Abstract
Glioma includes astrocytoma, oligodendroglioma, ependymoma and glioblastoma. We previously reported the epigenetic silencing of paternally expressed gene 3 (PEG3) in glioma cell lines. In this study, we investigated methylation of an exonic CpG island in the promoter region and the expression of PEG3 gene in 20 glioma and 5 non-tumor tissue samples. We found wide variations in the methylation level. Hypomethylaiton and hypermethylation was found in 3 and 4 glioma tissue samples, respectively. Monoallelic expression, which is an evidence of an imprinted gene, was maintained in eight out of nine informative cases which have T/C polymorphisms in PEG3. The lower gene expression, which suggested epigenetic silencing of PEG3, was confirmed statistically in glioblastoma using quantitative reverse-transcription polymerase chain reaction. Interestingly, we found higher expression of PEG3 in two out of three oligodendrogliomas. A negative correlation between the methylation level and gene expression was shown by regression analysis. These results suggest that the abnormal regulation of PEG3 is associated with several glioma subtypes and that it plays an important role in tumorigenesis.
Keywords: glioma, PEG3, DNA methylation, epigenetic silencing
Introduction
Paternally expressed gene 3 (PEG3) is an imprinted gene located on 19q13.4. PEG3 encodes a krüppel-type (C2H2) zinc-finger protein and is expressed predominantly in brain, ovary, testis and placenta in adult tissues.1) Although its function has not been completely characterized, PEG3 has been reported to play an important role in cell proliferation by activating NF-κB2) and in p53-mediated apoptosis by inducing Bcl2-associated X protein (Bax) translocation.3)–5) In female mice lacking Peg3, which is mouse homologue of human PEG3 gene,6) growth impairment and compromised nurturing behavior have been seen, although glioma genesis has not been reported.7)–9)PEG3 is believed to be involved in the development of various brain regions, such as the hypothalamus, through its role in apoptotic pathways.
Tumor suppressor activity of the human PEG3 gene has been reported in human glioma cell lines.10) In a previous study, we reported that the aberrant DNA methylation of the CpG island is associated with epigenetic silencing of PEG3 in glioma cell lines.11) We also identified a novel imprinted transcript gene called ITUP1, which is located upstream and oppositely oriented to PEG3 and is expressed on the paternal chromosome. The expression profile of ITUP1 was similar to that of PEG3 in glioma cell lines12) suggested methylation status of the CpG island is important for regulation of these genes.
To date, however, there are a few studies about PEG3 gene in tumor tissues of glioma. These studies are about oligodendroglioma.10),26) Therefore, relationship between PEG3 gene and other subtypes and malignancy grades of glioma is not clear. In this study, we examined the methylation and expression of PEG3 in glioma tissues to evaluate its availability of diagnosis and treatment of this tumor.
Materials and methods
Brain tissues
Samples of 20 gliomas (samples GT1–GT20) and 5 non-tumor brain tissues (samples B15–B19) were obtained from Tottori University Hospital, Yonago, Japan. They included 8 female and 13 male patients who ranged in age from 3 to 71 years. The study was approved by the Ethical Committee of the Faculty of Medicine, Tottori University, and informed consent was obtained from all patients.
Tumor samples were classified by a surgical pathologist using the World Health Organization (WHO) system.13) We found one sample of grade I (dysembryoplastic neuroepithelial tumor), 5 samples of grade II (2 astrocytomas, 2 ependymomas and 1 oligodendroglioma), 5 samples of grade III (2 anaplastic astrocytomas, 1 anaplastic ependymoma and 2 anaplastic oligodendrogliomas) and 9 samples of grade IV (9 glioblastomas) (Table 1). For this study, we unified the samples of grade I and II tumors into the “low grade” category. Non-tumor brain was obtained as normal control. Four of the five non-tumor brain samples B16, B17, B18 and B19 adjacent to the tumor were obtained from the patients with glioma GT11, GT20, GT3 and GT16, respectively. B15 was obtained from a patient of brain ischemia. All samples were snap frozen in liquid nitrogen and stored at −80 °C before the extraction of DNA or RNA.
Table 1.
Expression data for PEG3 in non-tumor and glioma brain samples
| Sample | Sex | Age | Diagnosis | WHO grade | Genotype | Expression |
|---|---|---|---|---|---|---|
| GT18 | M | 11 | dysembryoplastic neuroepithelial tumor | I | C/C | + |
| GT2 | F | 6 | Astrocytoma | II | C/T | + (C) |
| GT15 | F | 34 | Astrocytoma | II | C/T | + (T) |
| GT13 | M | 3 | Ependymoma | II | C/T | + (C) |
| GT4 | M | 22 | Ependymoma | II | C/C | + |
| GT16d | M | 32 | Oligodendroglioma | II | C/T | + (T) |
| GT12 | M | 43 | Anaplastic Astrocytoma | III | C/T | + (T) |
| GT20b | M | 70 | Anaplastic Astrocytoma | III | C/T | + (C) |
| GT10 | F | 11 | Anaplastic Ependymoma | III | C/T | + (T) |
| GT14 | F | 51 | Anaplastic Oligodendroglioma | III | T/T | + |
| GT7 | M | 45 | Anaplastic Oligodendroglioma | III | T/T | + |
| GT5 | F | 35 | Glioblastoma | IV | C/T | + (T) |
| GT11a | F | 39 | Glioblastoma | IV | T/T | + |
| GT6 | F | 71 | Glioblastoma | IV | T/T | + |
| GT17 | M | 17 | Glioblastoma | IV | T/T | + |
| GT19 | M | 25 | Glioblastoma | IV | C/C | + |
| GT3c | M | 35 | Glioblastoma | IV | C/T | ND |
| GT1 | M | 66 | Glioblastoma | IV | T/T | + |
| GT9 | M | 66 | Glioblastoma | IV | C/C | + |
| GT8 | M | 70 | Glioblastoma | IV | T/T | + |
| B16a | F | 39 | Non-tumor | — | T/T | + |
| B15 | F | 49 | Non-tumor | — | C/T | + (T) |
| B19d | M | 32 | Non-tumor | — | C/T | + (T) |
| B18c | M | 35 | Non-tumor | — | C/T | + (T) |
| B17b | M | 70 | Non-tumor | — | C/T | + (C) |
F: female; M: male; ND: not determined, were indicated.
the same alphabet means the same person respectively.
Extraction of nucleic acids and cDNA synthesis
Genomic DNA was extracted using standard phenol-chloroform extraction. Total RNA was extracted by using the RNeasy mini kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions and then treated with DNase I (Nippon Gene, Tokyo, Japan) to remove DNA. To investigate the possibility of DNA contamination, the first-strand cDNA was synthesized with (RT+) or without (RT-) M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) using random primers (Promega, Madison, WI, USA). Reaction conditions were 25 °C for 10 min, 42 °C for 50 min and 95 °C for 5 min. To confirm the synthesis of first-strand cDNA, the expression of human glyceraldehyde-3-phosphatase dehydrogenase (GAPDH) was detected using polymerase chain reaction (PCR) primers qGAPDH forward (5′-TGAACGGGAAG-CTCACTGG-3′) and qGAPDH reverse (5′-TCCA-CCACCCTGTTGCTGTA-3′). The PCR conditions were 30 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s.
Bisulfite treatment of genomic DNA and DNA methylation analysis
Extracted genomic DNA from tumor and non-tumor samples was treated with sodium bisulfite as previously described.14) Bisulfite treated genomic DNA-sequencing was then performed as previously described.12) For each DNA sample, ten clones were analyzed. We examined a total of 33 CpG sites in the CpG island. The sequencing was performed from both directions using the BigDye Terminator v3.1 cycle sequencing kit and ABI3130xl genetic analyzer (Applied Bio-systems, Foster City, CA, USA).
Methylation-specific PCR (MSP) assay
PEG3 is an imprinted gene and has both unmethylated and methylated alleles normally. Thus it is necessary to gauge ratio of both alleles to assess epigenetic difference. To measure relative methylated level, we performed MSP with assessable trait as below. Two sets of primers (U and M) designed for annealing to bisulfite-modified genomic DNA were used in this experiment. One primer set (U) annealed to unmethylated DNA that had undergone chemical modification. A second set (M) annealed to methylated DNA that had undergone chemical modification. Primer sets used for MSP were previously described11) (Fig. 1). For equalizing intensities of unmethylated and methylated bands in non-tumor brain tissues, the PCR conditions were set as follows: for unmethylated DNA, 35 cycles of 95 °C for 1 min, 60 °C for 1 min and 72 °C for 1 min; and for methylated DNA, 34 cycles of 95 °C for 1 min, 62 °C for 1 min and 72 °C for 1 min. PCR products were resolved in 2% agarose gel and visualized by ethidium bromide staining. Intensities of the U and M bands were analyzed using Densitograph version 4.0 software (ATTO, Tokyo, Japan) to measure band ratio of MSP.15)
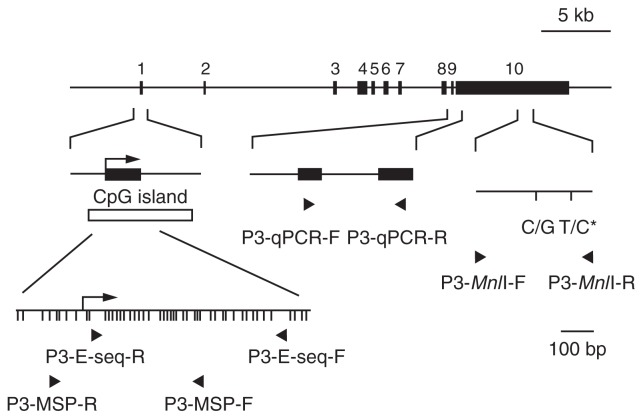
Fig. 1.
A schematic map of PEG3 gene.
Exons are indicated by black boxes. Numbering is followed by the information of GenBank accession no. AC006115. A CpG island surrounding exon 1 (open box), which is the transcriptional starting site of PEG3 (arrow), was identified by the ORNL GRAIL Form. Detailed CpG sites are indicated in lower left panel. DNA methylation was investigated by bisulfite genomic sequencing and MSP by using PCR primers designed on the CpG island indicated by arrowheads.11),12) Quantitative PCR analysis was performed with the primers P3-qPCR-F and P3-qPCR-R (arrowheads) designed on exon 9 and 10, respectively. Primer sequences are described in materials and methods. Imprinting test was performed with the primers P3-MnlI-F and P3-MnlI-R (arrowheads).11) Single nucleotide polymorphisms (SNPs) are shown as sequence polymorphism. The SNP analyzed for allelic expression (asterisk) is located at position 134718 on AC006115.
Genomic imprinting test
To distinguish parental alleles, we performed direct sequencing analysis to find T/C polymorphisms in exon ten of PEG3. DNA and cDNA from tumor and non-tumor brain tissue samples were amplified using PCR primers previously described11) (Fig. 1).
Quantitative gene-expression analysis
For quantitative reverse-transcription PCR (qRT-PCR), we used LightCycler fast-start DNA SYBR green I (Roche, Basel, Switzerland) and a Light-Cycler Real-Time PCR system (Roche). We used the primers P3-qPCR forward (5′-CAAGAGAAG-TGCCTACCCA-3′) and P3-qPCR reverse (5′-GA-ACTGCGTGACACATCC-3′) for PEG3 (Fig. 1) with the following PCR conditions: 45 cycles of 95 °C for 15 s, 60 °C for 5 s and 72 °C for 6 s. As an internal standard, GAPDH was detected with PCR primers qGAPDH forward and reverse. The PCR conditions were 45 cycles of 95 °C for 15 s, 55 °C for 5 s and 72 °C for 10 s. PCR products were resolved in 2% agarose gel and visualized by ethidium bromide staining. Gene-specific standard curves were generated using 10-fold serial dilutions of plasmid DNAs of a 147-bp PEG3 clone and a 307-bp GAPDH clone.
Data analysis
Data of MSP were pooled from three independent sets of experiments, and we calculated the methylation level (ML) using the following formula: methylated band/(intensity of methylated band + intensity of unmethylated band). An ML > 0.60 was defined as hypermethylation and an ML < 0.40 was defined as hypomethylation. Means ± 2 S.D. of the MLs of non-tumor brain tissues (except for sample B19) were included in these definitions. Data of qRT-PCR were pooled from three independent sets of experiments. These data were analyzed by one-way analysis of variance (ANOVA), Student’s t test and regression analysis.
Results
Methylation of the exonic CpG island in PEG3
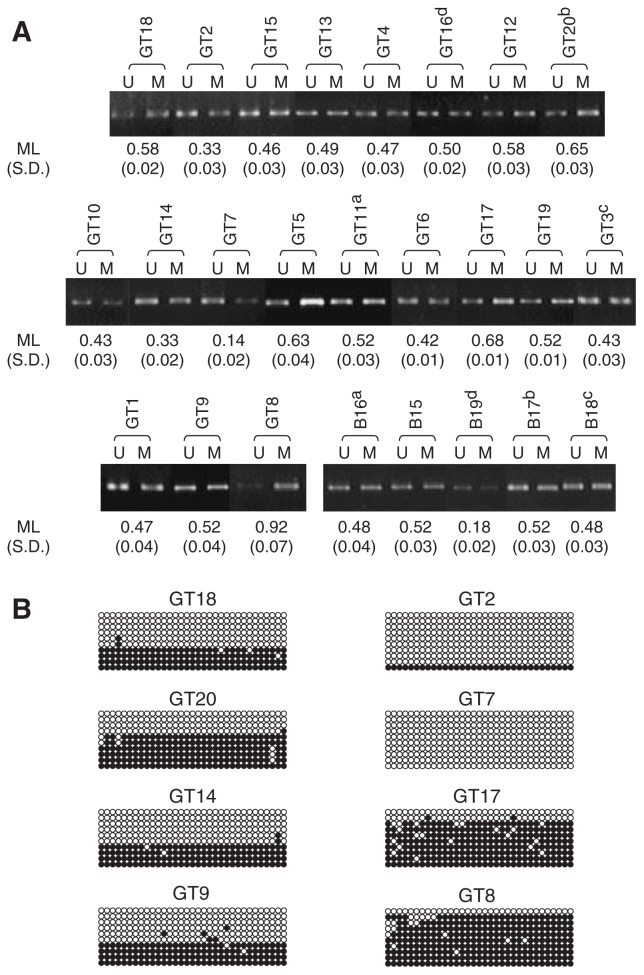
To determine the methylation level, we performed MSP. At first, we tested on non-tumor brain tissues. Approximately equivalent florescence intensities were detected from the U and M bands in four out of five non-tumor brain tissues (Fig. 2A). We performed this experiment three times and obtained similar results. Thus, MLs were calculated from the values of U and M band intensity as described in Materials and Methods. ML values in non-tumor brain tissues ranged from 0.48 to 0.52, except for that of sample B19. Then, we tested in glioma tissue samples (Fig. 2A). We detected both U and M bands from every sample in three sets of experiment. In samples GT18, GT4 and GT9, ML values were 0.58, 0.47 and 0.52, respectively. This implied methylation levels in these tumor samples were same as non-tumors. ML values in 13 out of 20 tumors were indicated normal range. On the other hand, we found wide variations in other tumor tissues. In samples GT2 and GT7, the U band was predominant and ML values were 0.33 and 0.14, respectively. These results implied that the CpG island was hypomethylated in these samples. In samples GT8, GT20 and GT17, the M band was predominant and MLs were 0.92, 0.65 and 0.68, respectively. These results implied hypermethylation of the CpG island. Finally, three samples indicated hypo- and four samples did hypermeth ylation. To confirm the MSP results, we performed bisulfite sequencing on all tissue samples (Fig. 2B). Although there was slight difference with fraction of clone number in each sample, the results were approximately consistent with those of MSP.
Fig. 2.
Methylation of the CpG island localized at the first exon of PEG3 gene.
(A) Methylation-specific PCR analysis of gliomas (from GT1 to GT20) and non-tumor brain tissues (from B15 to B19). U and M mean PCR products derived from unmethylated and methylated alleles, respectively. Methylation levels (ML) of each sample were calculated as described in materials and methods. Mean of ML and Standard deviation (S.D.) of each sample are indicated under the picture of electrophoresis. (B) Bisulfite sequencing analysis in the representative tissues. Circles that queue horizontally indicate a clone sequenced. Ten cloned from each samples were sequences. Thirty-three circles indicate CpG sites within the region were analyzed. Open and closed circles indicate unmethylated and methylated cytosines, respectively.
We attempted to identify differences in methylation between malignancy grades. Average ML values were 0:436 ± 0:144, 0:472 ± 0:080, 0:423 ± 0:202 and 0:568 ± 0:156 in the non-tumor and low-grade, grade III, and grade IV glioma brain tissues, respectively. The ML and tumor grade were statistically unrelated.
To clarify difference of methylation status between tumor and non-tumor, we investigated MLs in tissues from 4 patients who provided both tumor and non-tumor samples. MLs of tumor tissues were higher in samples GT16 (ML = 0:50, P = 0:0000162) and GT20 (ML = 0:65, P = 0:00701) than B19 (ML = 0:18) and B17 (ML = 0:52), respectively. However, there were no significant differences in MLs in samples GT11 (ML = 0:52, P = 0:265) and GT3 (ML = 0:43, P = 0:145) compared to B16 (ML = 0:48) and B18 (ML = 0:48), respectively.
Monoallelic expression of PEG3 in glioma
Hypomethylation of the CpG island suggests the possibility of biallelic expression of PEG3, which indicates deregulation of imprinting. To investigate the allelic pattern of PEG3 expression, we performed an imprinting test of glioma and non-tumor brain tissues by RT-PCR and direct sequencing of the PCR product. Almost all tissues except sample GT3 verified PCR products. We found, however, monoallelic PEG3 expression in eight out of nine gliomas and all four non-tumor brain tissues (Table 1).
PEG3 gene expression
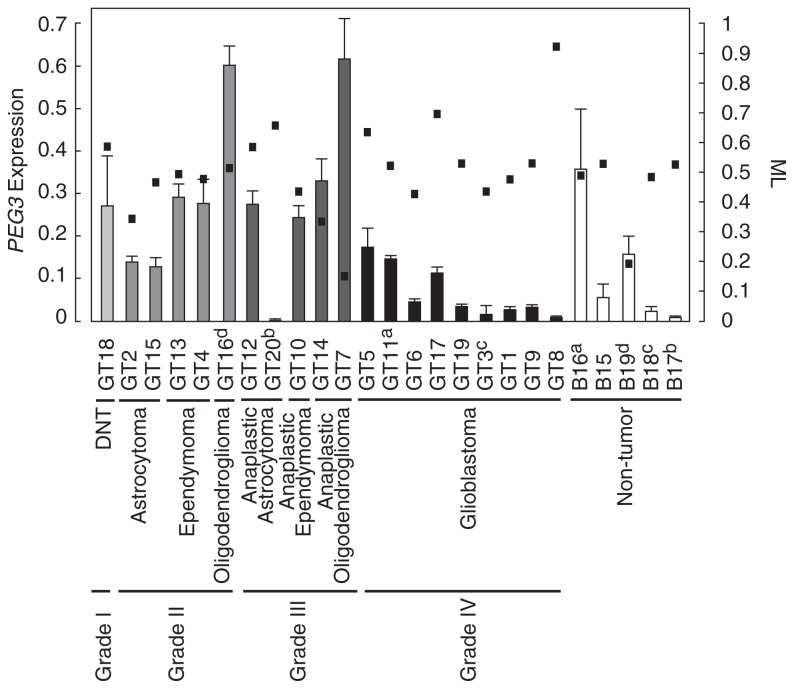
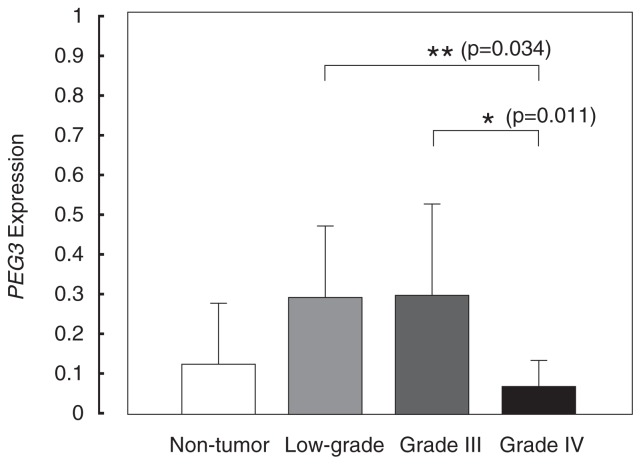
To determine the PEG3 expression level, we performed a qRT-PCR assay. After three sets of experiments, average values and SDs of each tissue were calculated (Fig. 3). We found lower expression in six (GT6, GT19, GT3, GT1, GT9 and GT8) out of nine glioblastomas compared with the average value of non-tumor brain tissues. Relatively higher expression of PEG3 was also found in two oligodendrogliomas (GT16 and GT7) but not in grade II astrocytomas or ependymomas. The average values of PEG3 expression were 0:124 ± 0:150, 0:285 ± 0:172, 0:294 ± 0:221, and 0:065 ± 0:061 in non-tumor and low-grade, grade III and grade IV glioma brain tissues, respectively. We found significant lower gene expression in grade IV than in low-grade and grade III tumors by ANOVA analysis (Fig. 4). Neither low-grade (p = 0:135), grade III (p = 0:193), nor grade IV (p = 0:313) glioma had the significant differences compared with non-tumor tissues.
Fig. 3.
PEG3 expression and methylation level in glioma and non-tumor tissues.
Relative expressions of PEG3 are shown as columns with standard deviation bars. Results of qRT-PCR were standardized by GAPDH. Each columns is indicated by the following colors, WHO grade I (
 ), grade II (
), grade II (
 ), grade III (
), grade III (
 ), grade IV (
), grade IV (
 ) and non-tumor (
) and non-tumor (
 ) brain tissues. Subtype and WHO grade of glioma were indicated under the sample name. Methylation level (ML) of each sample is shown by closed square.
) brain tissues. Subtype and WHO grade of glioma were indicated under the sample name. Methylation level (ML) of each sample is shown by closed square.
Fig. 4.
PEG3 expression and the grade of glioma.
Twenty gliomas and five non-tumor tissues were summarized into four groups by WHO grades. Grade I and grade II are included in the low-grade category. Statistic analysis was examined by ANOVA. Columnand bar indicate mean, and standard deviation, respectively. *; p < 0:05, **; p < 0:005.
Negative correlation between PEG3 expression and ML
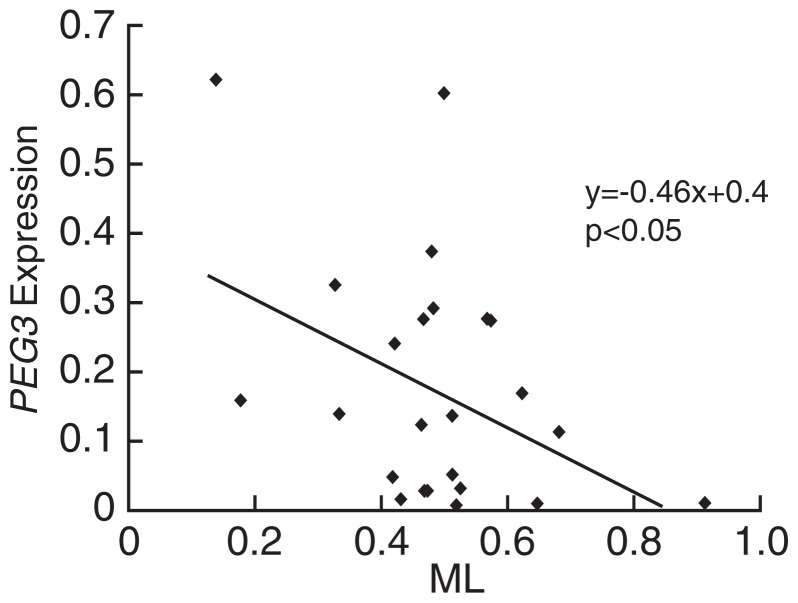
In glioma tissues showing hypomethylation (GT7 and GT14), PEG3 expression was high. In glioma tissues showing hypermethylation (GT8 and GT17), contrastingly, PEG3 expression was low. We also found a trend toward a negative correlation between PEG3 expression and ML (Fig. 3). To confirm this trend, we performed a regression analysis and found a weak negative correlation (r = −0:407, P < 0:05) (Fig. 5). Our result provided evidence that PEG3 expression is regulated by methylation of the CpG island.
Fig. 5.
The regression analysis of expression and methylation of PEG3.
PEG3 expression were plotted against methylation level. All data from tumor and non-tumor samples were plotted in the graph. Regression line was y = −0:46x + 0:4, p < 0:05.
It is thought that the age and sex of patients are associated with particular types of gliomas.16) To investigate whether there is any association between the age or sex of patients and PEG3 gene expression, we performed another regression analysis. No significant correlation was found (data not shown).
Discussion
In our previous report,11) downregulation of PEG3 was found in glioma cell lines. However, it was not clear whether a similar downregulation occurs in glioma tissues and whether there is any relationship between gene expression and tumor subtype or grade. In the current study, six out of nine glioblastomas showed lower mRNA levels than mean levels of non-tumor brain tissues (Fig. 3). The downregulation was statistically confirmed in grade IV glioblastomas, compared with low-grade and grade III gliomas (Fig. 4). These results suggest that PEG3 downregulation is associated with glioblastoma. Downregulation of PEG3 has been found in ovarian cancer cell lines and in nonendo-metrial and endometrial cancers and the promoter was hypermethylated.17)–19) Thus, PEG3 might play an important role in various types of cancer.
MLs of grade IV glioblastomas seem slightly higher than those in non-tumor brain tissues. However, there was no significance in static test. Nevertheless, hypermethylation of the CpG island was previously reported in glioma cells. Moreover, regression study demonstrated PEG3 expression has negative correlation with the methylation of the CpG island (Fig. 5). It is possible that the significance of this regression due to higher expression of PEG3 gene in oligodendroglioma. Although further study might be necessary to careful elucidation of this relationship between methylation and expression, our results indicated that PEG3 downregulation is due to methylation of the CpG island. Moreover, ML in a tumor GT20 was higher than matched non-tumor sample B17. The methylation is possibly increased in tumorigenesis.
Loss of imprinting, biallelic expression, and epigenetic silencing of an imprinted gene are frequently observed in various types of cancer —for example, biallelic expression of insulin-like growth factor 2 and epigenetic silencing of cyclin-dependent kinase inhibitor 1C.20),21) Previously, we reported the gene suppression of PEG3 in glioma cell lines.11) Biallelic PEG3 expression has also been reported in two choriocarcinoma cell lines.19) Furthermore, the mouse embryonic fibroblasts derived from imprint-free embryonic stem cells, which have loss of imprinting of multiple imprinted genes, including biallelic PEG3 expression, formed tumors in the immunodeficient mouse model.22) These results suggest that both loss of imprinting and epigenetic silencing of PEG3 are involved in tumorigenesis. We confirmed the monoallelic expression in eight out of nine informative cases, which suggests that genomic imprinting of PEG3 is strongly sustained in the brain and is not disrupted in glioma.
Interestingly, we found the highest PEG3 expression in one grade II and one grade III oligodendroglioma each. Recently, delta-like 1 and PEG10 gene expression were reported to be upregulated in hepatocellular carcinoma and B cell chronic lymphocytic leukemia, respectively.23),24) Additionally, ectopic c-Myc oncogene has been shown to induce H19 gene expression without affecting its imprinted status in breast epithelial cells.25) These implied upregulation of imprinted genes associate with tumor. However, loss of heterozygosity (LOH) of 19q has been frequently observed in oligodendrogliomas, and alterations in parental-origin specific copy number might also affect PEG3 expression levels.26) Furthermore, there has been another report that PEG3 silencing is associated with oligodendroglioma.10) These findings suggest the possibility that both upregulation and downregulation of PEG3 are involved in tumorigenesis.
Our study had several limitations. We detected wide variations in MLs and PEG3 expression not only in tumor but also in non-tumor brain tissues. PEG3 expression levels differed by more than 52-fold between control samples B16 and B17. In contrast, hypomethylation of the CpG island was observed in sample B19 (Fig. 3). These results may due to cell type and/or tissue-specific DNA methylation and epigenetic heterogeneity in the brain tissues among individuals. Furthermore, our non-tumor brain tissue samples may not have been appropriate as normal controls, given that they were obtained adjacent to tumor tissue. Low ML of B19 might be due to LOH of methylated allele. In further study, LOH analysis might be necessary to evaluate relationship between methylation level and allele copy number. Cells of tumor tissues may be heterogeneous, causing the gene to be variably expressed. It will be necessary to determine whether our observations are due to tumorigenesis in the peritumoral region or to natural variations in expression levels.
In our methylation analysis, a number of sample demonstrated normal range, thus its relationship with tumor grade was not clear. However, we observed a negative correlation between PEG3 expression and the ML (Fig. 5). This indicates the possibility that PEG3 gene expression is regulated by methylation of the CpG island, as previously reported.11) Silencing of the active PEG3 allele could be caused by multiple factors. Kim et al.27) reported that Gli-type transcription factor YY-1 binding motifs were localized at the first intron of PEG3 gene and differentially methylated between two parental alleles in vivo. However, an YY-1–deficient mouse model using shRNA exhibited increased PEG3 transcripts without alterations in promoter methylation or histone tail methylation.28) Furthermore, we found no significant difference in methylation on these motifs in glioma tissues (data not shown). More recently, macroH2A, a variant form of core histone H2A was reported to be deposited on the silencing allele at several imprinting control regions containing PEG3.29) However, in vivo knockdown of macroH2A showed not hypo-methylation of PEG3 but hypermethylation.30) Additionally, Lu et al.31) reported that the cyclophilin A gene protects parental PEG3 against DNA hypermethylation and trimethylation of histone H3 lysine 9 in mice. ITUP1, which we previously isolated,12) might be associated with regulation of 19q13.4 imprinted gene cluster. PEG3 regulation is complex, and the disruption of these mechanisms might be involved in tumorigenesis.
In conclusion, we found that the deregulation of PEG3 is associated with specific subtypes of glioma. However, it will be necessary to investigate this association in a larger cohort by narrow analysis and to determine other epigenetic mechanisms for the regulation of PEG3 gene expression in glioma. More recently, it was reported that downregulation of ARHI, which is an imprinted gene with tumor suppressor activity, was correlated with PEG3 downregulation in ovarian cancer by using pyrosequencing.32) This finding suggested coordinate downregulation of imprinted tumor suppressor genes might be associated with tumorigenesis. It is necessary to analyze expression of multiple imprinted genes and the association in glioma.
Acknowledgement
S.O. was supported by Japan Society for the Promotion of Science. The research was supported in part by grants from the Scientific Fund of the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Ministry of Health, Labor and Welfare of Japan.
Abbreviations
- PEG3
paternally expressed gene 3
- GAPDH
glyceraldehyde-3-phosphatase dehydrogenase
- PCR
polymerase chain reaction
- qRT-PCR
quantitative reverse transcription PCR
- MSP
methylation-specific PCR
- ML
methylation level
- ANOVA
analysis of variance
References
- 1).Kim J., Ashworth L., Branscomb E., Stubbs L. (1997) The human homolog of a mouse-imprinted gene, Peg3, maps to a zinc finger gene-rich region of human chromosome 19q13.4. Genome Res. 7, 532–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Relaix F., Wei X.J., Wu X., Sassoon D.A. (1997) Peg3/Pw1 is an imprinted gene involved in the TNF-NFκB signal transduction pathway. Nat. Genet. 18, 287–291 [DOI] [PubMed] [Google Scholar]
- 3).Deng Y., Wu X. (2000) Peg3/Pw1 promotes p53-mediated apoptosis by inducing Bax translocation from cytosol to mitochondria. Proc. Natl. Acad. Sci. USA 97, 12050–12055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Johnson M.D., Wu X., Aithmitti N., Morrison R.S. (2002) Peg3/Pw1 is a mediator between p53 and Bax in DNA damage-induced neuronal death. J. Biol. Chem. 277, 23000–23007 [DOI] [PubMed] [Google Scholar]
- 5).Yamaguchi A., Taniguchi M., Hori O., Ogawa S., Tojo N., Matsuoka N., et al. (2002) Peg3/Pw1 is involved in p53-mediated cell death pathway in brain ischemia/hypoxia. J. Biol. Chem. 277, 623–629 [DOI] [PubMed] [Google Scholar]
- 6).Kuroiwa Y., Kaneko-Ishino T., Kagitani F., Kohda T., Li L.L., Tada M., et al. (1996) Peg3 imprinted gene on proximal chromosome 7 encodes for a zinc finger protein. Nat. Genet. 12, 186–190 [DOI] [PubMed] [Google Scholar]
- 7).Li L.L., Keverne E.B., Aparicio S.A., Ishino F., Barton S.C., Surani M.A. (1999) Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science 284, 330–333 [DOI] [PubMed] [Google Scholar]
- 8).Szeto I.Y.Y., Barton S.C., Keverne E.B., Surani M.A. (2004) Analysis of imprinted murine Peg3 locus in transgenic mice. Mamm. Genome 15, 284–295 [DOI] [PubMed] [Google Scholar]
- 9).Curley J.P., Pinnock S.B., Dickson S.L., Thresher R., Miyoshi N., Surani M.A., et al. (2005) Increased body fat in mice with a targeted mutation of the paternally expressed imprinted gene Peg3. FASEB J. 19, 1302–1304 [DOI] [PubMed] [Google Scholar]
- 10).Kohda T., Asai A., Kuroiwa Y., Kobayashi S., Aisaka K., Nagashima G., et al. (2001) Tumour suppressor activity of human imprinted gene PEG3 in a glioma cell line. Genes Cells 6, 237–247 [DOI] [PubMed] [Google Scholar]
- 11).Maegawa S., Yoshioka H., Itaba N., Kubota N., Nishihara S., Shirayoshi Y., et al. (2001) Epigenetic silencing of PEG3 gene expression in human glioma cell lines. Mol. Carcinog. 31, 1–9 [DOI] [PubMed] [Google Scholar]
- 12).Maegawa S., Itaba N., Otsuka S., Kamitani H., Watanabe T., Tahimic C.G., et al. (2004) Coordinate downregulation of a novel imprinted transcript ITUP1 with PEG3 in glioma cell lines. DNA Res. 11, 37–49 [DOI] [PubMed] [Google Scholar]
- 13).Kleihues P., Cavenee K. (eds.) (2000) WHO Classificaton of Tumors: Pathology and Genetics of Tumours of the Nervous System. IARC Press, France [Google Scholar]
- 14).Clark S.J., Harrison J., Paul C.L., Frommer M. (1994) High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22, 2990–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Umbricht C.B., Evron E., Gabrielson E., Ferguson A., Marks J., Sukumar S. (2001) Hypermethylation of 14-3-3s (stratifin) is an early event in breast cancer. Oncogene 20, 3348–3353 [DOI] [PubMed] [Google Scholar]
- 16).Margaret W., Yuriko M., Terri C., Melissa B., Mitchel S.B. (2002) Epidemiology of primary brain tumors: Current concepts and review of the literature. Neuro-Oncology 4, 278–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Shridhar V., Sen A., Chien J., Staub J., Avula R., Kovats S., et al. (2002) Identification of underexpressed genes in early-and late-stage primary ovarian tumors by suppression subtraction hybridization. Cancer Res. 62, 262–270 [PubMed] [Google Scholar]
- 18).Risinger J.I., Maxwell G.L., Chandramouli G.V., Jazaeri A., Aprelikova O., Patterson T., et al. (2003) Microarray analysis reveals distinct gene expression profiles among different histologic types of endometrial cancer. Cancer Res. 63, 6–11 [PubMed] [Google Scholar]
- 19).Dowdy S.C., Gostout B.S., Shridhar V., Wu X., Smith D.I., Podratz K.C., et al. (2005) Biallelic methylation and silencing of paternally expressed gene 3 (PEG3) in gynecologic cancer cell lines. Gynecol. Oncol. 99, 126–134 [DOI] [PubMed] [Google Scholar]
- 20).Robertson K.D. (2005) DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610 [DOI] [PubMed] [Google Scholar]
- 21).Soejima H., Nakagawachi T., Zhao W., Higashimoto K., Urano T., Matsukura S., et al. (2004) Silencing of imprinted CDKN1C gene expression is associated with loss of CpG and histone H3 lysine 9 methylation at DMR-LIT1 in esophageal cancer. Oncogene 23, 4380–4388 [DOI] [PubMed] [Google Scholar]
- 22).Holm T.M., Jackson-Grusby L., Brambrink T., Yamada Y., Rideout W.M., 3rd, Jaenisch R. (2005) Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell 8, 275–285 [DOI] [PubMed] [Google Scholar]
- 23).Huang J., Zhang X., Zhang M., Zhu J.D., Zhang Y.L., Lin Y., et al. (2007) Up-regulation of DLK1 as an imprinted gene could contribute to human hepatocellular carcinoma. Carcinogenesis 28, 1094–1103 [DOI] [PubMed] [Google Scholar]
- 24).Kainz B., Shehata M., Bilban M., Kienle D., Heintel D., Krömer-Holzinger E., et al. (2007) Overexpression of the paternally expressed gene 10 (PEG10) from the imprinted locus on chromosome 7q21 in high-risk B-cell chronic lymphocytic leukemia. Int. J. Cancer 121, 1984–1993 [DOI] [PubMed] [Google Scholar]
- 25).Barsyte-Lovejoy D., Lau S.K., Boutros P.C., Khosravi F., Jurisica I., Andrulis I.L., et al. (2006) The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 66, 5330–5337 [DOI] [PubMed] [Google Scholar]
- 26).Trouillard O., Aguirre-Cruz L., Hoang-Xuan K., Marie Y., Delattre J.Y., Sanson M. (2004) Parental 19q loss and PEG3 expression in oligodendrogliomas. Cancer Genet. Cytogenet. 151, 182–183 [DOI] [PubMed] [Google Scholar]
- 27).Kim J., Kollhoff A., Bergmann A., Stubbs L. (2003) Methylation-sensitive binding of transcription factor YY1 to an insulator sequence within the paternally expressed imprinted gene, Peg3. Hum. Mol. Genet. 12, 233–245 [DOI] [PubMed] [Google Scholar]
- 28).Kim J., Kim J.D. (2008) In vivo YY1 knockdown effects on genomic imprinting. Hum. Mol. Genet. 17, 391–401 [DOI] [PubMed] [Google Scholar]
- 29).Choo J.H., Kim J.D., Chung J.H., Stubbs L., Kim J. (2006) Allele-specific deposition of macroH2A1 in imprinting control regions. Hum. Mol. Genet. 15, 717–724 [DOI] [PubMed] [Google Scholar]
- 30).Choo J.H., Kim J.D., Kim J. (2007) Macro-H2A1 knockdown effects on the Peg3 imprinted domain. BMC Genomics 8, 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Lu Y.C., Song J., Cho H.Y., Fan G., Yokoyama K.K., Chiu R. (2006) Cyclophilin - A protects Peg3 from hypermethylation and inactive histone modification. J. Biol. Chem. 281, 39081–39087 [DOI] [PubMed] [Google Scholar]
- 32).Feng W., Marquez R.T., Lu Z., Liu J., Lu K., Issa J.P., et al. (2008) Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer 112, 1489–1502 [DOI] [PubMed] [Google Scholar]