOVA–analysis according to FTO genotype.
| FTO SNP rs8050136 (N = 773) | |||
| Genotype | CC | CA | AA |
| N = 269 | N = 380 | N = 124 | |
| 35% | 49% | 16% | |
| BMI (kg· m −2 ) | 24.5±3.6 | 24.8±3.7 | 25.7±4.3* |
| Waist circumference (cm) | 85.8±10.3 | 86.0±9.7 | 88.8±12.2# |
| Fat per cent (%) | 17.5±7.2 | 17.9±7.0 | 19.4±8.0 |
| VO2max (mL·kg −1 ·min −1 ) | 42.0±8.3 | 41.8±7.8 | 40.2±8.6 |
| VO2max (L·min −1 ) | 3.29±0.56 | 3.30±0.58 | 3.30±0.62 |
| Maximal working capacity (Watts) | 244.2±44.7 | 245.0±46.5 | 242.7±49.1 |
| Maximal force of leg extensors (N) | 2927±872 | 2950±816 | 2942±992 |
| Plasma leptin (pg·mL −1 ) | 3598±3657 | 3702±3780 | 4289±4278 |
Earlier physical activity, smoking and age were set as covariates. Bonferroni correction was accounted for multiple post-hoc comparisons.
BMI: p = 0.007 for main effect, *p = 0.005 between the genotype groups CC and AA.
Waist circumference: p = 0.012 for main effect, #p = 0.012 between the genotype groups CC and AA.
p>0.0125 for main effect between genotype groups for rest of the variables.
Figure 1. Linear regression analysis.
The main effect of VO2max (mL·kg−1·min−1) on BMI was significant (p<0.001). The main effects of FTO genotype and FTO genotype-VO2max (mL·kg−1·min−1) interaction were not significant (p = 0.029 and p = 0.081, respectively).
Figure 4. Linear regression analysis.
The main effect of VO2max (L·min−1) on waist circumference was significant (p<0.001). The main effects of FTO genotype and FTO genotype-VO2max (L·min−1) interaction were not significant (p = 0.521 and p = 0.262, respectively).
Table 2. Showing results of the logistic regression analysis.
| OR | 95% C.I. | p-value | |
| BMI over 25 kg·m −2 adjusted for age, smoking, physical activity and relative aerobic fitness (mL·kg −1 ·min −1 ) | |||
| Genotype AA | 1.51 | 0.96–2.37 | 0.073 |
| VO2max (>46.9 mL·kg−1·min−1) | 0.15 | 0.09–0.25 | <0.001 |
| Genotype AA by VO2max (>46.9 mL·kg−1·min−1) | 1.31 | 0.43–3.96 | 0.639 |
| BMI over 25 kg·m −2 adjusted for age, smoking, physical activity and absolute aerobic fitness (L·min −1 ) | |||
| Genotype AA | 1.61 | 1.02–2.53 | 0.038 |
| VO2max (>3.7 L·min−1) | 1.10 | 0.75–1.63 | 0.626 |
| Genotype AA by VO2max (>3.7 L·min−1) | 1.02 | 0.41–2.58 | 0.959 |
| Waist circumference over 90 cm adjusted for age, smoking, physical activity and relative aerobic fitness (mL·kg −1 ·min −1 ) | |||
| Genotype AA | 1.92 | 1.22–3.03 | 0.005 |
| VO2max (>46.9 mL·kg−1·min−1) | 0.08 | 0.04–0.18 | <0.001 |
| Genotype AA by VO2max (>46.9 mL·kg−1·min−1) | 1.07 | 0.19–5.94 | 0.941 |
| Waist circumference over 90 cm adjusted for age, smoking, physical activity and absolute aerobic fitness (L·min −1 ) | |||
| Genotype AA | 1.96 | 1.22–3.15 | 0.005 |
| VO2max (>3.7 L·min−1) | 1.26 | 0.81–1.97 | 0.299 |
| Genotype AA by VO2max (>3.7 L·min−1) | 0.88 | 0.33–2.40 | 0.884 |
OR and 95% C.I. for the genotype AA (vs. genotype CA and CC) of the FTO rs8050136 according to overweight and abdominal obesity.
Figure 2. Linear regression analysis.
The main effect of VO2max (L·min−1) on BMI was significant (p<0.001). The main effects of FTO genotype and FTO genotype-VO2max (L·min−1) interaction were not significant (p = 0.964 and p = 0.937, respectively).
Figure 3. Linear regression analysis.
The main effect of VO2max (mL·kg−1·min−1) on waist circumference was significant (p<0.001). The main effects of FTO genotype and FTO genotype-VO2max (mL·kg−1·min−1) interaction were not significant (p = 0.040 and p = 0.093, respectively).
In ANCOVA analysis, LEP SNP rs7799039 did not associate with any variable (Table 3). No interactions were observed between aerobic fitness and genotype. LEPR SNP rs1137101 did not associate with BMI, body fat, leptin levels or physical performance in ANCOVA (Table 3), and no aerobic performance-genotype interactions were observed in the general linear model. In ANCOVA analysis, LEPR Lys656Asn SNP rs8179183 did not associate with any variable (Table 3). In addition, no interactions were observed between genotype and aerobic fitness in the general linear model.
Table 3. Showing the results of ANCOVA –analysis according to LEP and LEPR genotypes.
| LEP -2548 SNP rs7799039 (N = 713) | LEPR Lys656Asn SNP rs8179183 (N = 713) | LEPR Gln223Arg SNP rs1137101 (N = 713) | ||||||||
| Genotype | GG | GA | AA | Lys/Lys | Lys/Asn | Asn/Asn | AA | AG | GG | |
| N = 176 | N = 384 | N = 153 | N = 549 | N = 152 | N = 12 | N = 125 | N = 350 | N = 238 | ||
| 25% | 54% | 21% | 77% | 21% | 2% | 18% | 49% | 33% | ||
| BMI (kg·m −2 ) | 25.0±3.8 | 24.9±3.9 | 24.4±3.4 | 25.0±3.8 | 24.4±3.8 | 23.6±2.1 | 24.5±3.4 | 24.9±3.9 | 24.9±3.8 | |
| Waist circumference (cm) | 86.6±10.1 | 86.6±10.8 | 85.4±9.6 | 86.8±10.6 | 84.6±9.8 | 83.9±4.9 | 86.2±9.8 | 86.3±10.3 | 86.5±10.9 | |
| Fat per cent % | 18.5±7.2 | 18.1±7.4 | 17.1±7.0 | 18.2±7.3 | 17.2±7.4 | 16.8±6.0 | 18.1±6.9 | 17.6±7.5 | 18.4±7.1 | |
| VO2max (mL·kg −1 ·min −1 ) | 40.7±8.3 | 42.1±8.2 | 41.7±7.7 | 41.3±8.1 | 42.8±8.2 | 40.2±6.1 | 42.5±8.3 | 41.7±8.1 | 41.1±8.0 | |
| VO2max (L·min −1 ) | 3.26±0.55 | 3.33±0.60 | 3.27±0.60 | 3.29±0.58 | 3.33±0.60 | 3.09±0.42 | 3.34±0.58 | 3.31±0.58 | 3.27±0.58 | |
| Maximal working capacity (Watts) | 240.4±44.6 | 246.9±47.3 | 242.5±45.4 | 243.8±45.7 | 247.9±48.1 | 228.4±33.9 | 248.1±46.1 | 245.0±46.6 | 241.5±45.9 | |
| Maximal force of leg extensors (N) | 2952±835 | 2942±863 | 2920±901 | 2948±837 | 2937±967 | 2586±630 | 2913±758 | 2958±847 | 2928±937 | |
| Plasma leptin (pg·mL −1 ) | 3738±3686 | 3858±3961 | 3517±3635 | 3746±3679 | 3842±4 398 | 3069±2827 | 4161±4378 | 3781±4057 | 3513±3102 | |
Earlier physical activity, smoking and age were set as covariates. Bonferroni correction was accounted for multiple post-hoc comparisons.
p>0.0125 for main effect between genotype groups for every SNP.
Discussion
The main finding of the present study was that aerobic fitness does not modify the effect of FTO variation on BMI or waist circumference. However, relative aerobic capacity did associate with lower BMI and waist circumference regardless of the FTO genotype. We also report that the LEP promoter -2548 SNP and the LEPR Lys656Asn SNP did not associate with BMI, waist circumference or body fat percent.
Our observations that genotype AA of the FTO variant associated with higher BMI and greater waist circumference are in line with a recent meta-analysis [25]. Our novel finding, however, is that aerobic fitness does not modify the effect of FTO variation on BMI and waist circumference. In another recent meta-analysis, the level of physical activity was shown to modify the relationship between the FTO risk variant and body weight and risk of obesity [10]. Nevertheless, these studies are not fully comparable to ours because at present, this is the first study to report objectively measured aerobic fitness - FTO-interaction. The questionnaire-based assessment of physical activity may have some reporting bias [26]. In addition, the present findings may be explained by the fact that apart from physical activity, other factors may affect aerobic fitness. Approximately 20 to 40% of VO2max has been estimated to be heritable, and other factors, such as physical activity, intake of carbohydrates, smoking, body weight, blood haemoglobin levels, age and presence of chronic disease account for the remaining [4], [5]. In addition, the present and earlier studies differ in study design because in our study, all subjects were young males. Nevertheless, in the present study, aerobic fitness was associated with body composition traits regardless of the FTO genotype. Despite lack of FTO genotype-aerobic fitness interaction, the genotype AA carriers are more susceptible to obesity and may benefit from increased aerobic fitness to a greater extent due to its favourable effects on the regulation of body weight. In the present study, FTO variant was not found to associate with aerobic fitness, which is in line with previous studies [27], [28]. It can be speculated that low physical fitness combined with increased body weight in the allele A carriers of the FTO variant may further increase risk for lifestyle-associated disease, such as cardiovascular events [29]. This would in turn highlight the importance of physical activity to attenuate risk for obesity. Furthermore, the studied FTO variant did not associate with plasma leptin levels, which in line with other studies [30], [31].
In the present study, the leptin promoter SNP rs7799039 did not associate with BMI, which is supported by earlier reports [17], [18]. It has been speculated that lower leptin levels in allele G carriers of this SNP would be an underlying mechanism for increased accumulation of adipose tissue [32]. However, in the present study, leptin levels were similar between the genotype groups. Furthermore, no association with aerobic fitness, neuromuscular performance, or interaction with VO2max was found.
The LEPR Gln223Arg genotype showed no association with cardio-respiratory fitness, which is supported by our previous report [19]. Furthermore, in agreement with other studies [19], [33], no differences in leptin levels between the genotype groups were found, although controversial findings does exist [15], [34], [35]. It should be noted, however, that in many published studies, interpretation of the results is limited due to small sample size, differences in ethnicity of the subjects, and other factors related to the study settings.
The LEPR Lys656Asn SNP did not associate with BMI, waist circumference or body fat per cent, and these findings are also supported by others [36]–[38]. Moreover, subjects of the present study were not morbidly obese, and small prevalence of the minor Asn allele may also complicate evaluation of the effect of this SNP on body weight and body composition. Furthermore, no significant genotype-VO2max interactions were found.
There are certain limitations to the present study. First, the sample size was moderate for genetic association analyses. The relatively large confidence intervals may bias identification of a true interaction in logistic regression analysis. In addition, all subjects were males, which may affect generalization of the results to other populations. Our study design was cross-sectional, which may increase risk for selection bias. However, earlier physical activity status was assessed with IPAQ survey, and no associations with pre-study physical activity and genotypes were found. The bioimpedance method is both sensitive and specific for analysis of body fat mass, but may have limitations for accurate evaluation of muscle mass [39]. The method for assessing aerobic fitness was indirect, whereas direct gas exchange analysis would provide greater accuracy.
In conclusion, aerobic fitness may not modify the effect of FTO variation on body composition traits. However, relative aerobic capacity had a favourable effect on BMI and waist circumference, regardless of the FTO genotype. It is unlikely that FTO, LEP and LEPR genotypes associate with physical performance. These findings can be applied in sports medicine to decrease risk for diseases related to sedentary lifestyle and obesity, where improved aerobic fitness is beneficial regardless of the FTO-related predisposition to obesity.
Acknowledgments
We thank Elina Kokkonen for statistical assistance. Satu Marttila, Taina Vihavainen and Taija Hukkanen provided skilful technical assistance.
Funding Statement
This work was funded by the Research Council for Physical Education and Sports, Finnish Ministry of Education, TBGS Graduate School, Yrjö Jahnsson Foundation, the Scientific Advisory Board of Defense, Finland and COST actions B35 and BM0602. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sui X, Lee DC, Matthews CE, Adams SA, Hebert JR, et al. (2010) Influence of cardiorespiratory fitness on lung cancer mortality. Medicine and Science in Sports and Exercise 42: 872–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang Y (2010) Cardiovascular diseases in american women. Nutrition, Metabolism, and Cardiovascular Diseases : NMCD 20: 386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McPherson R (2007) Genetic contributors to obesity. The Canadian Journal of Cardiology 23 Suppl A: 23A–27A. [DOI] [PMC free article] [PubMed]
- 4. Laukkanen JA, Laaksonen D, Lakka TA, Savonen K, Rauramaa R, et al. (2009) Determinants of cardiorespiratory fitness in men aged 42 to 60 years with and without cardiovascular disease. The American Journal of Cardiology 103: 1598–1604. [DOI] [PubMed] [Google Scholar]
- 5. Bouchard C, Daw EW, Rice T, Perusse L, Gagnon J, et al. (1998) Familial resemblance for VO2max in the sedentary state: The HERITAGE family study. Medicine and Science in Sports and Exercise 30: 252–258. [DOI] [PubMed] [Google Scholar]
- 6. Huygens W, Thomis MA, Peeters MW, Vlietinck RF, Beunen GP (2004) Determinants and upper-limit heritabilities of skeletal muscle mass and strength. Canadian Journal of Applied Physiology = Revue Canadienne De Physiologie Appliquee 29: 186–200. [DOI] [PubMed] [Google Scholar]
- 7. Brookes AJ (1999) The essence of SNPs. Gene 234: 177–186. [DOI] [PubMed] [Google Scholar]
- 8. Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, et al. (2007) A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science (New York, N.Y.) 316: 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scuteri A, Sanna S, Chen WM, Uda M, Albai G, et al. (2007) Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genetics 3: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kilpelainen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, et al. (2011) Physical activity attenuates the influence of FTO variants on obesity risk: A meta-analysis of 218,166 adults and 19,268 children. PLoS Medicine 8: e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Friedman JM, Halaas JL (1998) Leptin and the regulation of body weight in mammals. Nature 395: 763–770. [DOI] [PubMed] [Google Scholar]
- 12. Kelesidis T, Kelesidis I, Chou S, Mantzoros CS (2010) Narrative review: The role of leptin in human physiology: Emerging clinical applications. Annals of Internal Medicine 152: 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shapiro A, Matheny M, Zhang Y, Tumer N, Cheng KY, et al. (2008) Synergy between leptin therapy and a seemingly negligible amount of voluntary wheel running prevents progression of dietary obesity in leptin-resistant rats. Diabetes 57: 614–622. [DOI] [PubMed] [Google Scholar]
- 14. Furusawa T, Naka I, Yamauchi T, Natsuhara K, Kimura R, et al. (2010) The Q223R polymorphism in LEPR is associated with obesity in pacific islanders. Human Genetics 127: 287–294. [DOI] [PubMed] [Google Scholar]
- 15. Ben Ali S, Kallel A, Sediri Y, Ftouhi B, Feki M, et al. (2009) LEPR p.Q223R polymorphism influences plasma leptin levels and body mass index in tunisian obese patients. Archives of Medical Research 40: 186–190. [DOI] [PubMed] [Google Scholar]
- 16. Fairbrother UL, Tanko LB, Walley AJ, Christiansen C, Froguel P, et al. (2007) Leptin receptor genotype at Gln223Arg is associated with body composition, BMD, and vertebral fracture in postmenopausal danish women. Journal of Bone and Mineral Research : The Official Journal of the American Society for Bone and Mineral Research 22: 544–550. [DOI] [PubMed] [Google Scholar]
- 17. Wang TN, Huang MC, Chang WT, Ko AM, Tsai EM, et al. (2006) G-2548A polymorphism of the leptin gene is correlated with extreme obesity in taiwanese aborigines. Obesity (Silver Spring, Md.) 14: 183–187. [DOI] [PubMed] [Google Scholar]
- 18. Mammes O, Betoulle D, Aubert R, Herbeth B, Siest G, et al. (2000) Association of the G-2548A polymorphism in the 5′ region of the LEP gene with overweight. Annals of Human Genetics 64: 391–394. [DOI] [PubMed] [Google Scholar]
- 19. Huuskonen A, Lappalainen J, Tanskanen M, Oksala N, Kyrolainen H, et al. (2010) Genetic variations of leptin and leptin receptor are associated with body composition changes in response to physical training. Cell Biochemistry and Function 28: 306–312. [DOI] [PubMed] [Google Scholar]
- 20. Loos RJ, Rankinen T, Chagnon Y, Tremblay A, Perusse L, et al. (2006) Polymorphisms in the leptin and leptin receptor genes in relation to resting metabolic rate and respiratory quotient in the quebec family study. International Journal of Obesity (2005) 30: 183–190. [DOI] [PubMed] [Google Scholar]
- 21. Wauters M, Considine RV, Chagnon M, Mertens I, Rankinen T, et al. (2002) Leptin levels, leptin receptor gene polymorphisms, and energy metabolism in women. Obesity Research 10: 394–400. [DOI] [PubMed] [Google Scholar]
- 22. Hakkinen A, Rinne M, Vasankari T, Santtila M, Hakkinen K, et al. (2010) Association of physical fitness with health-related quality of life in finnish young men. Health and Quality of Life Outcomes 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Craig CL, Marshall AL, Sjostrom M, Bauman AE, Booth ML, et al. (2003) International physical activity questionnaire: 12-country reliability and validity. Medicine and Science in Sports and Exercise 35: 1381–1395. [DOI] [PubMed] [Google Scholar]
- 24.Uitenbroek D (2004) Sample size. SISA. 2012. Available: (http://www.quantitativeskills.com/sisa/calculations/samsize.htm) Accessed 2012 Oct 8.
- 25. Peng S, Zhu Y, Xu F, Ren X, Li X, et al. (2011) FTO gene polymorphisms and obesity risk: A meta-analysis. BMC Medicine 9: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bouchard C (2008) Gene-environment interactions in the etiology of obesity: Defining the fundamentals. Obesity (Silver Spring, Md.) 16 Suppl 3S5–S10. [DOI] [PubMed] [Google Scholar]
- 27. Berentzen T, Kring SI, Holst C, Zimmermann E, Jess T, et al. (2008) Lack of association of fatness-related FTO gene variants with energy expenditure or physical activity. The Journal of Clinical Endocrinology and Metabolism 93: 2904–2908. [DOI] [PubMed] [Google Scholar]
- 28. Mitchell JA, Church TS, Rankinen T, Earnest CP, Sui X, et al. (2010) FTO genotype and the weight loss benefits of moderate intensity exercise. Obesity (Silver Spring, Md.) 18: 641–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ahmad T, Chasman DI, Mora S, Pare G, Cook NR, et al. (2010) The fat-mass and obesity-associated (FTO) gene, physical activity, and risk of incident cardiovascular events in white women. American Heart Journal 160: 1163–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mangge H, Renner W, Almer G, Weghuber D, Moller R, et al. (2011) Rs9939609 variant of the fat mass and obesity-associated gene and trunk obesity in adolescents. Journal of Obesity 2011: 186368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zimmermann E, Skogstrand K, Hougaard DM, Astrup A, Hansen T, et al. (2011) Influences of the common FTO rs9939609 variant on inflammatory markers throughout a broad range of body mass index. PloS One 6: e15958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoffstedt J, Eriksson P, Mottagui-Tabar S, Arner P (2002) A polymorphism in the leptin promoter region (-2548 G/A) influences gene expression and adipose tissue secretion of leptin. Hormone and Metabolic Research.Hormon- Und Stoffwechselforschung.Hormones Et Metabolisme 34: 355–359. [DOI] [PubMed] [Google Scholar]
- 33. Pyrzak B, Wisniewska A, Kucharska A, Wasik M, Demkow U (2009) No association of LEPR Gln223Arg polymorphism with leptin, obesity or metabolic disturbances in children. European Journal of Medical Research 14 Suppl 4201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Masuo K, Straznicky NE, Lambert GW, Katsuya T, Sugimoto K, et al. (2008) Leptin-receptor polymorphisms relate to obesity through blunted leptin-mediated sympathetic nerve activation in a caucasian male population. Hypertension Research : Official Journal of the Japanese Society of Hypertension 31: 1093–1100. [DOI] [PubMed] [Google Scholar]
- 35. van Rossum CT, Hoebee B, van Baak MA, Mars M, Saris WH, et al. (2003) Genetic variation in the leptin receptor gene, leptin, and weight gain in young dutch adults. Obesity Research 11: 377–386. [DOI] [PubMed] [Google Scholar]
- 36. Mergen H, Karaaslan C, Mergen M, Deniz Ozsoy E, Ozata M (2007) LEPR, ADBR3, IRS-1 and 5-HTT genes polymorphisms do not associate with obesity. Endocrine Journal 54: 89–94. [DOI] [PubMed] [Google Scholar]
- 37. Paracchini V, Pedotti P, Taioli E (2005) Genetics of leptin and obesity: A HuGE review. American Journal of Epidemiology 162: 101–114. [DOI] [PubMed] [Google Scholar]
- 38. Silver K, Walston J, Chung WK, Yao F, Parikh VV, et al. (1997) The Gln223Arg and Lys656Asn polymorphisms in the human leptin receptor do not associate with traits related to obesity. Diabetes 46: 1898–1900. [DOI] [PubMed] [Google Scholar]
- 39. Demura S, Sato S, Kitabayashi T (2004) Percentage of total body fat as estimated by three automatic bioelectrical impedance analyzers. Journal of Physiological Anthropology and Applied Human Science 23: 93–99. [DOI] [PubMed] [Google Scholar]