Abstract
In higher vertebrates, the expression of Sox2, a group B1 Sox gene, is the hallmark of neural primordial cell state during the developmental processes from embryo to adult. Sox2 is regulated by the combined action of many enhancers with distinct spatio-temporal specificities. DNA sequences for these enhancers are conserved in a wide range of vertebrate species, corresponding to a majority of highly conserved non-coding sequences surrounding the Sox2 gene, corroborating the notion that the conservation of non-coding sequences mirrors their functional importance. Among the Sox2 enhancers, N-1 and N-2 are activated the earliest in embryogenesis and regulate Sox2 in posterior and anterior neural plates, respectively. These enhancers differ in their evolutionary history: the sequence and activity of enhancer N-2 is conserved in all vertebrate species, while enhancer N-1 is fully conserved only in amniotes. In teleost embryos, Sox19a/b play the major pan-neural role among the group B1 Sox paralogues, while strong Sox2 expression is limited to the anterior neural plate, reflecting the absence of posterior CNS-dedicated enhancers, including N-1. In Xenopus, neurally expressed SoxD is the orthologue of Sox19, but Sox3 appears to dominate other B1 paralogues. In amniotes, however, Sox19 has lost its group B1 Sox function and transforms into group G Sox15 (neofunctionalization), and Sox2 assumes the dominant position by gaining enhancer N-1 and other enhancers for posterior CNS. Thus, the gain and loss of specific enhancer elements during the evolutionary process reflects the change in functional assignment of particular paralogous genes, while overall regulatory functions attributed to the gene family are maintained.
Keywords: Sox2, enhancers, neural primordium, paralogous genes, evolution
Introduction to the problem
Classic theories of molecular evolution of animals started from comparison of amino acid sequences of proteins and protein- or RNA-encoding DNA sequences. However, expanding knowledge of non-coding sequences in the genome, widely upstream, downstream or intragenic regions of various genes, owing to the whole genome sequencing of various animal species, indicated new and important aspects of the genomic basis of organismal evolution.
For successful derivation of a living organism from a fertilized egg through the developmental process, genes included in a genome must be regulated in precise spatio-temporal order. This regulation is accomplished by association to individual genes of many regulatory sequences, e.g. enhancers, to satisfy different spectra of regulations at individual stages and sites of embryonic development. However, in contrast to the coding sequences of the genome predictable from possession of long ORFs (open reading frames) or matching with ESTs (expressed sequence tags), the non-coding regulatory sequences are definable only by biological functional assays.
Genomic comparison of sequences of various vertebrate species has indicated the presence of many blocks of highly conserved non-coding sequences scattered around and between genes, and at least some of them have been shown to have regulatory functions acting as enhancers.1)–5) This observation provided a new routine to investigate regulatory sequences: to find phylogenetically conserved sequences first, then examine these sequences by functional assays. However, not all regulatory sequences are conserved across a wide range of phyla and the range of conservation depends on individual enhancers.3),6),7)
For embryonic development, regulatory functions attributed to a group of genes, which usually correspond to a set of paralogous genes generated through multiple rounds of genomic duplication, is more important than the activity of individual genes. The genomic duplication took place twice for many vertebrates and three times for teleosts, creating paralogous genes. The possible consequence of the creation of functionally analogous paralogue genes has been formulated in a model by Force et al.8) based on then available information; namely inactivation, subfunctionalization and neo-functionalization (gaining of new function by a paralogous gene with the original function being maintained by other paralogous genes). However, the correlation between variation of paralogous gene function and phylogenetical conservation has not been investigated in a systematic fashion. In fact, employment of paralogous genes in a particular developmental process varies considerably among animal species. As will be discussed in this article, regulatory sequences evolve dynamically within the framework of conservation of the overall regulatory function attributed to a paralogous gene set.
Our group has investigated the regulation of Sox2 and its related (paralogous) genes belonging to group B1 Sox genes, for their involvement in the regulation of neural primordia at various developmental stages. The Sox2 gene is regulated by an unexpectedly large number of distinct enhancers that are widely scattered in a genomic region centered around the gene. Analysis of interspecies conservation of these enhancer sequences and regulatory functions provide a global view of how Sox2 enhancers evolved in coordination with variation in paralogue employment, fulfilling regulatory functions attributed to group B1 Sox genes. This article aims to synthesize two aspects of genomic evolution, variable employment of paralogous genes and the variable extent of phylogenetic conservation of regulatory sequences.
Involvement of many enhancers in Sox2 gene regulation
The embryonic neural primordia, starting from neural plates and continuing to the ventricular zone of the neural tube express Sox genes belonging to group B19)–13) (Okuda et al., unpublished results). Neural primordial states are sustained by the shared functions of group B1 Sox genes.14),15) Group B1 Sox genes encode transcription factors with identical DNA binding specificity and very similar transactivation potentials,16)–19) but differ in the expression pattern in both spatial and temporal aspects, although their expression overlaps extensively in the neural primordia. In higher vertebrates represented by amniotes, three Sox genes, Sox1, Sox2 and Sox3, comprise group B1, among which Sox2 dominates over the other two in the expression domains, as well as in regulatory functions. Expression of Sox2 is coordinated with gastrulation events involving neural induction.20) It covers the entire domain of neural primordium,9)–13),21) and continues to neural stem cells of later stages.22) For these reasons, expression of the transcription factor gene Sox2 is considered a “panneural” marker in higher vertebrates. Functionally, the impact of knockout of individual group B1 Sox genes is distinct between Sox2 and the other two Sox genes. Homozygous knockout mice for Sox1 and Sox3 are somehow viable with minor neuro-sensory23)–25) or neuro-endocrinal defects,26) respectively, whereas downregulation of Sox2 alone in a CNS cell population causes a serious neurogenetic disorder.27) Thus, in higher vertebrates, Sox2 expression and function prevail over other group B1 Sox genes.
Shown in Fig. 1 is the expression pattern of Sox2 at various developmental stages for chicken embryo. At stage 4 when the organizer (Hensen’s node) is formed, Sox2 is activated in the organizer-surrounding region of epiblastic upper cell layer, and as the organizer moves posteriorly with development, new Sox2-expressing domains are posteriorly added in a continuous fashion. The initial domain of Sox2 expression continues to express the gene, becomes anterior neural plate and forms the cephalic part of CNS, namely brain, while the posteriorly added Sox2 expression domain mainly forms the spinal cord. After stage 10 of chicken embryogenesis, placodal precursors also express Sox2. Sox2 expression thus marks neural and sensory primordia in embryos.
Fig. 1.
Expression of Sox2 in chicken embryo at various developmental stages marking neural and sensory primordia, as indicated by in situ hybridization. Anterior is toward the top. Photographs were taken at the same scale. The position of organizer (Hensen’s node) is indicated by an arrowhead. Head ectoderm (E), lens placode (L) and otic vesicle (O) are indicated by arrows. Adapted from Fig. 1A in Uchikawa et al. (2003). Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell 4, 509–519, with permission from Elsevier.
Although the expression of Sox2 is thus continuous and persistent in time and space, we felt it unlikely that neural and sensory Sox2 expression depends on simple regulatory mechanisms, considering the dynamic change in tissue environment for neuro-sensory primordia across the embryo axis and during developmental progression. Therefore, many regulatory sequences must be involved in Sox2 gene regulation.
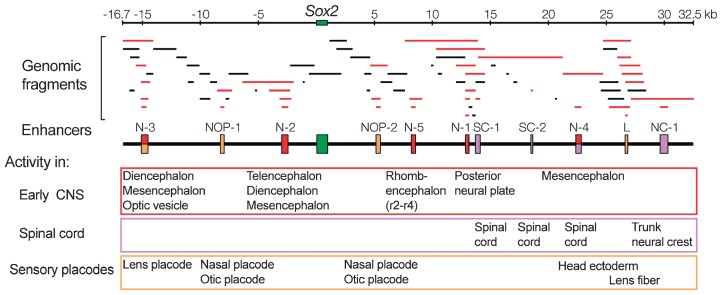
The experimental design we adopted took advantage of chicken embryo electroporation,4),28) a technique initiated by Tatsuo Muramatsu and others29) and refined by the group of Harukazu Nakamura.30) It was at a time before whole genome sequences were available. We cloned and determined the DNA sequence of the 50-kb chicken genomic region encompassing the Sox2 gene. We also constructed tk-EGFP vector to be used for chicken embryo electroporation.3) The advantage of this vector was that it would not express the fluorescent protein EGFP to any significant level unless an enhancer sequence was inserted. After insertion of an enhancer element, the vector activated EGFP expression sharply responding to the specificity and strength of the enhancer. Thus, we prepared a number of subgenomic fragments of the Sox2-surrounding region, inserted each in the tk-EGFP vector, and electroporated stage 4 embryo with these constructs on the epiblastic side or in stage 10 spinal cord, in order to examine if any enhancer activity is borne by the inserted DNA sequence. Once an enhancer activity was found to be associated with the inserted DNA, its subfragments were reexamined for enhancer activity using the same technique. By repeating this procedure, eleven different enhancers were identified in the 50 kb Sox2 genomic sequence (Fig. 2).3) Boundaries of the enhancers were roughly determined as the limits where smaller subfragments showed a decrease in enhancer activity. Five of the enhancers, N-1 to N-5, were active in the embryonic CNS (Fig. 3A).
Fig. 2.
Determination of Sox2 enhancers with activities in various and distinct domains of embryonic CNS and sensory placodes. Genomic fragments covering the 50 kb region of chicken Sox2 locus were individually tested for enhancer activity in electroporated chicken embryos. DNA fragments that demonstrated an enhancer activity are shown in red, and functionally determined enhancers are indicated by boxes on the map (middle). Adapted from Fig. 2 in Uchikawa et al. (2003). Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell 4, 509–519, with permission from Elsevier.
Fig. 3.
Conserved activity of enhancers N-1 to N-5 between chicken and mouse. (A) Domains of the embryonic CNS where chicken enhancers N-1 to N-5 show activity in electroporated embryo compared to Sox2 expression (in situ hybridization) of stage 11 embryo. Fluorescence images (green) representing enhancer activities are overlaid on darkened bright-field images to indicate respective embryonic domains. “r” indicates the rhombencephalic domain. The orange speckles in the enhancer N-5 specimen are due to DsRed fluorescence that was derived from co-electroporated control vector and not removed completely by optical filtration. (B) Alignment of chicken and mouse Sox2 enhancers with nucleotide sequence identity expressed as a percentage. (C) Activity of mouse enhancers at E9–9.5 in transgenic mouse embryos, compared to Sox2 in situ hybridization. Transgenes for Sox2 enhancers, except for enhancer N-1, were constructed by introducing each tetrameric Sox2 enhancer of mouse upstream of the hsp68 promoter-LacZ cassette,44) and primary transgenic embryos were examined. Tetrameric N-1 enhancer in a tk-LacZ vector was used to generate transgenic lines.
Enhancer N-1 was active in the embryonic domain surrounding the organizer, and the domain moved posteriorly together with the organizer. Enhancer N-2 was active in the wide anterior domain of the CNS and covered most of the future brain. Enhancer N-3 was active in the diencephalon, optic vesicle (future retina) and mesencephalon. Enhancer N-4 showed its activity posterior to the mesencephalon with a gap of activity in the rhombencephalic region, with the activity of enhancer N-5 in the rhombencephalic region filling this gap. Thus, by combining the activity of all these enhancers, original Sox2 expression patterns earlier than stage 12 were reconstructed (Fig. 3A), indicating that enhancers N-1 to N-5 fully account for the regulation of Sox2 in these early developmental stages.
Electroporation of spinal cord of later stage embryo allowed identification of SC-1 and SC-2, enhancers active in the spinal cord and NC-1 active in dorsal spinal cord-derived neural crest cells (Fig. 2).
Enhancer N-4 was also active in the cephalic ectoderm that derivates sensory placodes, and enhancer N-3 showed activity in the lens placode. In addition, three enhancers showed activity in the developing sensory systems: NOP-1 and NOP-2 in the nasal and otic placodes, and enhancer L in the lens fibers (Fig. 2).
Correspondence of functionally defined enhancers with phylogenetically conserved sequence blocks
Comparison of the Sox2 locus sequence of the chicken with those of human and mouse indicated the following interesting points (Fig. 4A).3) (1) Twenty-five highly conserved sequence blocks were clearly recognized (Three of the blocks are not conserved in the mouse sequence, but this represents a unique situation for the mouse). (2) Ten of the functionally defined enhancers matched very well to those of conserved sequence blocks. (Enhancer L is missing in the mammalian sequences, consistent with the lack of Sox2 expression in mammalian lens fibers, as will be discussed below) (Fig. 4A). (3) The correspondence between the functionally defined enhancers and conserved blocks of sequences were clear when chicken and mammalian sequences were compared, however the enhancer sequences were completely embedded in much longer stretches of non-functional sequence conservation when two mammalian genomes were compared (Fig. 4B). These observations indicated that the phylogenetic distance between birds and mammals is just appropriate for predicting functionally significant sequences, such as enhancers, as highly conserved non-coding sequences.
Fig. 4.
Correspondence of the functionally defined Sox2 enhancers with blocks of highly conserved sequences found by comparison of chicken and mammalian Sox2 locus sequences. (A) Conserved sequence blocks found in the Sox2 locus sequences of amniotes. Blocks of sequences that show > 60% identity over the stretch of 100 base pairs are indicated by boxes. The 25 sequence blocks No. 1 to No. 25 conserved between chicken and mammalian sequences are indicated on the top. Blocks No. 3, 21 and 25 marked by asterisks are not conserved in the mouse sequence. Sequences conserved between human and mouse genomic sequences but not strongly conserved in the chicken sequence are indicated in blue. (B) Dot matrices comparing DNA sequences of the three animal sequences encompassing enhancer N-5 (conserved sequence block No. 14). A dot indicates a 10 bp sequence with > 60% matching. Between the chicken and mammalian sequences, only the enhancer sequence is significantly conserved, however between human and mouse, the enhancer sequence is embedded in a broader region of possibly non-functional sequence conservation. Adapted from Fig. 6 in Uchikawa et al. (2003). Functional analysis of chicken Sox2 enhancers highlights an array of diversity regulatory elements that are conserved in mammals. Dev. Cell 4, 509–519, with permission from Elsevier.
The early neural enhancers N-1 to N-5 of Sox2 were conserved well in the mouse not only in the DNA sequence, but also in their regulatory functions (Fig. 3B, C). The mouse versions of enhancers N-1 to N-5 were regulated similar to chicken counterparts in electroporated chicken embryos, and in transgenic mouse embryos where the lacZ gene was expressed under the regulation of the mouse version of Sox2 enhancers (Fig. 3C).
Distinct regulation of enhancers N-2 and N-1 governing development of anterior and posterior neural plates
It is not that all enhancers N-1 to N-5 are activated simultaneously, but that enhancers N-1 and N-2 are activated first, followed by the addition of activities of N-3, N-4 and N-5 in this sequence.3) Thus, when the developmental stages are taken much earlier than those shown in Fig. 3, the active Sox2 enhancers are only N-1 and N-2. As indicated by two-color fluorescence representing the unover-lapping activities of enhancer N-1 (red) and N-2 (green) in electroporated chicken embryo (Fig. 5A), the future brain and future spinal cord portions of the CNS are already distinct at this stage. Regulation of these enhancers has been studied in great detail31) (Iwafuchi et al., unpublished). Enhancer N-1 is actually active in the primordial cell population called the “stem zone” located in the anterior-most domain of the primitive streak, which serves itself as the common precursor for posterior neural plate and paraxial mesoderm.32),33) As summarized in Fig. 5B, enhancer N-1 is activated by the combined action of Wnt and FGF signals and repressed when the stem zone cells are positioned in the mesodermal lineage, possibly by the action of T-box factors, through respective target regulatory sites present in the N-1 core sequence.31) Enhancer N-2 is not affected by any of these signals or factors, confirming independent regulation of anterior and posterior neural plates.
Fig. 5.
Distinct characteristics of enhancers N-2 and N-1 that regulate Sox2 in anterior and posterior neural plates, respectively. A. In early stage chicken embryos, N-2 and N-1 are the only active Sox2 enhancers, covering un-overlapping anterior (N-2, red fluorescence of mRFP1) and posterior domains (N-1, green fluorescence of EGFP) of the developing CNS. B. Regulatory modules of enhancer N-1 core sequence determined by Takemoto et al.31) C. When a tk-mCherry vector carrying chicken enhancer N-2 was injected into zebrafish embryo, it was regulated to have activity in the anterior neural plate, whereas analogous tk-Venus vector carrying dimeric chicken N-1 was not activated with regional restriction.
Difference in the range of cross-species conservation between enhancers N-2 and N-1
Given that the regulation of anterior and posterior neural plates are distinct, as exemplified by the independence of enhancers N-2 and N-1, it is interesting to learn how these regulations are conserved across phyla. Recent extension of whole genome sequencing to various vertebrate species opened up a new perspective concerning this issue.
The sequence of enhancer N-2, in particular the essential core region (Iwafuchi et al., unpublished), is strongly conserved through mammals, chicken, Xenopus to fish (zebrafish, tetraodon, medaka), as shown in Fig. 6A. This holds true for other neural enhancers N-3,34) N-4 and N-5 showing activities in the anterior neural plate. Indeed, when tk-mCherry vector carrying the chicken N-2 enhancer sequence was injected into fertilized zebrafish egg, this vector was activated in the anterior neural plate of zebra-fish embryo (Fig. 5C, top), demonstrating conservation of the overall regulatory system involved in the anterior neural plate Sox2 expression.
Fig. 6.
Differential range of phylogenetic conservation of Sox2 enhancers. A. Comparison of highly conserved sequence blocks (boxes) and enhancers functionally assessed in higher vertebrates (colored boxes) among six animal species, human, mouse, opossum, chicken, Xenopus and zebrafish. Enhancers showing activity in the brain-forming anterior CNS are connected by red lines, while those in the posterior CNS are connected by blue lines. Most of the enhancers showing activity in the posterior CNS are conserved in amniotes, however conservation is limited in Xenopus and absent in fish. B. Alignment of enhancer N-1 sequences between five animal species. Although the entire sequence is highly conserved among amniotes, sequence conservation is limited to the core-proximal sequences in Xenopus; even in the core sequence only one Lef1 binding sequence among three essential functional elements31) is conserved in the Xenopus sequence.
The situation for enhancer N-1 is very different. Although the entire enhancer N-1 sequence is strongly conserved within amniotes (Fig. 6B, for human, mouse, opossum and chicken), sequence conservation is only partial in Xenopus, and is not found in any fish species. In the core sequence of 56 bp,31) the genetic elements for the activation of this enhancer, two Lef1 binding sites and an FGF signal-responsive sequence, are perfectly conserved among amniote species, however only one Lef1 site is conserved in Xenopus (Fig. 6B).
When chicken enhancer N-1-bearing expression vector (tk-Venus) was injected into zebrafish eggs, the vector was expressed very weakly throughout the embryo possibly in response to FGF and Wnt signals, but without much posterior prominence (Fig. 5C, bottom). This observation indicates that zebrafish embryos are not furnished with a defined system for posterior activation of enhancer N-1, in addition to lacking the N-1 sequence in the Sox2 genomic region.
Minor position of Sox2 among group B1 Sox genes in fish
Although Sox2 plays the role of major “pan-neural” Sox gene in amniotes, it is a minor player and bears an anterior CNS-limited function among group B1 Sox genes in fish species. In contrast to higher vertebrates having three group B1 Sox genes, fish species, which have undergone an extra round of genome duplication, have six group B1 Sox genes, Sox1a, Sox1b, Sox2, Sox3, Sox19a and Sox19b. Of these, Sox2, Sox3, Sox19a and Sox19b are expressed during early embryonic stages, as shown in Fig. 7A. In zebrafish, expression of Sox2 is preceded by Sox3, Sox19a and Sox19b, and from the expression pattern, the “pan neural Sox” role is undertaken by Sox19a rather than Sox2 (Fig. 7B). Expression of Sox2 is limited to the anterior neural plate and its derivatives, which are accounted for by activity of the conserved enhancer N-2 (Fig. 7B).
Fig. 7.
Expression of group B1 Sox genes in early stage zebrafish embryos. A. RT-PCR analysis of transcripts of Sox1a, Sox1b, Sox2, Sox3, Sox19a and Sox19b in embryos at various stages. β-actin was used as control for the reaction. B. Expression pattern of the genes at various developmental stages indicated by in situ hybridization (blue). Hybridization with no tail probe (orange) was used to mark mesodermal precursors in gastrulating embryos. Developmental fates of early embryonic stages are illustrated for comparison: NNE, non-neural ectoderm; NE, neural ectoderm; D, dorsal; V, ventral; F, forebrain; M, midbrain; H, hindbrain; SC, spinal cord. Arrowheads indicate the site of shield. In 12-somite stage embryos, optic vesicle (ov), otic placode (otp), fore-midbrain boundary (fmb) and mid-hindbrain boundary (mhb) are indicated. Reprinted from Figs. 3, 4 and 6A in Okuda et al. (2006). Comparative genomic and expression analysis of group B1 sox genes in zebrafish indicates their diversification during vertebrate evolution. Dev. Dyn. 235, 811–825, with permission from Wiley-Blackwell.
Sox2 is not expressed to a significant level in the spinal cord of zebrafish embryos (Fig. 7B). This is likely a reflection of the lack of conservation of enhancer N-1 sequence and some other spinal cord-related enhancers of Sox2, SC-2 and NC-1, in the fish genome (Fig. 6A).
Evolution of group B1 Sox genes
The above observations may be retrospectively summarized as “enhancer N-1 and spinal cord related regulations of Sox2 are not conserved in lower vertebrates”, but considering the sequence of events underlying genomic evaluation, the actual situation may be that Sox2 gradually gained its dominance among group B1 Sox genes by acquiring additional regulatory sequences for expression of Sox2 in the posterior CNS. As the set of enhancers for vertebrate group B Sox prototype genes (Fig. 8, below) is not known, it is difficult to distinguish whether a particular enhancer is really acquired through the evolutionary process or has remained ‘unlost’. Nevertheless, enhancer acquiring should be considered an important process for evolution of genome function.
Fig. 8.
Evolutionary history of genesis of group B1 Sox paralogues as a consequence of multiple rounds of genomic duplication. “Vertebrate group B Sox prototypic composition” is hypothetical. Linkages of “Sox14 and Sox2” and “Sox21 and Sox1” are conserved in human and other primates, cow and chicken, but disrupted in mouse after extensive chromosomal rearrangements (Ensembl 50, July 2008; http://www.ensembl.org/index.html).35) Linkage of “Sox21 and Sox1” is found in a broader range of vertebrate species, e.g., dog, opossum, and medaka and other fish species, although this is disrupted in zebrafish. Linkages of “Sox14 and Sox2” is reported for platypus.45) Linkages of these genes in Xenopus genome have not been confirmed.
To gain insight into this issue, evolution of the all paralogous group B1 Sox genes needs to be considered. Group B1 Sox genes code for transcriptional activators that act on group B1 SOX-specific target elements, such as δ-crystallin DC5 enhancer or Nestin core Nes30 enhancer.17),18) Based on various criteria, group B1 SOX proteins have equivalent activities once expressed (Okuda et al., unpublished results). Group B1 Sox genes have their relatives Sox14 and Sox21, classified as group B2 Sox genes, the protein products of which bind to the same target sites as group B1 SOX proteins but repress gene transcription.11)
As illustrated in Fig. 8, a pair of prototypic B1 and B2 class Sox genes appears to have arisen by tandem duplication of a genomic locus, as (1) Sox14-Sox2 and Sox21-Sox1 pairs are conserved, including their local synteny in the chicken35) and human genomes,19) with the Sox21-Sox1 linkage observed in wider vertebrate species (see legend to Fig. 8), and (2) at least a pair of group B1 and group B2 Sox genes are recognizable in the genomes of lower animals, sponge,36) sea urchin,37) ascidian38) and amphioxus,39) although their linkages have not been confirmed.
Subsequently, two rounds of genomic duplication occurred, during which two group B2 Sox genes appear to have been lost, and the prototype of the vertebrate group [B1 + B2] Sox gene set must have been established (comprising the Sox14-Sox2 pair, Sox21-Sox1 pair plus Sox3 and Sox19). The Sox19-type genes are characterized by their possession of an intron at a conserved position in the coding sequence, as in Sox19a and Sox19b in fish, in contrast to other intron-less group B1/B2 Sox genes.
As mentioned above, higher vertebrates have only three genes of group B1 Sox, Sox1, Sox2 and Sox3. Thus, the evolutionary fate of Sox19 in higher vertebrates has been an intriguing problem. An analysis of conserved synteny among animal species clearly indicates that the Sox15 gene, found only in mammals and classified into the singleton G group, is derived from Sox19. Sox15 indeed has an intron sequence in the same position as fish Sox19a/b. The genomic segment of zebrafish chromosome 5 encompassing the Sox19a gene, the zebrafish chromosome 7 segment including Sox19b, and the human chromosome 17 segment including Sox15 are highly syntenic to one another. Despite this evolutionary history of its derivation, Sox15-encoded protein lacks the activity as a group B1 SOX protein, as assessed by activation of DC5 or Nes30 target sequences.19) Therefore, the derivation of Sox15 in higher vertebrates from Sox19 represents an example of “neofunctionalization”8) of a paralogous gene.
In Xenopus, the composition of group B1 Sox genes appears to reflect the vertebrate prototype, where SoxD reported by Yoshiki Sasai’s group40) corresponds to Xenopus Sox19. The SoxD sequence is slightly more divergent compared to other group B1 Sox genes, but its expression in the neural primordia is typical of group B1 Sox genes (Fig. 9B, below).
Fig. 9.
Evolutionary shift of the major “pan-neural” group B1 Sox genes and of posterior coverage of the CNS by Sox2, Sox3 and Sox19 paralogues, as indicated by expression patterns in embryos at comparable developmental stages. (A) Expression pattern in zebrafish embryo of 3 somite stage. Sox19a is the most prevalent, while strong Sox2 expression is mostly confined to the anterior CNS, as in other stages (Fig. 7). Reprinted from Fig. 5B in Okuda et al. (2006). Comparative genomic and expression analysis of group B1 sox genes in zebrafish indicates their diversification during vertebrate evolution. Dev. Dyn. 235, 811–825, with permission from Wiley-Blackwell. Reported Sox3 expression pattern in medaka embryo is analogous to zebrafish.46) (B) Expression pattern in Xenopus embryos at stage 13 and later, where Sox3 is the major Sox gene expressed. Expression patterns of Sox2 and Sox3 are reproduced from Fig. 1 of Schlosser and Ahrens (2004). Molecular anatomy of placode development in Xenopus laevis. Dev. Biol. 271, 439–466, and that of SoxD is reproduced from Fig. 2 of Mizuseki et al. (1998). SoxD: an essential mediator of induction of anterior neural tissues in Xenopus embryos. Neuron 21, 77–85, with permission from the authors and Elsevier. (C) Expression pattern in chicken embryo at stage 8, where Sox2 expression is dominating. In the chicken, Sox19-paralogous gene has not been identified. Arrows indicate the shift of major group B1 Sox among the species. A and P indicate anterior and posterior, respectively.
Evolutionary change of regulatory dominance and regional expression specificity among group B1 Sox genes
Comparison of the expression patterns of representative group B1 Sox genes in the embryos of zebrafish, Xenopus and chicken (Fig. 9) indicates a dramatic alteration of the dominating Sox gene among the B1 group during the evolutionary process. In zebrafish embryo, Sox19 is the most prevailing in expression level and regional coverage, and Sox2 is active only in the anterior CNS (Fig. 9A).19) In Xenopus embryo, however, expression of Sox3 appears to be dominating,41) while strong expression of Sox19-derived SoxD is confined to the anterior neural plate, although weak expression continues to the posterior neural plate.40) Very interestingly, in Xenopus, Sox2 expression extends to the posterior neural plates, in good correspondence with possession of enhancer N-1 like sequence (Fig. 6B). In embryos of chicken and other amniotes, the Sox19 activity is lost, and Sox2 expression prevails over the entire neural plate (Fig. 9C).
This variation of the major Sox genes on service is not limited to the CNS. While activation of group B1 Sox genes is essential for lens development,17),23),42) employment of group B1 Sox genes for lens development is highly variable among vertebrate species. In the mouse embryo only Sox1 is expressed in lens fibers,23),43) but chicken embryonic lens has expression of Sox1, Sox2 and Sox3 in the fibers.43)Xenopus embryo lens fibers express Sox2 and Sox3,41) and lens fibers of zebrafish embryo express Sox1a, Sox1b and Sox3.19) This variation of group B1 Sox expression must reflect the variation of enhancers associated with individual Sox genes. In fact, lens fiber enhancer L of Sox2 is conserved only between chicken and Xenopus (Fig. 6A). This kind of variation in the paralogous gene employment between different animal species must occur in a vast variety of organogenic processes.
The phylogenetic change of the expression pattern of Sox2 will largely be explained by the evolutionary gain of new enhancers or enhancer loss, as indicated in Fig. 6A. Other group B1 Sox genes must have gone through analogous gain and/ or loss of regulatory sequences to generate each species-specific expression pattern of individual genes. An important point however, is that regardless of a gain or loss of the expression domain of individual Sox genes in a given species, overall balance of the activity of the group B1 SOX proteins appears to be conserved.
It is very often assumed that signaling systems to generate an organ rudiment in an embryo are conserved across wide vertebrate species without experimental verification. However, this is not a correct concept, according to the arguments and experience elaborated in this article. In the regulation of posterior neural plate development, signaling systems for posterior extension of Sox19a expression in zebrafish and that of Sox2 (activating enhancer N-1) are likely different. Involvement of variable combinations of paralogous genes under distinct signaling systems in a defined organogenic process must always be considered when comparing results from different animal species.
Acknowledgements
This article summarizes our team effort over several years. We thank Drs. Gerhard Schlosser and Yoshiki Sasai for their provision of Xenopus data, and members of the Kondoh Laboratory for stimulating discussions. This study was supported by Grants-in-Aid for Scientific Research 18017019 and 18570197 to YK and 17107005 to HK. MI is a recipient of a fellowship from the Japan Society for the Promotion of Sciences for Japanese Junior Scientists.
Profile of corresponding author
Hisato Kondoh was born in 1949, and received his B.Sc. and Ph.D. from Kyoto University, Japan. He then served as Postdoctoral Fellow in the Department of Biochemistry, University of Wisconsin-Madison, USA. He returned to Kyoto University to take up the position of Assistant Professor in 1978 in the Department of Biophysics, when he started investigation of developmental gene regulation. He moved on to assume Professorships at Nagoya University Department of Molecular Biology (1988–1993) and Osaka University Institute for Molecular and Cellular Biology (1993–2003). He served as the Director of the Institute during 1999–2003. He is currently Professor at Osaka University Graduate School of Frontier Biosciences and also served as the Dean of the same institution from 2006–2008. To promote molecular studies in developmental biology and related areas, he organized several research projects and assumed the leadership, including those of Human Frontier Science Programs (1993–1996), Exploratory Research for Advanced Technology (1998–2004, Japan Science and Technology Agency), and Specially Promoted Research (COE) (2000–2005, MEXT Japan).

References
- 1).Hardison R. C. (2000) Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 16, 369–372 [DOI] [PubMed] [Google Scholar]
- 2).Levy S., Hannenhalli S., Workman C. (2001) Enrichment of regulatory signals in conserved non-coding genomic sequence. Bioinformatics 17, 871–877 [DOI] [PubMed] [Google Scholar]
- 3).Uchikawa M., Ishida Y., Takemoto T., Kamachi Y., Kondoh H. (2003) Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell 4, 509–519 [DOI] [PubMed] [Google Scholar]
- 4).Uchikawa M., Takemoto T., Kamachi Y., Kondoh H. (2004) Efficient identification of regulatory sequences in the chicken genome by a powerful combination of embryo electroporation and genome comparison. Mech. Dev. 121, 1145–1158 [DOI] [PubMed] [Google Scholar]
- 5).Woolfe A., Goodson M., Goode D. K., Snell P., McEwen G. K., Vavouri T., et al. (2005) Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3, e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Matsumata M., Uchikawa M., Kamachi Y., Kondoh H. (2005) Multiple N-cadherin enhancers identified by systematic functional screening indicate its Group B1 SOX-dependent regulation in neural and placodal development. Dev. Biol. 286, 601–617 [DOI] [PubMed] [Google Scholar]
- 7).Kondoh H., Uchikawa M. (2008) Dissection of chick genomic regulatory regions. Methods Cell Biol. 87, 314–337 [DOI] [PubMed] [Google Scholar]
- 8).Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., Postlethwait J. (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Uwanogho D., Rex M., Cartwright E. J., Pearl G., Healy C., Scotting P. J., et al. (1995) Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech. Dev. 49, 23–36 [DOI] [PubMed] [Google Scholar]
- 10).Rex M., Orme A., Uwanogho D., Tointon K., Wigmore P. M., Sharpe P. T., et al. (1997) Dynamic expression of chicken Sox2 and Sox3 genes in ectoderm induced to form neural tissue. Dev. Dyn. 209, 323–332 [DOI] [PubMed] [Google Scholar]
- 11).Uchikawa M., Kamachi Y., Kondoh H. (1999) Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech. Dev. 84, 103–120 [DOI] [PubMed] [Google Scholar]
- 12).Collignon J., Sockanathan S., Hacker A., Cohen-Tannoudji M., Norris D., Rastan S., et al. (1996) A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development 122, 509–520 [DOI] [PubMed] [Google Scholar]
- 13).Wood H. B., Episkopou V. (1999) Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 86, 197–201 [DOI] [PubMed] [Google Scholar]
- 14).Graham V., Khudyakov J., Ellis P., Pevny L. (2003) SOX2 functions to maintain neural progenitor identity. Neuron 39, 749–765 [DOI] [PubMed] [Google Scholar]
- 15).Bylund M., Andersson E., Novitch B. G., Muhr J. (2003) Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nat. Neurosci. 6, 1162–1168 [DOI] [PubMed] [Google Scholar]
- 16).Kamachi Y., Sockanathan S., Liu Q., Breitman M., Lovell-Badge R., Kondoh H. (1995) Involvement of SOX proteins in lens-specific activation of crystallin genes. EMBO J. 14, 3510–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Kamachi Y., Uchikawa M., Tanouchi A., Sekido R., Kondoh H. (2001) Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 15, 1272–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Tanaka S., Kamachi Y., Tanouchi A., Hamada H., Jing N., Kondoh H. (2004) Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol. Cell. Biol. 24, 8834–8846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Okuda Y., Yoda H., Uchikawa M., Furutani-Seiki M., Takeda H., Kondoh H., et al. (2006) Comparative genomic and expression analysis of group B1 sox genes in zebrafish indicates their diversification during vertebrate evolution. Dev. Dyn. 235, 811–825 [DOI] [PubMed] [Google Scholar]
- 20).Darnell D. K., Stark M. R., Schoenwolf G. C. (1999) Timing and cell interactions underlying neural induction in the chick embryo. Development 126, 2505–2514 [DOI] [PubMed] [Google Scholar]
- 21).Streit A., Sockanathan S., Perez L., Rex M., Scotting P. J., Sharpe P. T., et al. (1997) Preventing the loss of competence for neural induction: HGF/SF, L5 and Sox-2. Development 124, 1191–1202 [DOI] [PubMed] [Google Scholar]
- 22).Cai J., Wu Y., Mirua T., Pierce J. L., Lucero M. T., Albertine K. H., et al. (2002) Properties of a fetal multipotent neural stem cell (NEP cell). Dev. Biol. 251, 221–240 [DOI] [PubMed] [Google Scholar]
- 23).Nishiguchi S., Wood H., Kondoh H., Lovell-Badge R., Episkopou V. (1998) Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev. 12, 776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Malas S., Postlethwaite M., Ekonomou A., Whalley B., Nishiguchi S., Wood H., et al. (2003) Sox1-deficient mice suffer from epilepsy associated with abnormal ventral forebrain development and olfactory cortex hyperexcitability. Neuroscience 119, 421–432 [DOI] [PubMed] [Google Scholar]
- 25).Ekonomou A., Kazanis I., Malas S., Wood H., Alifragis P., Denaxa M., et al. (2005) Neuronal migration and ventral subtype identity in the telencephalon depend on SOX1. PLoS Biol. 3, e186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Rizzoti K., Brunelli S., Carmignac D., Thomas P. Q., Robinson I. C., Lovell-Badge R. (2004) SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat. Genet 36, 247–255 [DOI] [PubMed] [Google Scholar]
- 27).Ferri A. L., Cavallaro M., Braida D., Di Cristofano A., Canta A., Vezzani A., et al. (2004) Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 131, 3805–3819 [DOI] [PubMed] [Google Scholar]
- 28).Uchikawa M. (2008) Enhancer analysis by chicken embryo electroporation with aid of genome comparison. Dev. Growth Differ. 50, 467–474 [DOI] [PubMed] [Google Scholar]
- 29).Muramatsu T., Mizutani Y., Ohmori Y., Okumura J. (1997) Comparison of three nonviral transfection methods for foreign gene expression in early chicken embryos in ovo. Biochem. Biophys. Res. Commun. 230, 376–380 [DOI] [PubMed] [Google Scholar]
- 30).Nakamura H., Watanabe Y., Funahashi J. (2000) Misexpression of genes in brain vesicles by in ovo electroporation. Dev. Growth Differ. 42, 199–201 [DOI] [PubMed] [Google Scholar]
- 31).Takemoto T., Uchikawa M., Kamachi Y., Kondoh H. (2006) Convergence of Wnt and FGF signals in the genesis of posterior neural plate through activation of the Sox2 enhancer N-1. Development 133, 297–306 [DOI] [PubMed] [Google Scholar]
- 32).Delfino-Machin M., Lunn J. S., Breitkreuz D. N., Akai J., Storey K. G. (2005) Specification and maintenance of the spinal cord stem zone. Development 132, 4273–4283 [DOI] [PubMed] [Google Scholar]
- 33).Cambray N., Wilson V. (2002) Axial progenitors with extensive potency are localised to the mouse chordoneural hinge. Development 129, 4855–4866 [DOI] [PubMed] [Google Scholar]
- 34).Inoue M., Kamachi Y., Matsunami H., Imada K., Uchikawa M., Kondoh H. (2007) PAX6 and SOX2-dependent regulation of the Sox2 enhancer N-3 involved in embryonic visual system development. Genes Cells 12, 1049–1061 [DOI] [PubMed] [Google Scholar]
- 35).Kuroiwa A., Uchikawa M., Kamachi Y., Kondoh H., Nishida-Umehara C., Masabanda J., et al. (2002) Chromosome assignment of eight SOX family genes in chicken. Cytogenet. Genome Res. 98, 189–193 [DOI] [PubMed] [Google Scholar]
- 36).Larroux C., Luke G. N., Koopman P., Rokhsar D. S., Shimeld S. M., Degnan B. M. (2008) Genesis and expansion of metazoan transcription factor gene classes. Mol. Biol. Evol. 25, 980–996 [DOI] [PubMed] [Google Scholar]
- 37).Howard-Ashby M., Materna S. C., Brown C. T., Chen L., Cameron R. A., Davidson E. H. (2006) Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus. Dev. Biol. 300, 90–107 [DOI] [PubMed] [Google Scholar]
- 38).Satou Y., Satoh N. (2005) Cataloging transcription factor and major signaling molecule genes for functional genomic studies in Ciona intestinalis. Dev. Genes Evol. 215, 580–596 [DOI] [PubMed] [Google Scholar]
- 39).Meulemans D., Bronner-Fraser M. (2007) The amphioxus SoxB family: implications for the evolution of vertebrate placodes. Int. J. Biol. Sci. 3, 356–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Mizuseki K., Kishi M., Shiota K., Nakanishi S., Sasai Y. (1998) SoxD: an essential mediator of induction of anterior neural tissues in Xenopus embryos. Neuron 21, 77–85 [DOI] [PubMed] [Google Scholar]
- 41).Schlosser G., Ahrens K. (2004) Molecular anatomy of placode development in Xenopus laevis. Dev. Biol. 271, 439–466 [DOI] [PubMed] [Google Scholar]
- 42).Kondoh H., Uchikawa M., Kamachi Y. (2004) Interplay of Pax6 and SOX2 in lens development as a paradigm of genetic switch mechanisms for cell differentiation. Int. J. Dev. Biol. 48, 819–827 [DOI] [PubMed] [Google Scholar]
- 43).Kamachi Y., Uchikawa M., Collignon J., Lovell-Badge R., Kondoh H. (1998) Involvement of Sox1, 2 and 3 in the early and subsequent molecular events of lens induction. Development 125, 2521–2532 [DOI] [PubMed] [Google Scholar]
- 44).Sasaki H., Hogan B. L. (1996) Enhancer analysis of the mouse HNF-3 beta gene: regulatory elements for node/notochord and floor plate are independent and consist of multiple sub-elements. Genes Cells 1, 59–72 [DOI] [PubMed] [Google Scholar]
- 45).Kirby P. J., Waters P. D., Delbridge M., Svartman M., Stewart A. N., Nagai K., et al. (2002) Cloning and mapping of platypus SOX2 and SOX14: insights into SOX group B evolution. Cytogenet Genome Res. 98, 96–100 [DOI] [PubMed] [Google Scholar]
- 46).Koster R. W., Kuhnlein R. P., Wittbrodt J. (2000) Ectopic Sox3 activity elicits sensory placode formation. Mech. Dev. 95, 175–187 [DOI] [PubMed] [Google Scholar]