Abstract
Mammalian immune response can be divided into innate and acquired immunity. Furthermore, much evidence has demonstrated that activation of innate immunity is a prerequisite to induction of acquired immunity. This paradigm shift has changed our thinking on the pathogenesis and treatment of infections, immune diseases, allergy, and cancers.
Keywords: Toll-like receptor, innate immunity, pathogen, interferon, signaling, cytokine
1. History of immunology
Immunology originated from smallpox vaccination by Edward Jenner. Based on the common observation that milkmaids who had experienced cowpox protected them from smallpox, he tested the possibility of using the coxpox vaccine as an immunization for smallpox in humans. In 1796 Jenner inoculated James Phipps, a young boy with cowpox pus in the arm. James became mildly ill with cowpox but was well again soon. Later the boy was challenged with smallpox but showed no sign of infection. This finding showed that infection with cowpox would give immunity to smallpox. “Vaccination”, the word Jenner invented for his treatment (from Latin, vacca, a cow) was adopted by Pasteur for immunization against any disease. In 1980, the World Health Assembly officially declared smallpox an eradicated disease.
In the end of 19th Century, Shibasaburo Kitasato studied under Dr. Robert Koch in Germany, and developed a serum therapy for tenanus together with Emil von Behring. Behring and Kitasato demonstrated the value of antitoxin in preventing disease by producing a passive immunity to tetanus in an animal that received graded injections of blood serum form another animal infected with the disease. This was the discovery of antibody. Subsequently, Paul Ehrlich proposed his famous side-chain theory of immunity. He believed that toxic substances produced by bacteria bind to the cells through side chain molecular structures expressed on the cell surface and thereby cause disease, and the body produces in the blood abundant side chains (antibodies) to react with the specific bacterial toxins, and thus prevent the toxins from reacting with the side chains of cells. This theory exactly predicted the mechanism of antibody production from B cells in response to antigens.
Around the same period, while studying on starfish larvae, Elie Metchnikoff observed that certain white blood cells could engulf and destroy harmful bodies such as bacteria, and he called these cells phagocytes and name the process phagocytosis. This work formed the basis of innate immunity. Innate immunity constitutes a basic and phylogenetically ancient defense system and the first line of defense against infection. The characteristics of the innate immune response include non-specific killing of microbes by phagocytosis, and luekocyote recruitment to sites of infection and initiation of inflammation. Mechnikov received the Nobel Prize for Medicine together with Ehrlich from discovery of the two different aspects of immune responses, innate immunity and humoral (acquired) immunity, respectively. However, humoral immunologists including Behring claimed that serum but not cells destroyed invading organisms, and innate immunity could not explain specificity of immunity. While the humoral immunity has remarkable progresses later until recently, innate immunity had been discarded for a long time as the ancient and non-specific immunity which works in lower animal kingdom (Table 1).
Table 1.
History of immunology research
| Innate Immunity | Adaptive Immunity |
|---|---|
| Discovery of phagocytosis by Metchnikoff (1845–1916) founder of innate immunity(1900) | Serum Therapy by Kitasato and Behring (1890) |
| Side-chain theory of antibody by Ehrlich | |
| Clonal selection by Burnet (1957) | |
| After 1960’s | |
| Primary structure of antibodies | |
| Discovery of IgE and mechanism of allergy | |
| Discovery of T and B cells | |
| Discovery of histocompatibility antigens | |
| Development of monoclonal antibodies | |
| Mechanism of antibody diversity | |
| After 1996 | Cloning of T cell receptor |
| Role of Drosophila Toll in antifungal response | Cloning of cytokines involved in antibody production |
| Discovery of mammalian Toll-like receptors (TLRs) and identification of their function | Th1 and Th2 responses |
| Identification of molecules involved in antibody diversity |
2. Principle of acquired immunity
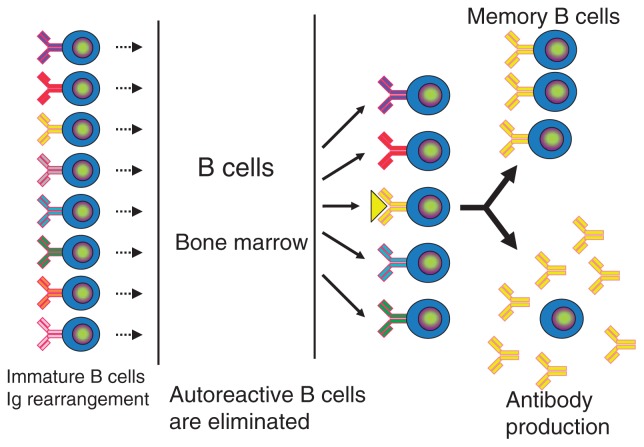
Acquired immunity is largely categorized in humoral and cellular immunity. Humoral immunity is involved in eradication of microbes present in the blood or fluid by generating antibodies, which are produced by B cells. On the other hand, cellular immunity is responsible for eradication of cancer cells and microbes hidden inside the cells, and is mediated by killer T cells. T and B cells express unique T cell receptors (TCRs) and B cell receptors (BCRs), and recognize a vast amount of different antigens. TCRs and BCRs are generated by DNA recombination during differentiation of T and B cells. A huge repartoire of TCRs and BCRs are generated, and initially includes receptors which react with the components of the host. Lymphocytes harboring self-reacting receptors are then excluded during differentiation. When the pathogen invades the body, T and B cells with the corresponding receptors are activated, and killer T cells development as well as antibody production is induced. At the same time, memory T and B cells are generated, allowing the host to respond more rapidly when the same pathogen invades the body again (Fig. 1).
Fig. 1.
Principal of immune response.
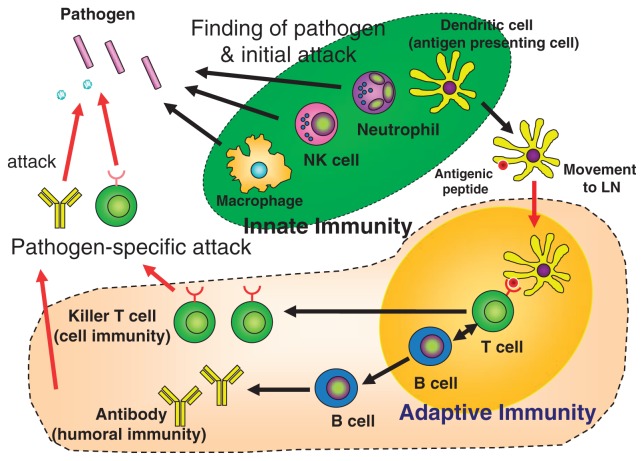
On the other hand, innate immunity is mediated by leukocytes, macrophages, and dendritic cells, collectively called phagocytes because these cells engulf and kill microbes. In the case of dendritic cells, they have an additional role of presentation of antigenic peptides derived from microbes to T cells (Fig. 2).
Fig. 2.
Innate immunity and adaptive immunity. NK: natural killer; LN: lymphnode.
Until recently, far less attention had been paid to the study of innate immunity due to lack of specificity and diversity in microbe recognition. However, recent studies have shown that the innate immune system possesses a greater degree of specificity than previously believed, and is highly developed in its ability to discriminate self versus foreign pathogens. This discrimination relies to a greater extent on a family of evolutionarily conserved receptors, named the Toll-like receptors (TLRs).1) Furthermore, accumulating evidence indicates that activation of innate immunity is a prerequisite to the induction of acquired immunity. This paradigm shift is changing our thinking on the pathogenesis and treatment of infectious diseases, immune diseases, allergic diseases, and cancers.
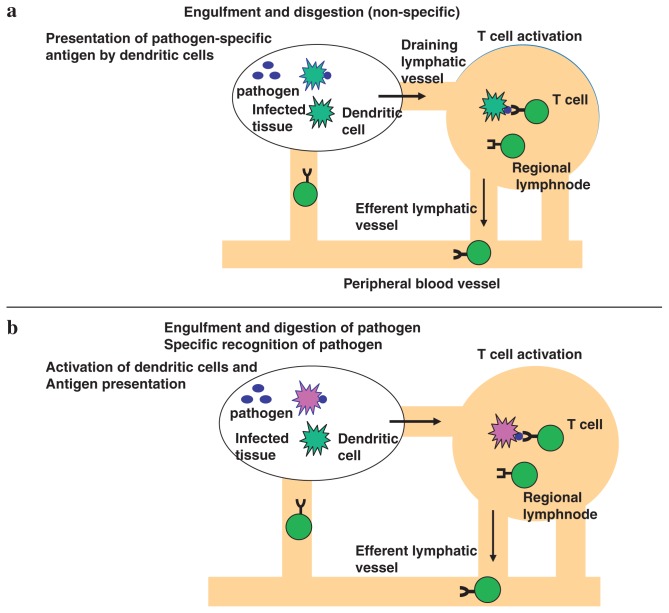
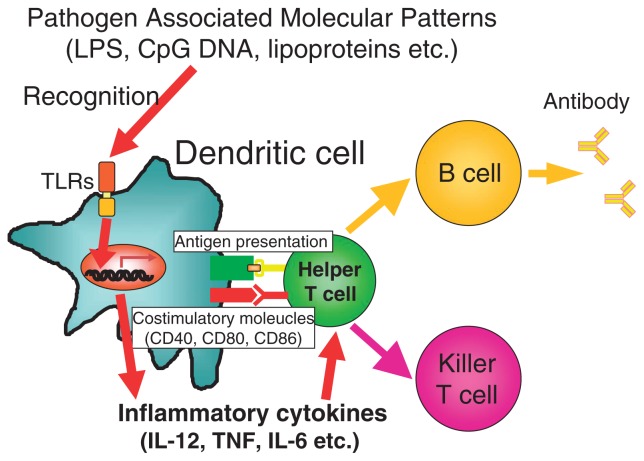
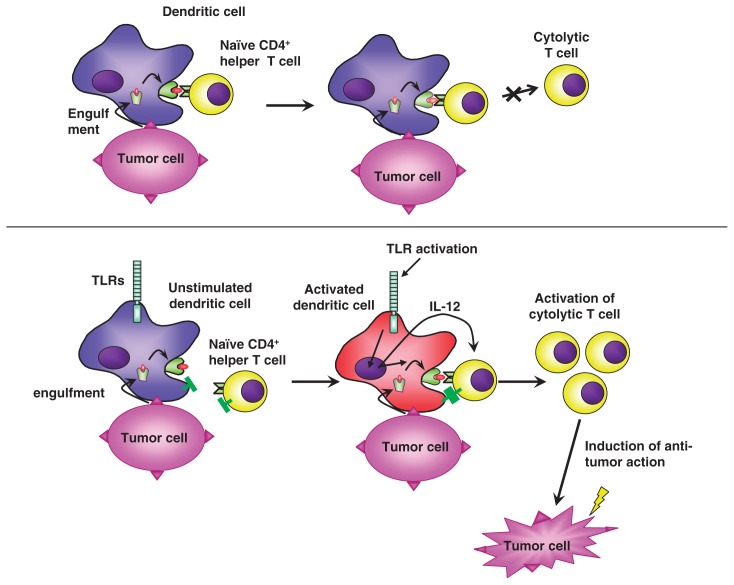
According to the old theory of the immune response, dendritic cells engulf invading pathogens, and digest them into small peptides, and express the peptide antigens on the cell surface. Then dendritic cells migrate from the infected tissue to the regional lymphnode, where dendritic cells present the antigen to naïve T cells with the corresponding receptor. In this model, pathogen recognition takes place at the stage of T cell activation in the lymphnode (Fig. 3a). On the other hand, in the new theory, in addition to phagocytosis and digestion of pathogens, dendritic cells must be activated for T cell activation. T cells are activated only when the antigen is presented by activated dendritic cells (Fig. 3b). In normal tissue, cells die continuously, and dead cells are treated by dendritic cells. Self antigens are presented to T cells by dendritic cells, however, in such process T cells cannot be activated because dendritic cells are not activated. Dendritic cells present antigens to naïve T cells. However, just antigen presentation does not activate T cells, rather resulting in unresponsiveness or anergy. For efficient T cell activation, dendritic cells must be activated, which accompanies induction of costimulatory molecules and cytokine production (Fig. 4). Such dendritic activation is mediated by Toll-like receptors. This principle is also applicable to tumor rejection by host (Fig. 5). Although dead tumor cells are engulfed by dendritic cells, and the tumor antigen is presented to T cells by the dendritic cells, anti-tumor immunity cannot be generated because tumor cells of self origin do not activate dendritic cells. However, addition of microbial extracts containing TLR ligands enable the tumor antigen present to T cells by activated dendritic cells, which results in induction of effective anti-tumor immunity. Indeed, several reports show that anti-tumor action by inoculation of several bacterial cell wall components such as BCG-CWS is due to TLR-dependent activation of dendritic cells.
Fig. 3.
Pathogen recognition and immune responses.
Fig. 4.
Interaction between dendritic cells and T cells.
Fig. 5.
Induction of tumor immunity by TLRs stimulation.
3. My road to TLR research
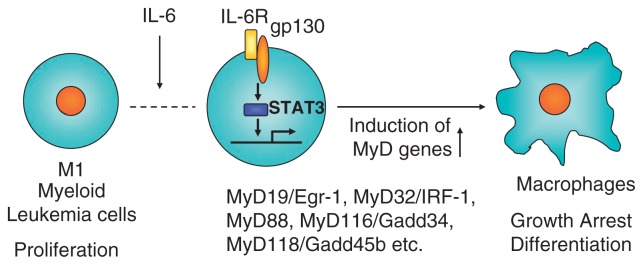
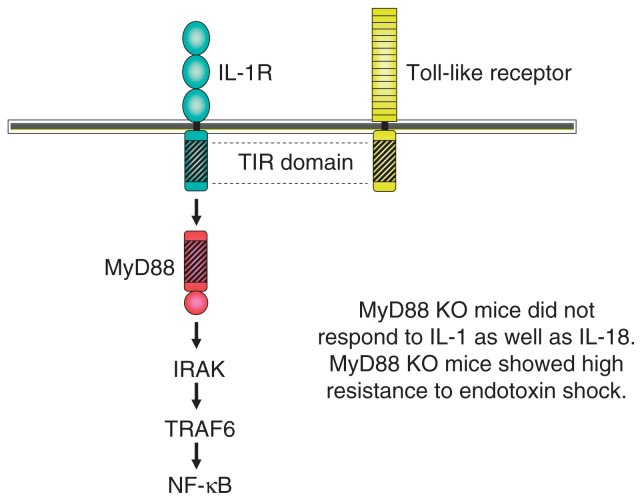
Before I became involved in TLR research, I had studied IL-6 gene regulation and IL-6 signaling pathway in the laboratory of Professor Kishimoto, who discovered interleukin 6 (IL-6). During my stay in Kishimoto’s lab, I cloned NF-IL6, also called C/EBPβ as a critical transcription factor involved in LPS-dependent IL-6 induction, and STAT3 as a critical signal transducer of IL-6 response as well.2),3) In 1996 I left professor Kishimoto’s lab and became a professor of Biochemistry at Hyogo College of Medicine. I started a new project mainly by knockout mice generation. One project was the molecular mechanism of macrophage differentiation. A myeloid leukemic cell line, M1 cells differentiate into macrophages in response to IL-6 (Fig. 6). I previously showed that loss of STAT3 action completely abolished the macrophage differentiation in response to IL-6.4) I thought that the genes regulated by STAT3 are responsible for macrophage differentiation. Several early immediate genes induced in response to IL-6 were already identified in M1 cells, but the function of them was not known at that time. I started to knockout several genes including the MyD88 gene. During the process of MyD88 KO mice generation, a paper came out, showing that MyD88 might be involved in IL-1 receptor signaling.5) Upon stimulation, IL-1-IL-1 receptor complex recruits IRAKs via MyD88, and finally induces NF-κB activation. As expected, MyD88 knockout mice did not respond to IL-1 or IL-1 related cytokine, IL-18.6) In addition, we soon realized that MyD88 was unresponsive to LPS as well.7) MyD88 KO mice were completely resistant to high doses of LPS. This finding indicated that the LPS signaling receptor uses MyD88 as adaptor. In 1996, Jules Hoffmann’s group demonstrated that Toll is essential for anti-fungal immunity in the fly.8) In the next year human Toll was reported by Medzhitov and Janeway.9) Additional four TLR members were published by DNAX group in United States.10) The TLR family harbors an extracellular leucine-rich repeat (LRR) domain as well as a cytoplasmic domain that is homologous to that of the IL-1R family (Fig. 7). Therefore, I imagined that a member of Toll-like receptors is a candidate of LPS receptor. We searched in database and found several unpublished TLR members and started to knockout all of TLR family members. Although the first publication of TLR4 as LPS receptor was done by Bruce Beutler through positional cloning with LPS unresponsive natural mutant mice,11) we confirmed the essential role of TLR4 in LPS response using TLR4 KO mice and synthetic lipid A.12) In addition, we had advantage of identification of ligands of other TLR members because of a full stock of TLR knockout mice.
Fig. 6.
Myeloid differentiation (MyD) genes in hematopoietic differentiation.
Fig. 7.
Toll-like receptors comprise a superfamily with IL-1Rs.
4. Identification of TLR ligands
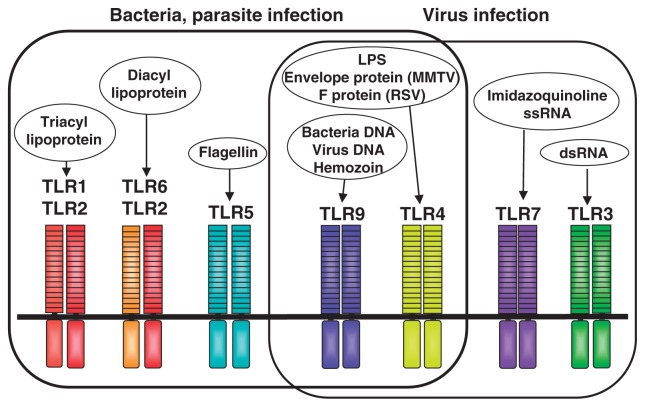
We soon knew that MyD88-deficient cells are unresponsive to whole mycobacterium tuberculosis lysates, indicating that many immunostimulatory components from pathogens may be recognized by TLRs.13) I searched in publication the bacterial components or synthetic compounds that stimulated mammalian immune cells, and compared the response to each molecule between wild-type and MyD88-deficient cells. This screening identified the candidates for TLR ligands. In searching the ligands, we tried to use the pure or synthetic compounds because we wanted to exclude the contamination. We demonstrated that TLR2 and TLR4 recognize different ligands each other, TLR2 recognizes peptidoglycan and lipoprotein whereas TLR4 recognizes LPS.14),15) By comparing the response between TLR2, TLR1 and TLR6 kockout mice, we demonstrated that together with TLR2, TLR1 and TLR6 recognize the subtle difference in the lipid portion of lipoproteins.16),17) TLR1 recognizes triacylated lipoprotein by forming a heterodimer with TLR2 whereas TLR6 recognizes diacylated lipoportein by forming a heterodimer with TLR4.
Besides the cell wall components, several other components including bacterial DNA and flagellin are shown to have an immunostimulatory activity. The finding that CpG DNA derived from bacteria has immunostimulatory activity was first reported by Dr. Tokunaga’s group in the early 80’s.18) They were studying the anti-tumor action of bacillus Calmette-Guerin (BCG) and found that DNA purified from BCG is responsible for the immunostimulatory activity. CpG motif is 20 times more common in bacteria than in mammals. CpG pairs in mammals are methylated, and methylated CpG motif does not activate mammalian immune cells. So this structural difference is discriminated by the host and, therefore bacterial DNA can provoke strong immune responses. Synthetic oligonucleotides containing this motif also possess similar immunostimulatory activity. However, the molecular mechanisms by which CpG DNA triggers immune stimulatory responses were at that time unknown, although clinical trials for treatments of infectious disease, cancers, and allergy were underway due to its strong immunostimulatory activities but low toxicity. In 2000 we generated the mice lacking a novel TLR named TLR9, and found that TLR9 deficient mice were unresponsive to CpG DNA.19) This result clearly showed that TLR9 discriminates between pathogen-derived DNA (CpG DNA) and mammalian DNA, and thereby detects bacterial invasion. This finding also established the link between the two arms of innate and acquired immunities. Furthermore, the identification of CpG receptor further accelerated the clinical application of immunostimulatory DNA. TLR9 is shown to recognize DNA viruses, including Herpes simplex virus type 1 (HSV-1), HSV-2, and murine cytomegalovirus (MCMV), whose genomes that are abundant in CpG-DNA motifs. Signaling via TLR9 initiates at an endosomal compartment. Different from TLR4 and TLR2, TLR9 is not expressed on the cell surface but located in the endosome. Activation of immune cells via CpG DNA requires cellular uptake by DNA sequence-independentt endocytosis, and endosomal maturation.
In 2001 Alan Aderem in collaboration with our group showed that TLR5 recognizes flagellin, a major constituent of flagella.20) Utilizing flagella, bacteria can move in aqueous environment. TLR5 is expressed on the basolateral, but not apical surfaces of intestinal epithelia. Therefore, flagellin activates the intestinal epithelia only when bacteria invade across intestinal epithelia. Dendritic cells present in the lamina propria express TLR5 abundantly, respond to flagellin, and produce inflammatory cytokines such as IL-6 and IL-12.21) The lamina propria dendritic cells have been shown to play an important role in induction of acquired immunity such as IgA class-switching of B cells and Th17 cell development as well.22)
Viral replication within infected cells results in the generation of dsRNA which can stimulate immune cells. In the same year Flavell and Medzhitov showed that TLR3 recognizes double-stranded RNA.23)
The TLR7 and TLR8 genes are located on X chromosome and shows high homology with each other. In 2002 we demonstrated that TLR7 recognizes imidazoquinoline derivative imquimod, an antiviral chemical which is now used for treatment of genital warts caused by papilloma virus.24) We also showed that another imidazoquinoline derivative R848, also called resquimod is recognized by TLR7. In human, both TLR7 and TLR8 recognize R848. Anticancer synthetic chemicals, including loxoribine (7-allyl-8-oxoguanosine) and bropirimine (2-amin-5-bromo-6-phenyl-4 (3)-pyrimidinone) and certain guanosine analogs have been also identified as TLR7 ligands.25) Later, it has been shown that as natural ligand TLR7 recognizes single-stranded RNAs (ssRNA) or RNA viruses including influenza virus and vascular stomatitis virus.26),27)
The Toll-like receptor family is now composed of twelve members, but such relatively small members may be enough to detect all pathogens from bacteria to viruses (Fig. 8). One reason is that TLR targets the components common to pathogens as well as essential for life of pathogens, therefore difficult for the pathogen to change the structure. A second reason is that a single TLR recognizes several pathogen components that are structurally unrelated each other. For example, besides LPS TLR4 recognizes Taxol, and several viral envelop glycoproteins. TLR9, a CpG DNA receptor also recognizes hemozoin, a malarial pigment.
Fig. 8.
TLR family and its ligands.
Recent findings have shown the existence of endogenous TLR ligands28) (Table 2). Heat shock proteins (HSPs) are intracellular molecular chaperones of naïve, aberrantly folded, or mutated proteins and their expression and release are increased when cells are exposed to stress. In addition to their role in protein folding, assembly and transport, HSPs released from cells by necrosis, such as HSP60 and HSP70 induce inflammation through their interactions with TLRs. HMGB1, a nuclear protein is also released from dying cells during late stage apoptosis. Upon release into the tissue microenvironment HMGB1 can signal tissue injury and initiate an inflammatory response through binding TLR4.
Table 2.
Endogenous ligands of TLRs
| TLR | Ligands |
|---|---|
| TLR2 | HSP60, HSP70, Gp96, Minimally modified LDL |
| HMGB1 | |
| TLR3 | mRNA |
| TLR4 | HSP60, HSP70, Gp96 |
| Fibronectin, Heparan sulphate | |
| Oligosaccharides of hyaluronic acid | |
| Surfactant protein-A | |
| HMGB1 | |
| TLR7 | mRNA |
| TLR9 | DNA, Chromatin-IgG complexes |
HSP: heat shock protein; LDL: low density lipoprotein; HMGB1: high mobility group B1; Gp: 96 kDa glycoprotein of the endoplasmic reticulum
During inflammation associated with traumatic injury, proteolytic enzymes release biological active soluble fragments from extracellular matrix components, some of which are reported to activate innate immune responses via TLRs. They include heparin sulfate, hyaluronan-derived oligosaccharides, the extra domain A of fibronectin, and bioglycan.
5. MyD88-dependent and -independent pathways in TLR signaling
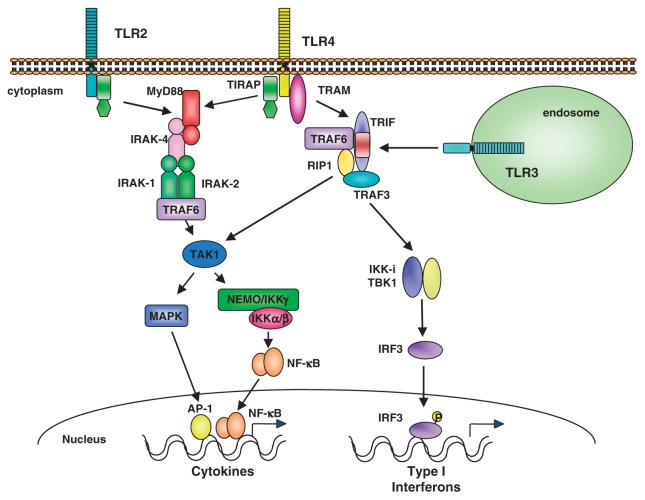
MyD88-dependent pathway activates intracellular signaling molecules such as IL-1RI-associated protein kinases (IRAKs), the TGF-β-activated kinase (TAK1), TAK1-binding protein 1 (TAB1) and 2 (TAB2), and the tumor necrosis factor receptor-associated factor 6 (TRAF6) (Fig. 9). Triggering of the IL-1R or TLR induces recruitment of MyD88 to the receptor complex, which in turn promotes association with the IL-1R-associated kinases IRAK4 and IRAK1. During the formation of this complex, activated IRAK4 phosphorylates IRAK1 and IRAK2, which then induces the interaction of TRAF6 with the complex. IRAK1 and IRAK2 are sequentially activated; IRAK1 is involved in activation of the downstream signaling during the initial one hour, followed by its rapid degradation whereas IRAK2 activity is sustained and is responsible for cellular activation at the later phase.29) The IRAKs/TRAF6 complex next interacts with another complex consisting of TAK1, TAB1 and TAB2. This interaction induces phosphorylation and activation of TAK1, leading to the activation of IKKs. Inactive IKK sequesters NF-κB in the cytoplasm, but phosphorylation by IKKs leads to degradation of Iκb, and subsequent release of NF-κB. Activation of TAK1 also results in the activation of MAP kinases and JNK.
Fig. 9.
MyD88-dependent and TRIF-dependent pathways in TLR signaling.
MyD88 KO mice fail to respond to LPS and other microbial cell wall components such as PGN and lipoproteins, or to IL-1 or IL-18 in terms of cytokine production. However there is a difference in the signaling pathways triggered by LPS and other microbial components. Mycoplasmal lipopeptide activation of NF-κB and MAP kinases, which is mediated by TLR2, is completely abolished in TLR2-deficient or MyD88-deficient macrophages. However, LPS activation of MAP kinases and NF-κB remains intact in MyD88-deficient macrophages, although the activation of these molecules was delayed when compared with wild-type mice.7) This indicates that the LPS response is mediated by both MyD88-dependent and -independent pathways, each of which leads to the activation of MAP kinases and NF-κB. Although the MyD88-dependent pathway is essential for the inflammatory response mediated by LPS, MyD88-deficient macrophages have the ability to induce interferon-κ and interferon-inducible genes in response to LPS. We also showed that the MyD88-independent pathway activates the transcription factor IRF-3.30) Following LPS treatment, IRF-3 undergoes phosphorylation and translocates from the cytoplasm to the nucleus, resulting in the transcriptional activation of type I interferons.31) Another adaptor molecule named TIR domain containing adaptor inducing interferon-β (TRIF) or TIR-containing adapter molecule-1 (TICAM-1) has been identified.32),33) TRIF deficient mice showed its essential role in the MyD88-independent pathways of TLR3 and TLR4 signaling.34) Interestingly, TRIF deficient mice also abolished the response to LPS in terms of cytokine production that was mediated by the MyD88-dependent pathway as well. The adaptor molecule named TRIF-related adaptor molecule (TRAM) has been identified. TRAM is shown to be essential to TLR4-TRIF dependent pathway for recruiting TRIF to the cytoplasmic portion of TLR4.35) Additional adaptor TIR-domain containing adaptor protein (TIRAP), also named MyD88 adaptor-like (Mal) is demonstrated essential to MyD88-dependent signaling pathways downstream of TLR2 and TLR4 for recruiting MyD88 to the cytoplasmic portion of these receptors.36),37)
TLR3 recognizes dsRNA present as viral genome or generated in virally infected cells in the course of virus replication. TLR3 is unique among the TLRs in that it lacks a highly conserved proline residue in its cytoplasmic portion, which in other TLRs has been shown to be essential for signaling. There is an alanine residue in this position in TLR3, which is conserved between human and mouse TLR3. TLR3 does not use the MyD88-dependent pathway, but requires TRIF.
The signaling pathway downstream of TRIF has been clarified. TRIF associates with TNF receptor-associated factor 3 (TRAF3), TRAF6 and receptor-interacting protein-1 (RIP1) as downstream signaling molecules.38)–41) Among them, TRAF6 and RIP1 activate NF-κB, whereas TRAF3 is responsible for inducing type I IFNs. TRAF3 activates two IκB kinase-related kinases, TANK-binding Kinase 1 (TBK1) and inducible IκB Kinase (IKKi), both of which are involved in the activation of IRF3 and/or IRF7 (Fig. 10).42)–44)
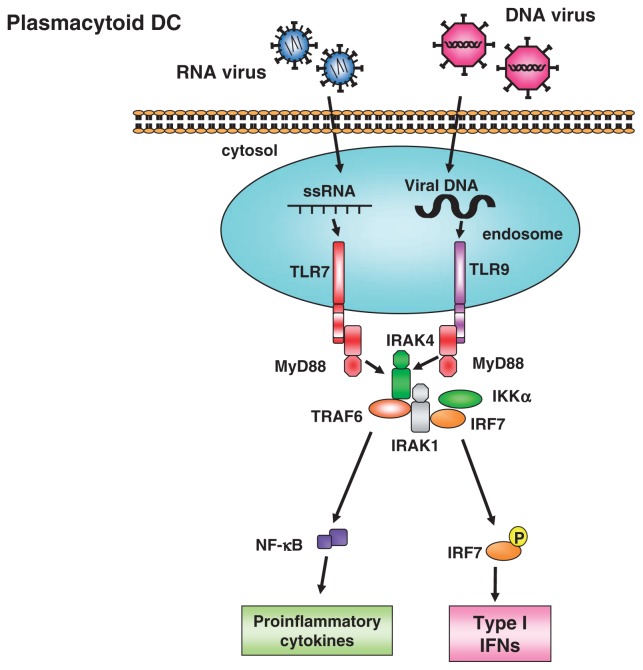
Fig. 10.
MyD88-dependent type I interferon production in plasmacytoid dendritic cells.
In various cell types, the MyD88-dependent signaling is associated with inflammatory responses via activation of NF-κB and MAP kinases. In contrast, in pDC TLR7 or TLR9 activation induces production of type I IFNs via direct association of MyD88 with IRF7 (Fig. 10).45),46) pDCs lacking both TBK1 and IKKi are able to produce IFNα in response to CpG-DNA stimulation or RNA virus infection, indicating that pDCs activate IRF-7 independent of TBK1 and IKKi in contrast to TRIF-dependent type 1 interferon production.
6. Cytoplasmic helicases for sensing of viral infection
Although the involvement of TLRs in virus-induced cytokines and IFN production was well established, fibroblasts lacking both MyD88 and TRIF were still capable of expressing IFN-inducible genes in response to RNA virus infection, indicating the existence of TLR-independent virus detectors.
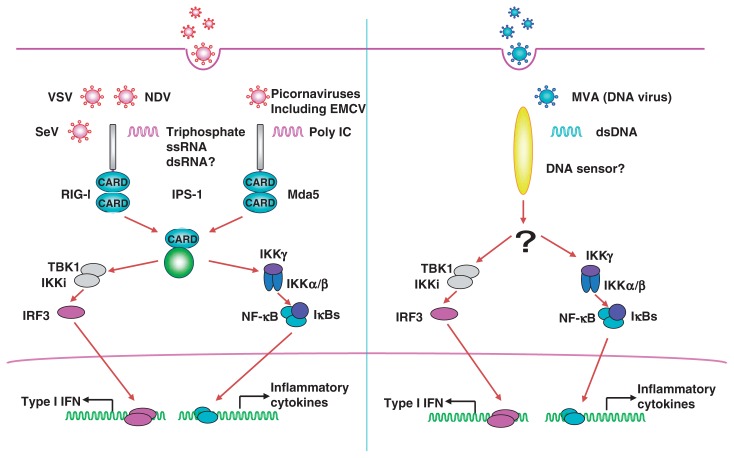
An RNA helicase retinoic-acid-inducible gene I (RIG-I) was identified as a cytosolic sensor for viral invasion, which induces type I IFNs in a TLR-independent manner.47) Melanoma differentiation-associated gene 5 (MDA5) is homologous to RIG-I. Both possess two N-terminal caspase-recruitment domains (CARDs) followed by an RNA helicase domain. The CARDs are responsible for the signal transduction leading to activation of NF-κB and IRF3/7 via their adaptor molecule IFN-β promoter stimulator 1 (IPS-1), also called Cardif, MAVS or VISA,48)–51) which is located on the outer membrane of mitochondria. TBK1 and IKKi are also responsible for the activation of RIG-I or MDA5-dependent IFN production, indicating that the signaling pathways triggered by TLR stimulation and RIG-I converges at the level of TBK1/IKKi (Fig. 11).
Fig. 11.
Cytoplasmic pathogen sensors and anti-viral responses.
The functional and differential role of RIG-I and MDA5 was revealed by gene targeting.52) RIG-I−/− mouse embryonic fibroblasts (MEFs) did not produce IFNα or cytokines in response to infections with RNA viruses including Newcastle disease virus (NDV), Sendai virus or Vesicular Stomatitis virus (VSV). NDV-induced activation of NF-κB and IRF3 was abrogated in RIG-I−/− cells. Furthermore, cDCs derived from RIG-I−/− mice also failed to induce type I IFNs and cytokines upon exposure to NDV. Interestingly, pDCs from RIG-I−/− mice produced a comparable amount of IFNα to pDCs from wild-type mice upon NDV stimulation, indicating that cDCs and pDCs utilize different mechanisms for the induction of IFNs. In contrast, MyD88−/− pDCs showed severely impaired IFNα production in response to NDV infection, confirming that TLRs are essential for the recognition of viruses in pDCs.
In contrast to RIG-I−/− mice, MDA5−/− MEFs normally produced IFNβ in response to several RNA viruses, such as NDV, Sendai virus and Vesicular Stomatitis virus (VSV) infection. However, type I IFN production in response to encephalomyocarditis virus (EMCV) was abrogated in MDA5−/− cells, whereas the response was not impaired in RIG-I−/− cells. EMCV belongs to the picornavirus family with the positive sense ssRNA genome. The picornavirus family viruses including theiler’s virus and Mengo virus were also recognized by MDA5, but not RIG-I, suggesting that MDA5 is responsible for the recognition of picornaviruses. These data indicate that RIG-I and MDA5 sense distinct RNA viral infection.
It is well known that viruses evade the host innate immune system by various ways, and the RIG-I/MDA5 pathway is one of the targets for viruses. Signaling molecules such as IPS1 and TBK1 are shown to be inactivated by proteins encoded in the viral genome. For example, IPS1 can be cleaved by hepatitis C virus (HCV) nonstructural protein (NS) 3/4A for disrupting its ability to induce type I IFNs. Not only the signaling molecules, but also the receptors themselves are inhibited by RNA viruses. The influenza virus NS1 protein is shown to associate with RIG-I, and inhibit RIG-I-mediated influenza virus recognition.
Not only RNA viruses, but also synthetic RNAs are differentially recognized by RIG-I and MDA5.53) For instance, Poly I:C induced type I IFNs in the RIG-I-independent and MDA5-dependent manner. In contrast, in vitro transcribed dsRNAs, generated with the T7 RNA polymerase, induced type I IFNs normally in MDA5−/− MEFs, but not in RIG-I−/− MEFs. The reason why these two synthetic dsRNAs are differentially recognized is now clarified. SsRNA generated by in vitro transcription with T7 RNA polymerase possesses triphosphate at 5′ end. Recently ssRNA with 5′-triphosphate has been demonstrated as RIG-I ligand.54),55) RIG-I−/− MEFs failed to express type I IFNs in response to 5′-triphosphate ssRNA. Indeed, it was reported that 5′-end of the influenza genome harvors triphosphate. Genomic RNA purified from influenza virus is shown to be sensed by RIG-I. Treatment of 5′ triphosphate RNA and influenza genomic RNA with a phosphatase abolished the ability of these RNAs to stimulate cells, implicating the importance of 5′ phosphate of RNAs. Eukaryotic mRNAs have a 5′ 7-methylguanosine residue called cap structure, which suppresses nuclease-mediated mRNA degradation and facilitates translation. Since eukaryotic mRNA prevent recognition by host immune cells, it is possible that 5′ cap structure also functions to prevent surveillance by the innate immune system.
The length of dsRNA is also critical for the differential recognition.56) Commercially available poly I-C is around 4–8 kbp in length, and specifically recognized by MDA5. When poly I-C was digested with dsRNA digesting enzyme, poly I-C shorter than 2 kbp became a RIG-I ligand. We showed that conversion of RIG-I ligand to MDA5 ligand takes place around 3–4 kbp. Renovirus is a double-strand RNA virus, and the genomic RNA contains both RIG-I and MDA5 ligands. Renovirus genome RNA consists of 10 segments in three different classes called L (3.9 kbp), M (2.2–2.3 kbp), and S (1.2–1.4 kbp) correspondingly to their size. When these fragments were separately transfected into MEFs, the production of type 1 interferon in response to S segments was dependent on RIG-I, whereas IFN production in response to L segements was reduced in both RIG-I- and MDA5-deficient MEF, suggesting that MDA5 contributes more to the recognition of longer segments of reovirus genomic dsRNA. In the case of cells infected with EMCV, long dsRNAs are generated in the cytoplasm and recognized by MDA5, while short dsRNAs are generated in cells infected with VSV and predominantly recognized by RIG-I. In VSV-infected cells, 5′ triphosphated ssRNAs are also generated in the cytoplasm. Thus, VSV recognition by RIG-I may involve both short dsRNA and 5′ triphosphate of ssRNA.
7. Clinical application of innate immunity research
The discovery of Toll-like receptors (TLRs) and their ligands has opened the possibility to develop new approaches to treat a wide spectrum of diseases, including infectious, malignant, auto-immune and allergic diseases. TLR ligands are promising for improvement of viral and bacterial vaccines, cancer immunotherapy, and allergic disesease, while TLR antagonists or inhibitors of TLR responses may be effective for treatment of endotoxin shock as well as inflammatory and auto-immune diseases. Table shows clinical trials now on-going. Importantly, several ligands had been already in clinical trials before the action of these drugs was shown to be mediated by TLR activation. For example, the TLR7 agonist imiquimod is marketed for topical use against warts, caused by papillomavirus. Topical application of resiquimod and analogs is safe and effective at activating the local immune response without side effects such as induction of systemic cytokines. LPS is highly toxic and a cause of endotoxin shock. However, LPS derivative, 3-0-descyl-4′-monophosphoryl lipid A (MPL) is devoid of high toxicity but retains adjuvant activity. MPL absorbed on aluminum hydroxide or aluminum phosphate (AS04, GSK Biologics) has been tested in vaccines against human hepatitis virus (HBV), human papilloma virus (HPV), herpes simplex virus (HSV), respiratory syncytial virus (RSV) and Epstein-Barr virus (EBP). A vaccine with L1 virus-like particles from HPV-16 and HPV-18 formulated with AS04 (Cervarix) is now clinically used.
Table 3.
Clinical Trials of TLR Drugs
| TLR | Drug | Action | Application |
|---|---|---|---|
| TLR3 | poly I:C analogs | agonist | HIV, HBV |
| TLR4 | Monophosphoryl lipid A, | agonist | HBV vaccine adjuvant |
| RC-529 | agonist | HBV vaccine adjuvant | |
| E5564 | antagonist | Sepsis | |
| TAK-242 | antagonist | Sepsis | |
| TLR7 | Imiquimod | agonist | Genital warts |
| Resiquimod | agonist | Genital herpes | |
| Loxoribine | agonist | Cancer | |
| TLR9 | CpG 7909 | agonist | Cancer |
| CpG-ODN | agonist | HCV | |
| CpG-ODN | agonist | Asthma, Allergies |
HIV: human immunodeficiency virus; HBV: hepatitis B virus; HCV: hepatitis C virus
Cancer immunotherapy has its roots in the work of Dr. William B. Coley, an American surgeon who practiced in New York. In the early 1880’s, he noticed that cancer disappeared when the patients experienced bacterial infections after operation. So he started to treat patients by injecting live Streptococcus pyogenes, later a mixture of dead Streptococcus pyogenes and dead Serratia marcescens bacteria directly into inoperable tumors. Although his clinical trial achieved significant therapeutic effects, such treatment was not officially approved, and replaced by radiation therapy. In Japan, similar treatment is still trialed. They include BCG-CWS which is the extract of the cell wall skeleton of Bacillus Calmette-Guerin from Mycobacterium bovis and was originally developed by Ichiro Azuma and Yuichi Yamamura, Osaka University, and Maruyama Vaccine, the extract from human tubercle bacilli, and OK-432(Picibanil), a penicillin-treated lyophilized preparation of Streptococcus pyogenes. Recent evidence indicates that these substances induce anti-tumor immunity by activating TLRs in dendrititic cells. In fact, BCG-CWS is shown to activate TLR2 and TLR4. It has been reported that OK-432 contains TLR2 and TLR9 stimulants. A variety of bacterial extracts and synthetic compounds that activate TLRs are now going on in clinical trial for a variety of cancers. CpG-ODN-based immunotherapy was shown to be beneficial in controlling micrometastasis after surgical removal of tumors as well as breaking tumor-induced tolerance.
TLR ligands generally skew immunity toward a Th1 response, and therefore they may be useful for treatment of allergic diseases such as pollen allergy and asthma. Particularly, CpG DNA is a potent Th1 stimulant, and clinical trials using CpG DNA alone, a mixture of CpG DNA and allergen, or allergen conjugated CpG DNA for treatment of variety of allergic diseases are currently under way.
Sepsis is a systemic response to infection, caused by LPS from Gram-negative bacteria. Interaction of LPS with TLR4 causes the release of cytokines and other inflammatory mediators from monocytes and macrophages, resulting in fever, hypotension, organ failure and death. Recently several TLR4 antagonists including Eritoran (E5564) and TAK-242 have been developed. These compounds may be promising for the treatment of spepsis.
Inappropriate activation of TLRs by endogenous ligands will lead to sterile inflammation or autoimmunity. TLR7 and TLR9 activation by endogenous RNA and DNA has been reported to be involved in development of autoimmune diseases such as systemic lupus erythematosus (SLE) and psoriasis. In SLE, pDCs are activated by circulating immune complexes consisting of self-nucleic acids and autoantibodies against nucleic acids or nucleoproteins. Through FcgammaRII-dependent endocytosis, autoantibodies in the immune complex transport self-DNA to the endosome, where TLR9 activated by self-DNA produces type 1 IFNs, promoting dendritic cell maturation and activation of autoreactive T cells, resulting in autoimmunity. Also the antimicrobial peptide LL37 produced from the epithelium, forms a complex with self DNA released by damaged cells and triggers TLR9-mediated pDC activation to produce type 1 IFNs, which lead to the formation of psoriatic skin lesions. Different from immunostimulatory DNA containing unmethylated CpG motifs, DNA containing multiple TTAGGG motifs harbor immunosuppressive activity. Preclinical studies suggest that these suppressive DNA may slow or prevent autoimmune diseases. Thus, the TLR system is a double-edge sword, which needs a proper care in clinical trials of TLR stimulating compounds.
Acknowledgements
I thank Dr. Osamu Takeuchi for figures and all members who participated in the research works presented here.
Profile
Shizuo Akira was born in Osaka Prefecture, Japan in 1953. He graduated from Osaka University School of Medicine. He is a director and professor of WPI Immunology Frontier Research Center, and also a professor in Institute for Microbial Diseases at Osaka University, Japan. He received Ph.D. in 1984 from Osaka University. After two years of postdoctoral working in Department of Immunology, University of California at Berkeley, he started to study on IL-6 gene regulation and signaling under the direction of Professor Tadamitsu Kishimoto at the Institute for Molecular and Cellular Biology, Osaka University, and cloned transcription factors, NF-IL6 and STAT3. He was a professor in Department of Biochemistry, Hyogo College of Medicine from 1996 to 1999, where he became involved in Toll-like receptor research. His current research interests are molecular mechanisms of host defense and innate immunity, which are studied mainly by generating knockout mice. He has authored over 700 papers, and is the top cited researcher in immunological field in a 10 year period since 2004. In 2006 and 2007 he was twice recognized the top hottest scientist who had published the greatest number of Hot Papers over the preceding two years. He is the recipient of several international awards including the Robert Koch Prize and William B. Coley Award.

References
- 1).Akira S., Uematsu S., Takeuchi O. (2006) Pathogen recognition and innate immunity. Cell 24, 783–801 [DOI] [PubMed] [Google Scholar]
- 2).Akira S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., et al. (1990) A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 9, 1897–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Akira S., Nishio Y., Inoue M., Wang X., Wei S., Matsuzaka T., et al. (1994) Molecular cloning of APRF, a novel ISGF3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 77, 63–71 [DOI] [PubMed] [Google Scholar]
- 4).Minami M., Inoue M., Wei S., Takeda K., Matsumoto M., Kishimoto T., et al. (1996) STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc. Natl. Acad. Sci., USA 93, 3963–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Muzio M., Ni J., Feng P., Dixit V.M. (1997) IRAK(Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science 278, 1612–1615 [DOI] [PubMed] [Google Scholar]
- 6).Adachi O., Kawai T., Takeda K., Matsumoto M., Tsutsui H., Sakagami M., et al. (1998) Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9, 143–150 [DOI] [PubMed] [Google Scholar]
- 7).Kawai T., Adachi O., Ogawa T., Takeda K., Akira S. (1999) Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 11, 115–122 [DOI] [PubMed] [Google Scholar]
- 8).Lemaitre B., Nicolas E., Michaut L., Reichhart J.M., Hoffmann J.A. (1996) The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983 [DOI] [PubMed] [Google Scholar]
- 9).Medzhitov R., Reston-Hurlburut P., Janeway C.A., Jr (1997) A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388, 394–397 [DOI] [PubMed] [Google Scholar]
- 10).Rock F.L., Hardiman G., Timans J.C., Kastelein R.A., Bazan J.F. (1998) A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci., USA 95, 588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Poltorak A., He X., Smirnova I., Liu M.Y., Huffel C.V., Du X., et al. (1998) Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 12).Hoshino K., Takeuchi O., Kawai T., Sanjo H., Ogawa T., et al. (1999) Cutting edge: TLR4-deficient mice is hyporesponsive to LPS: evidence for TLR4 as the Lps gene product. J. Immunol. 162, 3749–3752 [PubMed] [Google Scholar]
- 13).Takeuchi O., Takeda K., Hoshino K., Adachi O., Ogawa T., Akira S. (2000) Cellular responses to bacterial cell wall components are mediated through MyD88-dependent signaling cascades. Int. Immunol. 12, 113–117 [DOI] [PubMed] [Google Scholar]
- 14).Takeuchi O., Hoshino K., Kawai T., Sanjo H., Takada H., Ogawa T., et al. (1999) Differential roles of Toll-like receptor (TLR) 2 and TLR4 in recognition of Gram-negative and Gram-positive bacterial cell wall components. Immunity 11, 443–451 [DOI] [PubMed] [Google Scholar]
- 15).Takeuchi O., Grotel K., Kaufmann A., Kawai T., Hoshino K., Morrl M., et al. (2000) Cutting edge: Preferentially the R-stereoisomer of the mycoplasmal lipopeptide MALP-2 activates immune cells through a TLR-2 and MyD88-dependent signaling pathway. J. Immunol. 164, 554–557 [DOI] [PubMed] [Google Scholar]
- 16).Takeuchi O., Kawai T., Muhlradt P.F., Morr M., Radolf J.D., Zychlinsky A., et al. (2001) Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int. Immunol. 13, 933–940 [DOI] [PubMed] [Google Scholar]
- 17).Takeuchi O., Sato S., Horiuchi T., Hoshino K., Takeda K., Dong Z., et al. (2002) Cutting edge: Role of Toll-Like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 169, 10–14 [DOI] [PubMed] [Google Scholar]
- 18).Tokunaga T., Yamamoto T., Yamamoto S. (1999) How BCG led to the discovery of immunostimulatory DNA. Jpn. J. Infect. Dis. 52, 1–11 [PubMed] [Google Scholar]
- 19).Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., et al. (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 [DOI] [PubMed] [Google Scholar]
- 20).Hayashi F., Smith K.D., Ozinsky A., Hawn T.R., Yi E.C., Goodlett D.R., et al. (2001) The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 [DOI] [PubMed] [Google Scholar]
- 21).Uematsu S., Jang M.H., Chevrier N., Guo Z., Kumagai Y., Yamamoto M., et al. (2006) Detection of pathogenic intestinal bacteria y Toll-like receptor 5 on intestinal CD11c(+) lamina propria cells. Nat. Immunol. 7, 868–874 [DOI] [PubMed] [Google Scholar]
- 22).Uematsu S., Fujimoto K., Jang M.H., Yang B.G., Jung Y.J., Nishiyama M., et al. (2008) Nat. Immunol. 9, 769–776 [DOI] [PubMed] [Google Scholar]
- 23).Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. (2001) Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 24).Hemmi H., Kaisho T., Takeuchi O., Sato S., Sanjo H., Hoshino K., et al. (2002) Small antiviral compounds activate immune cells via the TLR7-MyD88-dependent signaling pathway. Nat. Immunol. 3, 196–200 [DOI] [PubMed] [Google Scholar]
- 25).Akira S., Hemmi H. (2003) Recognition of pathogen-associated molecular patterns by TLR family. Immunol. Lett. 85, 85–95 [DOI] [PubMed] [Google Scholar]
- 26).Heil F., Hemmi H., Hochrein H., Ampenberger F., Kirschning C., Akira S., et al. (2004) Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 505, 1526–1529 [DOI] [PubMed] [Google Scholar]
- 27).Dinebold S.S., Kaisho T., Hemmi H., Akira S., Reis E., Sousa C. (2004) Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531 [DOI] [PubMed] [Google Scholar]
- 28).Rafkin I.R., Leadbetter E.A., Busconi L., Viglianti G., Marshak-Rothstein A. (2005) Toll-like receptors, endogenous ligands, and systemic autoimmune disease. Immunol. Rev. 204, 27–42 [DOI] [PubMed] [Google Scholar]
- 29).Kawagoe T., Sato S., Matsushita K., Kato H., Matsui K., Kumagai Y., et al. (2008) Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat. Immunol. 9, 684–691 [DOI] [PubMed] [Google Scholar]
- 30).Kawai T., Takeuchi O., Fujita T., Inoue J.-I., Muhlradt P.F., Sato S., et al. (2001) Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IRF-3 and the expression of a subset of LPS-iducible genes. J. Immunol. 167, 5887–5894 [DOI] [PubMed] [Google Scholar]
- 31).Tamura T., Yanai H., Savitsky D., Taniguchi T. (2008) The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 26, 535–584 [DOI] [PubMed] [Google Scholar]
- 32).Yamamoto M., Sato S., Mori K., Hoshino K., Takeuchi O., Takeda K., et al. (2002) Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-β promoter in the Toll-like receptor signaling. J. Immunol. 169, 6668–6672 [DOI] [PubMed] [Google Scholar]
- 33).Oshiumi H., Matsumoto M., Funami K., Akazawa T., Seya T. (2003) TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-β induction. Nat. Immunol. 4, 161–167 [DOI] [PubMed] [Google Scholar]
- 34).Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., et al. (2003) Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 301, 640–643 [DOI] [PubMed] [Google Scholar]
- 35).Yamamoto M., Sato S., Hemmi H., Uematsu S., Hoshino K., Kaisho T., et al. (2003) TRAM is specifically involved in thte Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat. Immunol. 4, 1144–1150 [DOI] [PubMed] [Google Scholar]
- 36).Yamamoto M., Sato S., Hemmi H., Sanjo H., Uematsu S., Kaisho T., et al. (2002) Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature 420, 324–329 [DOI] [PubMed] [Google Scholar]
- 37).Horng T., Barton G.M., Flavell R.A., Medzhitov R. (2002) The adaptor molecule TIRAP provides signaling specificity for Toll-like receptors. Nature 420, 329–333 [DOI] [PubMed] [Google Scholar]
- 38).Sato S., Sugiyama M., Yamamoto M., Watanabe Y., Kawai T., Takeda K., et al. (2003) Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) associates with TNF receptor-associated factor 6 and TANK-Binding kinase 1, and activates two distinct transcription factors, NF-kappaB and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 171, 4304–4310 [DOI] [PubMed] [Google Scholar]
- 39).Meylan E., Burns K., Hofmann K., Blancheteau V., Martinon F., Kelliher M., et al. (2004) RIP1 is an essential mediator of Toll-like receptor 3-induced NFkappaB activation. Nat. Immunol. 5, 503–507 [DOI] [PubMed] [Google Scholar]
- 40).Häcker H., Redecke V., Blagoev B., Kratchmarova I., Hsu L.C., Wang G.G., et al. (2006) Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439, 204–207 [DOI] [PubMed] [Google Scholar]
- 41).Oganesyan G., Saha S.K., Guo B., He J.Q., Shahangian A., Zarnegar B., et al. (2006) Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439, 208–211 [DOI] [PubMed] [Google Scholar]
- 42).Fitzgerald K.A., McWhirter S.M., Faia K.L., Rowe D.C., Latz E., Golenbock D.T., et al. (2003) IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 43).Perry A.K., Chow E.K., Goodnough J.B., Yeh W.C., Cheng G. (2004) Differential requirement for TANK-binding kinase-1 in type I interferon responses to toll-like receptor activation and viral infection. J. Exp. Med. 199, 1651–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Hemmi H., Kawai T., Hoshino K., Takeda K., Akira S. (2004) The roles of two IkB kinase-related kinases in lipopolyusaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 199, 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Kawai T., Sato S., Ishii K.J., Coban C., Hemmi H., Yamamoto M., et al. (2004) Interferon-α induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 5, 1061–1068 [DOI] [PubMed] [Google Scholar]
- 46).Honda K., Yanai H., Mizutani T., Negishi H., Shimada N., Suzuki N., et al. (2004) Role of a transductional-transcriptional processor complex involving MyD88 and IRF-7 in Toll-like receptor signaling. Proc. Natl. Acad. Sci., USA 101, 15416–15421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., et al. (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5, 730–737 [DOI] [PubMed] [Google Scholar]
- 48).Kawai T., Takahashi K., Sato S., Covan C., Kumar H., Kato H., et al. (2005) IPS-1, an adaptor triggering RIG-I and Mda5-mediated type I interferon induction. Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 49).Seth R.B., Sun L., Ea C.K., Chen Z.J. (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 50).Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., et al. (2005) Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172 [DOI] [PubMed] [Google Scholar]
- 51).Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z., Shu H.B. (2005) VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19, 727–740 [DOI] [PubMed] [Google Scholar]
- 52).Kato H., Sato S., Yoneyama M., Yamamoto M., Uematsu S., Matsui K., et al. (2005) Cell type-specific involvement of RIG-I in antiviral response. Immunity 23, 19–28 [DOI] [PubMed] [Google Scholar]
- 53).Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., et al. (2006) Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 54).Hornung V., Ellegast J., Kim S., Brzozka K., Jung A., Kato H., et al. (2006) 5′-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997 [DOI] [PubMed] [Google Scholar]
- 55).Pichlmair A., Schulz O., Tan C.P., Naslund T.I., Liljestrom P., Weber F., et al. (2006) RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314, 997–1001 [DOI] [PubMed] [Google Scholar]
- 56).Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., et al. (2008) Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205, 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]