Abstract
Mitochondrial protein phosphorylation is a well-recognized metabolic control mechanism, with the classical example of pyruvate dehydrogenase (PDH) regulation by specific kinases and phosphatases of bacterial origin. However, despite the growing number of reported mitochondrial phosphoproteins, identity of the protein kinases mediating these phosphorylation events remains largely unknown. The detection of mitochondrial protein kinases is complicated by the low concentration of kinase relative to target protein, lack of specific antibodies and contamination from associated, but non-matrix, proteins. In this study we used blue native gel electrophoresis (BN-PAGE) to isolate rat and porcine heart mitochondrial complexes for screening of protein kinase activity. To detect kinase activity, 1D BN-PAGE gels were exposed to γ-32P-ATP and then followed by SDS gel electrophoresis (2D BN/SDS-PAGE). 32P-labeled dozen of mitochondrial proteins in this setting, including all five complexes of oxidative phosphorylation and several citric acid cycle enzymes. The near ubiquitous 32P protein labeling demonstrates protein kinase activity within each mitochondrial protein complex. The validity of this 2D-BN-PAGE method was demonstrated by detecting the known PDH kinases and phosphatases within the PDH complex band using Western blots and mass spectrometry. Surprisingly, these same approaches detected only a few additional conventional protein kinases suggesting a major role for autophosphorylation in mitochondrial proteins. Studies on purified Complex V and creatine kinase confirmed that these proteins undergo autophosphorylation, and, to a lesser degree, tenacious 32P-metabolite association. In-gel Complex IV activity was shown to be inhibited by ATP, and partially reversed by phosphatase activity, consistent with an inhibitory role for protein phosphorylation in this complex. Collectively, this study proposes that many of the mitochondrial complexes contain an autophosphorylation mechanism, which may play a functional role in the regulation of these multi-protein units.
Keywords: Mitochondria, Kinases, Autophosphorylation, 32P, Proteomics
Mitochondria are dynamically modified organelles that perform several vital functions in the eukaryotic cell. In addition to their role in energy metabolism, mitochondria are also involved in intermediary metabolism, biochemical synthesis, calcium signaling, redox regulation, and apoptosis 1,2. The complexity of mitochondria requires a sophisticated system of intracellular communication, capable of responding rapidly to changes in cellular energy-metabolism as well as many other processes. In recent years, reversible protein phosphorylation has emerged as a potential ubiquitous regulatory mechanism in mitochondria. Several proteomic screening studies, using a combination of SDS gel electrophoresis and mass spectrometry, have revealed an extensive network of phosphoproteins in mitochondria 3–10. Additionally, our lab recently coupled 32P-labeling in intact mitochondria with 2D gel electrophoresis to identify dozens of mitochondrial proteins with potential regulatory (i.e., 32P-turnover) phosphorylation sites 3,5. While data continues to accumulate for the functionality of the mitochondrial phosphoproteome 5,11,12, the exact mechanisms governing reversible phosphorylation remain poorly defined for a majority of mitochondrial proteins. In fact, unlike pyruvate dehydrogenase (PDH) and branched chain α-ketoacid dehydrogenase (BCKDH) complexes, which have their own matrix-localized kinases 13,14, most mitochondrial phosphoproteins have not been linked to specific kinases. To date, no experimental studies have directly examined the localization of active kinases to mitochondria. protein complexes outside of the PDH complex. Thus, identifying the matrix kinases remains an important challenge for resolving the mechanism and regulation of mitochondrial phosphorylation events.
The relatively low abundance of mitochondrial kinases in complex mixtures limits the usefulness of mass spectrometry and gel electrophoresis approaches for detecting these proteins. That is, the ratio of target protein to kinase is on the order of 10 to 100 or even greater, resulting in a very low concentration of kinase protein. Furthermore, current techniques are unable to distinguish mitochondrial kinases from co-purifying contaminants (i.e., cytosolic proteins associated with the mitochondrial outer-membrane). This is especially problematical when a kinase is present in both the cytosol and mitochondrial matrix, as this makes enhancement via mitochondrial purification difficult to interpret 15, 16. For instance, more than ~30% of the heart’s cellular protein is mitochondrial, which means that the enhancement ratio can only approach 4 under the best of conditions.
To better screen for active mitochondrial kinases, we coupled direct ATP-dependent 32P-labeling in isolated protein complexes with blue native gel electrophoresis (BN)-PAGE. Specifically, the non-denaturing conditions of BN-PAGE were used to resolve mitochondrial enzyme complexes in their near native conformation 17 in the pig and rat heart mitochondria. To screen for protein phosphorylation events, these mitochondrial complexes were then incubated in vitro with γ-32P-ATP and resolved into individual protein subunits by SDS gel electrophoresis (2D BN/SDS-PAGE). Autoradiography of the 2D BN/SDS-PAGEs directly evaluated protein phosphorylation and other covalent modifications, since the presence of SDS removed a majority of weak metabolite associations.
This study revealed dozens of kinase-mediated phosphorylations for the proteins associated with all five complexes of oxidative phosphorylation However, despite this extensive network of mitochondrial protein phosphorylation, few kinases—with the exception of the well characterized mitochondrial PDH kinases—were detected. These data suggest a significant role for autophosphorylation within the mitochondrial oxidative phosphorylation protein complexes.
Materials and Methods
Materials
Salts and inorganic phosphate (Pi) were purchased from Sigma (St. Louis, MO). 32P (10 mCi/ml), α-32P-ATP (10 mCi/ml), γ-32P-ATP (10 mCi/ml), and γ-32P-GTP (10 mCi/ml) were purchased from PerkinElmer (Boston, MA). BN-PAGE buffers, reagents and gels were purchased from Invitrogen (Carlsbad, CA). Unless otherwise stated, 2D gel electrophoresis reagents, equipment and software were purchased from GE Healthcare (Piscataway, NJ).
Mitochondrial isolation and incubation conditions
All procedures were performed in accordance with the guidelines described in the Animal Care and Welfare Act (7 U.S.C. 2142 § 13) and approved by the NHLBI Animal Care and Use Committee. Porcine heart mitochondria were isolated from tissue that was cold-perfused in situ to remove blood and extracellular calcium as well as to prevent any warm ischemia, as previously described 3. Isolated rat heart mitochondria were prepared under identical conditions. Mitochondrial preparations were tested for viability by measuring both the respiratory control ratio (RCR) (>8fold) and the ability to maintain matrix ATP (>2nmole/mg mitochondria protein) content, as previously described 3. Protein concentration was determined using a Bradford assay (USB quantification reagent, Cleveland, OH), against a standard curve of BSA.
Our previous study revealed strong 32P-incorporation for several mitochondrial proteins that were de-phosphorylated during isolation as a means of restoring their native, steady-state phosphorylation status 3. Since the current study sought to screen for kinase-mediated 32P-turnover, the mitochondrial protein-phosphate pools were rebuilt by incubating mitochondria (1 mg/ml) in oxygen-saturated buffer A (125 mM KCl, 15 mM NaCl, 20 mM HEPES, 5 mM MgCl2, 1 mM K2EDTA, 1 mM K2EGTA, pH 7.1) containing 5 mM potassium-glutamate, 5 mM potassium-malate and 5 mM inorganic phosphate at 37°C for 20 min. To assure adequate oxygen for oxidative phosphorylation, 100% oxygen was passed over the incubation medium. Immediately following incubation, mitochondria were centrifuged at 10,000 rpm for 5 min at 4°C, and the pellet was used for BN-PAGE.
In vitro 32P-Labeling of Mitochondria Separated by 2D BN/SDS-PAGE
BN-PAGE was used to maintain mitochondrial protein complexes in their intact form 17. BN-PAGE relies on the binding of Coomassie Blue dye to the protein complexes to enhance the differential migration of the protein complexes during the PAGE process. BN-PAGE was performed according to the Invitrogen protocol for the NativePAGE Novex Bis-Tris Gel system 18, with Dodecyl-maltoside as the solubilizing detergent. Four to 16% 1 mm Bis-Tris gels were used, with 75 μg of mitochondrial protein was loaded per well. Electrophoresis was performed at 4°C at 150 V for 1 hr and then at 250 V for 1.3 hr.
Immediately following electrophoresis, BN-PAGE gel slices were cut and incubated with buffer A for 10 min at room temperature to remove excess Coomassie Blue. Each BN-PAGE gel slice was then placed in a 15 ml tube, containing 3.5 ml of buffer A and 350 μCi of α-32P-ATP, γ-32P-ATP, γ-32P-GTP, or 32P. We reasoned that keeping the level of radioactivity was a better normalizing factor than concentration since the specific activities of the metabolites were close to each other. The absolute activity used was primarily based on radiation safety concerns. Based on the specific activity of these metabolites the final concentration of each metabolite was 33 μM α-32P-ATP, 33 μM γ-32P-ATP, 17 μM γ-32P-GTP, or 11 μM 32P BN-PAGE lanes were incubated for 4.5 hr at room temperature, with agitation, and then washed in buffer A four times, for 15 min each to remove exogenous metabolite. BN-PAGE gel slices were incubated for 5 min in 5 ml of buffer A, first containing 50 mg of DTT and then 125 mg of iodoacetemide. Each BN-PAGE gel slice was rinsed briefly with 1x TGS buffer (25 mM Tris (pH 8.3), 192 mM glycine and 0.1% SDS) and applied to a 10–15% SDS gel (Nextgen Sciences, Ann Arbor, MI) and sealed with 1% agarose, containing bromophenol blue. Electrophoresis was performed in an Ettan DALT-12 tank (GE Healthcare, Piscataway, NJ) at 20°C in electrophoresis buffer containing 25 mM Tris (pH 8.3), 192 mM glycine, and 0.2% SDS for ~2100 Vhr. After electrophoresis, 2D BN-PAGE gels were stained in a Coomassie Blue Solution, containing 50% methanol, 3% phosphoric acid and 0.075% (w/v) Coomassie Blue G-250 (Bio-Rad Laboratories, Hercules, CA) for 2 hr. Gels were destained for 30 min with 50% methanol and 3% phosphoric acid, followed by 1.5 hr with 30% methanol and 3% phosphoric acid. Radio-labeled gels were briefly rehydrated with deionized water, placed on filter paper, and dried in a large format dryer (Bio-Rad), before exposing to a phosphor-screen (GE Healthcare) for 72 hrs and scanning on a Typhoon 9410 imager (GE Healthcare), as previously described 3. Non radio-labeled gels were scanned using the Typhoon 9400 scanner at an excitation wavelength of 532 nm, with an emission filter of 560LP.
Mass Spectrometry Identifications
Protein bands from 1D BN-PAGE gels were excised and de-stained overnight, 50% MeOH, 50mM NH4HCO3. In gel digestion was performed as previously described using trypsin or chymotrypsin 19. Peptides were extracted using 50/50/0.1 (v/v/v) water/ACN/HCOOH in a water bath sonicator (25C, 10min), three times. The desalted digests were LC-MS/MS analyzed with an LTQ-Orbitrap Velos (Thermo-Fisher Scientific, LLC) interfaced with an Eksigent nanoLC-Ultra 1D plus system (Dublin, CA). Peptides were first loaded onto an Zorbax 300SB-C18 trap column (Agilent, Palo Alto, CA) at a flow rate of 6 μL/min for 6 min, and then separated on a reversed-phase PicoFrit analytical column (New Objective, Woburn, MA) using a 90-min linear gradient of 5–40% acetonitrile in 0.1% formic acid at a flow rate of 250 nL/min. LTQ-Orbitrap Velos settings were as follows: spray voltage 1.5 kV; full MS mass range: m/z 230 to 2000. The LTQ-Orbitrap Velos was operated in a data-dependent mode; (i.e., one MS1 high resolution (60,000) scan for precursor ions followed by six data-dependent MS2 scans for precursor ions above a threshold ion count of 2000 with collision energy of 35%.
To obtain protein identifications from 2D BN/SDS-PAGE gels, proteins spots were excised, washed and trypsin digested (37C, 4 hours) using an Ettan Spot Handling Workstation (GE Healthcare), and identified with the LTQ-Orbitrap Velos with the same settings as earlier described except for the linear gradient was for 20-min instead. In order to assign protein identifications to the radio-labeled 2D BN/SDS-PAGE gels, radio-labeled and non-radio-labeled gels were aligned, as previously described 3.
All files generated from the LTQ-Orbitrap Velos (.RAW files) were processed with Proteome Discoverer, v1.1 (Thermo-Fisher Scientific, LLC) prior to submitting to our six-processor Mascot cluster at NIH (http://biospec.nih.gov, version 2.3) using the following criteria: database: SwissProt; taxonomy: human+pig+bovine; enzyme: trypsin; miscleavages: 2; fixed modifications: carbamidomethylation (+57 Da); variable modifications: methionine oxidation (+16 Da); acetyl (Prot N-term) and deamidation (NQ), precursor tolerance set to 15 ppm; MSMS tolerance set to 0.8 Da. The combined databases pig+human and bovine were used in this case because porcine protein sequences are not fully available at the present time. For the rat heart mitochondria the rat database was used exclusively. The automatic decoy database search option was selected and the high confidence (FDR, 0.01) peptides were only accepted for protein identification. Briefly, every time a peptide sequence search is performed on a target database a random sequence of equal length is automatically generated and tested. The statistics for matches are calculated and a peptide significance is generated, an in depth explanation can be found at the Matrix Science website (www.matrixscience.com).
Targeted Mass Spectrometry Kinase/Phosphatase Screen
To screen for kinases and phosphatases from both pig and rat, 10 bands of each mitochondrial Complexes I, II, III, IV, V, and PDH were extracted from BN-PAGE lanes, pooled, digested with trypsin and another set with chymotrypsin. The complexes were prepared for mass spectrometry as described above. Liquid chromatography-tandem mass spectrometry was then performed using an Eksigent nanoLC-Ultra 1D plus system (Dublin, CA) coupled to an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, San Jose, CA) using CID fragmentation. Peptides were first loaded onto an Zorbax 300SB-C18 trap column (Agilent, Palo Alto, CA) at a flow rate of 6 μL/min for 6 min, and then separated on a reversed-phase PicoFrit analytical column (New Objective, Woburn, MA) using a 90-min linear gradient of 5–40% acetonitrile in 0.1% formic acid at a flow rate of 250 nL/min. LTQ-Orbitrap Velos settings were as follows: spray voltage 1.5 kV; full MS mass range: m/z 230 to 2000. The LTQ-Orbitrap Velos was operated in a data-dependent mode; (i.e., one MS1 high resolution (60,000) scan for precursor ions followed by six data-dependent MS2 scans for precursor ions above a threshold ion count of 2000 with collision energy of 35%). To generate complementary fragment ions the higher energy collision dissociation (HCD) cell was used to yield different fragmentation patterns and to achieve more identifications. The settings for HCD were the same except for the MS1 resolution (30,000) scan for precursor ions and the collision energy was set to 45% and the MS2 resolution was set to 7500. The raw files generated were processed the same as above except for the chymotrypsin digest samples. The enzyme option was set to chymotrypsin and when the HCD mode was used the MSMS tolerance was set to 0.1 Da.
Data Repository
All positively identified proteins were deposited into Peptidome, National Center for Biotechnology Information (NCBI) peptide data resource center. A positive protein identification consist of the following criteria; 1) Two or more unique peptides with a Mascot peptide rank of 1, an IonScore above the threshold set for high confidence (FDR≤0.01); or 1 unique peptide that was sampled more then once with the same scoring criteria as above. The public repository can be found at www.ncbi.nlm.nih.gov/peptidome (accession pending). These data include all of the proteins indentified as in the 1D BN-PAGE and 1D BN/SDS PAGE gels of the rat and pig.
Western Analysis
Western blots were performed on porcine mitochondrial proteins separated by 2D BN/SDS-PAGE, as described above with the following modifications: BN-PAGE gel slices were applied to 8–16% SDS gels (Bio-Rad) and run in a Criterion Cell (Bio-Rad) for ~210 Vhr. Proteins were blotted to a nitrocellulose membrane and pyruvate dehydrogenase kinase 1 (PDK1) was detected with anti-PDK1 (Stressgen, Ann Arbor, MI) as the primary antibody and Alexa Fluor® 488 anti-rabbit IgG (Invitrogen) as the secondary antibody. Blots were visualized using a Typhoon 9400 imager (GE Healthcare). In addition to the PDK1, several other kinases were screened, including: PDK2, PDK3, PDK4 (see below), JAK1, JAK2, PKCβ1, PKCδ, and PKCε (Santa Cruz Biotechnology, Santa Cruz, CA), PKA Cα and MEK1 (Cell Signaling Technology, Danvers, MA), and MEK6 (Genway Bio, San Diego, CA). Purified kinases were obtained from Invitrogen for MEK6, PKCδ and PKCε, to further validate the antibodies. For studies probing 2D BN/SDS-PAGE gels alongside purified proteins and isolated mitochondria, 0.05 μg of protein and 100 μg of mitochondria were loaded. When probing for PDK4, an antibody from Abgent (San Diego, CA, catalog # AP7041b) was initially used. After conducting several experiments (Supplemental Figure S3), we believe that the dominate interaction of this PDK4 antibody in mitochondrial proteins is with the α-subunit of Complex V, and recommend caution for any laboratories using this antibody to probe for PDK4 in mitochondria.
Auto-catalytic Reactions in Purified Proteins
To screen for auto-catalytic processes, 100 μg of Complex V were purified via immunocapture (Mitosciences, Eugene, OR) and incubated in enzyme buffer (60 mM sucrose, 50 mM Tris-HCl, 50 mM KCl, 4 mM MgCl2, 2mM EGTA, pH 8.0) at a concentration of 0.5 μg/μl for 5 min at 37°C. The enzymatic reaction was started by adding a mixture of 2 mM ATP and 150 μCi [α- or γ-32P-ATP], and incubating for an additional 30 min at 37°C. Cold ATP was added for two reasons: 1) ATP was necessary to achieve proper isoelectric focusing of Complex V and 2) to approach the specific activity of 32P-ATP in the matrix of intact mitochondria. The reaction was stopped by adding 50 μl of lysis buffer (15 mM Tris-HCl, 7 M urea, 2 M thiourea, 4% CHAPS (w/v), pH 8.5) and placing on ice for 15 min. 32P-labeled Complex V was then analyzed by 2D gel electrophoresis, as previously described 3.
Creatine kinase (CK) was also screened for auto-catalytic processes using the method described above, with slight modifications. Ten μg of purified CK (Sigma-Aldrich, St. Louis, MS; catalogue number C9858) were incubated in buffer B (100 mM glycylgycine, 2 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, pH 8.4; adapted from 20) at a concentration of 1 μg/μl for 5 min at 37°C. The reaction was started by a adding a mixture of 2 mM ATP and 50 μCi [α- or γ-32P-ATP], and incubating for an additional 30 min at 37°C. The reaction was stopped by adding 50 μl of lysis buffer and placing on ice for 15 min. 32P-labeled CK was then analyzed by 2D gel electrophoresis, as described above.
Complex IV activity
To determine if ATP altered enzyme activity, gel strips were incubated for 1 hr at room temperature in pre-incubation buffer (125 mM KCl, 15 mM NaCl, 20 mM HEPES, 1 mM EGTA, 1 mM EDTA, 5 mM MgCl2, pH 7.1) with 0 mM ATP to 5 mM ATP. Complex IV activity was then measured for 30 min using an in-gel assay, as previously described 21. To screen for a recovery of Complex IV activity, gel strips were incubated with 0 mM ATP or 5 mM ATP for 2hr or with 5 mM ATP for 1 hr and then with 0 mM ATP or protein phosphatase 1 (PP1) (Sigma, St. Louis, MO) for the subsequent hour.
The linearity of this assay was conducted over protein concentrations of 2.5 to 75 ug of mitochondria protein per lane as well as from 15 to 60 minutes. The assay was linear over all of these concentration and temporal ranges (See Supplemental Figure S1).
Statistical analysis
Data are reported as mean ± standard deviation and were analyzed using a student’s t-test. A p-value less than 0.01 was considered statistically significant, and is denoted by (*).
Results
Protein identifications for one- and two-dimensional BN-PAGE
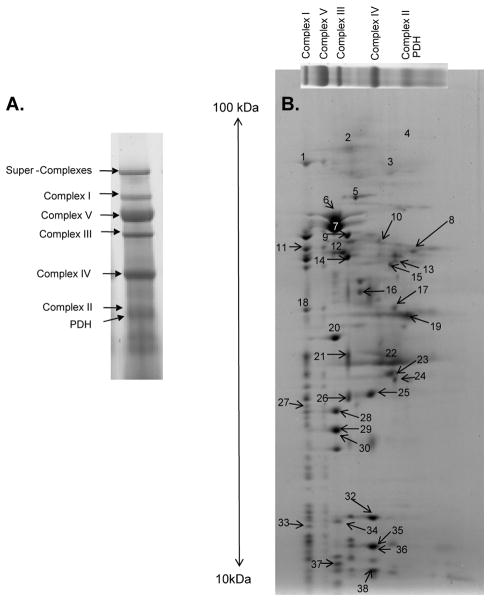
BN-PAGE relies on the separation of intact mitochondrial protein complexes in the first dimension, and 2D BN/SDS-PAGE relies on the separation of the complexes into their individual subunits via SDS gel electrophoresis. In both dimensions, separation is primarily dependent on the molecular weight of the complexes and individual proteins, respectively. A key advantage of 2D BN/SDS-PAGE is that it provides information about protein interactions and the multi-enzymatic processes (i.e., post-translational modifications) that occur at a natural level of cellular compartmentalization 22. Figure 1 shows the dominant mitochondrial protein complexes in each 1D BN-PAGE band and provides identifications for the individual protein subunits separated via 2D BN/SDS-PAGE in the pig. A complete list of mass spectrometry identifications for the 1D BN-PAGE gel are provided in Supplemental Figure S2 and Table S1, and the corresponding identifications for the 2D BN/SDS-PAGE are given in Table 1. The complete spectra and identification criterion have been deposited online as discussed in the Methods section.
Figure 1.
Mitochondrial protein identifications obtained by mass spectrometry. The mitochondrial complex identifications for a 1D BN-PAGE lane (A) and 2D BN/SDS-PAGE gel (B). Numbers in panel B refer to the individual subunit identifications, which are listed in Table 1. Following 1D BN-PAGE, proteins are separated in the vertical direction by molecular weight, from ~150 to 10 kDa.
Table 1.
Mitochondrial protein identifications obtained from a 2D BN/SDS-PAGE gel by mass spectrometry. These 38 proteins correspond to those labeled in Figure 1 and is referenced in Figures 2 and 5.
| 2D BN-PAGE Spot Number | Protein Annotation | Protein Accession Number | # peptides |
|---|---|---|---|
| 1 | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa | Q66HF1 | 5 |
| 2 | Trifunctional enzyme, alpha subunit | P40939 | 2 |
| 3 | Complex II, flavoprotein subunit | Q8HXW3 | 4 |
| 4 | Aconitase | P16276 | 3 |
| 5 | Heat shock protein 60, 60kDa | P18687 | 2 |
| 6 | Complex V, alpha subunit | P25705 | 11 |
| 7 | Complex V, beta subunit | P06576 | 7 |
| 8 | Citrate synthase | O75390 | 3 |
| 9 | Ubiquinol-cytochrome c reductase core protein I | P31800 | 2 |
| 10 | Fumarase | P10173 | 3 |
| 11 | NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa | P17694 | 7 |
| 12 | Creatine kinase | Q3ZBP1 | 3 |
| 13 | Pyruvate dehydrogenase, E1 alpha subunit | P08559 | 4 |
| 14 | Ubiquinol-cytochrome c reductase core protein II | P23004 | 2 |
| 15 | Long-chain specific acyl Coenzyme A dehydrogenase | P51174 | 2 |
| 16 | Isocitrate dehydrogenase | Q28480 | 3 |
| 17 | Pyruvate dehydrogenase, E1 beta subunit | P11177 | 2 |
| 18 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa | Q0MQB3 | 2 |
| 19 | Malate dehydrogenase | P00346 | 6 |
| 20 | Complex V, gamma subunit | P05631 | 5 |
| 21 | Ubiquinol-cytochrome c reductase, heme protein | Q9D0M3 | 2 |
| 22 | ADP/ATP translocase 1 | P48962 | 2 |
| 23 | Succinate dehydrogenase complex, subunit B | P21912 | 5 |
| 24 | Electron transfer flavoprotein, beta subunit | Q6UAQ8 | 2 |
| 25 | Cytochrome c oxidase subunit II | P50667 | 3 |
| 26 | Ubiquinol-cytochrome c reductase, iron-sulfur subunit | P20788 | 4 |
| 27 | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa | P04394 | 4 |
| 28 | Complex V, subunit b | P24539 | 3 |
| 29 | Complex V, O subunit | P13621 | 2 |
| 30 | Complex V, subunit d | AAI04565 | 2 |
| 31 | Peroxiredoxin-3 ** | P35705 | 2 |
| 32 | Cytochrome c oxidase subunit IV | Q95283 | 3 |
| 33 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa | Q9CQC7 | 2 |
| 34 | Complex V, delta subunit | P05630 | 2 |
| 35 | Cytochrome c oxidase subunit Va | P12787 | 3 |
| 36 | Cytochrome c oxidase subunit Vb | Q5S3G4 | 2 |
| 37 | Complex V, subunit f | Q95339 | 2 |
| 38 | Cytochrome c oxidase subunit VIb | P00429 | 3 |
Demonstration of mitochondrial kinase activity by radio-labeled 2D BN/SDS-PAGE
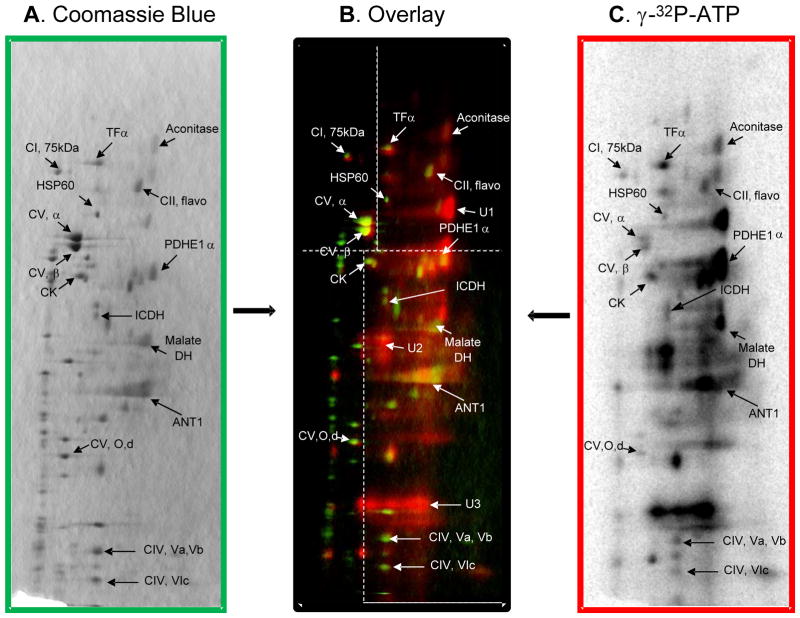
In order to screen for kinase-dependent phosphorylations within intact mitochondrial protein complexes, BN-PAGE gels were incubated with 350 μCi of γ-32P-ATP for 4.5 hours at room temperature, and subsequently resolved into individual subunits by SDS PAGE. As shown in Figure 2C, significant 32P-labeling was observed for dozens of mitochondrial proteins. The overlay image of the γ-32P-ATP-labeled gel and the Coomassie blue stained gel (Figure 2) reveals 32P-incorporation into mitochondrial proteins involved with intermediary metabolism, energy transfer, and all five complexes of oxidative phosphorylation. Notably, intense 32P-labeling was observed for several PDH subunits, malate dehydrogenase, creatine kinase, adenine nucleotide translocase 1 and several subunits of Complexes IV and V. The presence of the PDH complex (PDHC) in these gels is surprising since the molecular weight of PDHC at 10MDa far exceeds that of the normal mitochondrial oxidative phosphorylation Complexes23. Thus, the consistent location of PDH subunits in this region of the gel likely represents reproducible partial breakdown of the PDHC. There were also a few low concentration proteins that had very strong 32P-incorporation, but were below the detection limits of mass spectrometry (i.e., unidentified proteins U1, U2 and U3 in Figure 2B). Importantly, these proteins do not likely represent subunits of the PDH or oxidative phosphorylation complexes, since their lack of Coomassie blue signal implies that they are not stoichiometric with the other subunits in the complexes. Additionally, a few proteins that we previously shown to label with 32P (i.e., branched chain keto-acid dehydrogenase and the E2 and E3-binding protein subunits of PDH3) were not detected in the current experiment, possibly because they were masked by other abundant proteins or not part of a larger protein complex and ran off the 1D BN-PAGE.
Figure 2.
Overlay of a 2D BN/SDS-PAGE following incubation with γ-32P-ATP. The Coomassie blue stained gel (A) is colored green in the overlay (B) and the γ-32P-ATP-labeled gel (C) is colored red. The overlay image in Panel B applies different contrast levels across four regions of the gel image, as indicated by the white dotted lines. Following 1D BN-PAGE, proteins are separated in the vertical direction by molecular weight, from ~150 to 10 kDa. Figure abbreviations correspond to Table 1 as follows: CI, 75kDa, protein #1; TFα, protein #2; CII, flavor, protein #3; Aconitase, protein #4; HSP60, protein #5; CV, α, protein #6; CV, β, protein #7; CK, protein #12; PDHE1α, protein #13; ICDH, protein #16; Malate DH, protein #19; ANT1, protein #22; CV, O, protein #29; CIV, Va, protein #35; CIV, Vb, protein #36; and CIV, VIc, protein #38. Additionally, proteins marked U1, U2 and U3 represent unidentified proteins 1, 2 and 3.
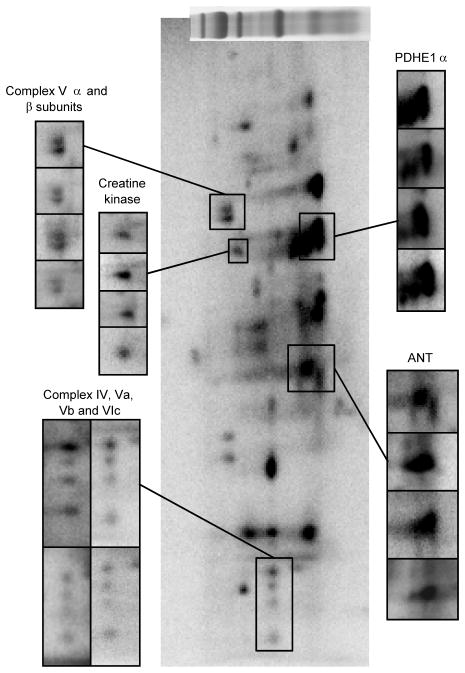
The reproducibility of this technique is demonstrated in Figure 3, where four γ-32P-ATP-labeled 2D BN/SDS-PAGE gels, from four different animals, are presented. Representative examples for the α- and β-subunits of Complex V, creatine kinase, Complex IV subunits IV, Va, Vb, and VIb, ANT1, and PDHE1α show that 32P-labeling is consistent between experiments. The extent of 32P-labeling observed with this strategy implies that active kinases are present in the mitochondrial complexes following resolution by BN-PAGE.
Figure 3.
Reproducibility of a γ-32P-ATP-labeled 2D BN/SDS-PAGE gel. Individual panels show replicates from four different animals in different regions of the gel to demonstrate reproducibility. Each panel is labeled for the dominant 32P-labled protein in the magnified area. Following 1D BN-PAGE, proteins are separated in the vertical direction by molecular weight, from ~150 to 10 kDa.
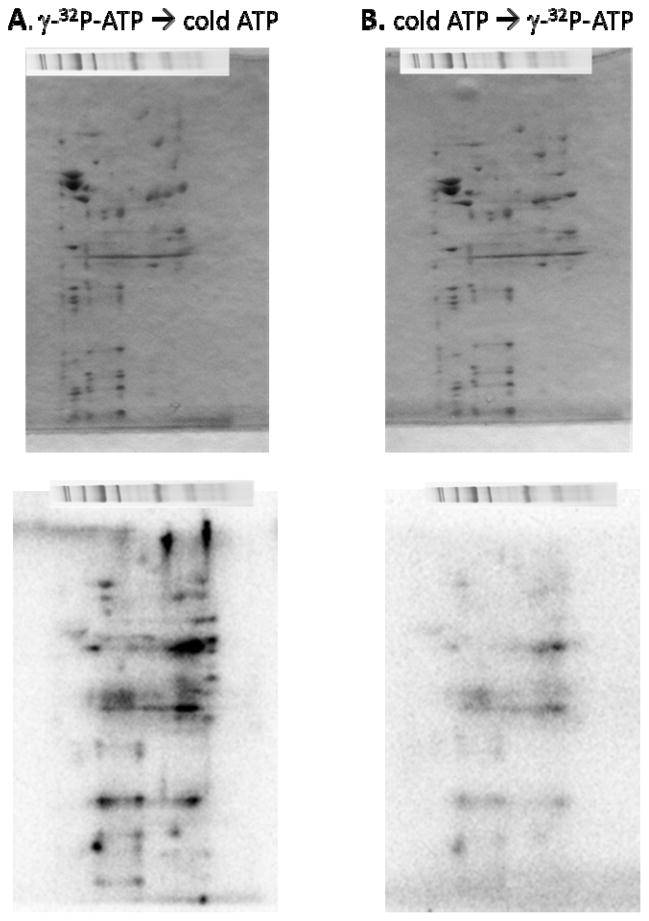
To further characterize the ATP dependent 32P labeling, we conducted competition and chase experiments with cold ATP. In these experiments porcine heart mitochondria BN-PAGE gels were incubated with γ-32P-ATP for 4.5 hours, followed by a 4.5 hour incubation with 1 mM cold (non-radioactive A) or incubated in cold ATP for 4.5 hours, followed by a 4.5 hour incubation with g-32P-ATP. The results of these studies are shown in Figure 4. In a very similar fashion as observed for inorganic 32P additions to intact mitochondria 24, chasing the hot ATP with cold ATP had little effect on the 32P labeling of the proteins. In contrast, adding excess cold ATP before the γ-32P-ATP resulted in a significant and wide spread decrease in 32P labeling. This behavior is consistent with an initial pool building of 32P association sites that once established do not turnover and that if cold ATP occupies these sites first, the 32P from ATP cannot displace it. These data suggest that the kinase activity, or on rate, far exceeds the phosphatase activity, or off rate, under these experimental conditions. An identical situation was reported for the addition of inorganic 32P to isolated mitochondria being energized for the first time in vitro24, likely analogous with the addition of ATP to a blue native gel.
Figure 4.
2D BN/SDS-PAGE chase labeling experiment. The top panel shows the Coomassie blue stained gel and the bottom panel shows the phosphor-image. Panel A shows a 2D BN/SDS-PAGE gel labeled with g-32P-ATP for 4.5 hours, followed by a 4.5 hour incubation with 1 mM cold (non-radioactive ATP). Panel B shows the reverse, a 2D BN/SDS-PAGE gel labeled with cold ATP for 4.5 hours, followed by a 4.5 hour incubation with g-32P-ATP.
Attempted Verification of mitochondrial kinase identification and localization
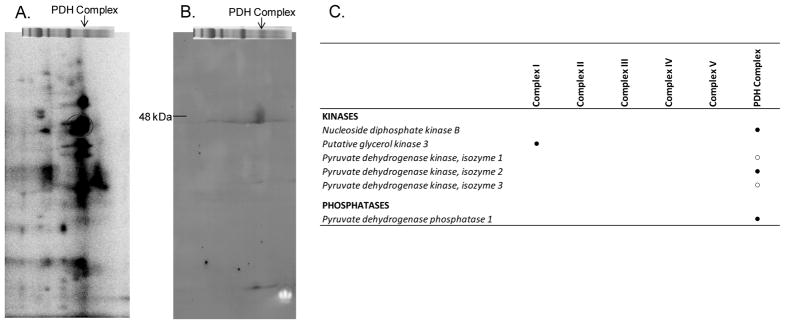
Although the extent of 32P-labeling observed for mitochondrial proteins incubated with γ-32P-ATP provides strong evidence for kinase-mediated protein phosphorylation, the mitochondrial kinases resulting in the observed 32P-labeling must be identified to reach such a conclusion. Thus, we coupled 2D BN/SDS-PAGE with antibody screening to identify specific kinases. PDK1 is a known mitochondrial kinase that phosphorylates PDHE1α. Figure 5A reveals strong 32P-labeling for the PDHE1α subunit, which implies that a functional PDK1 exists within the PDH complex. To confirm, a 2D BN/SDS-PAGE gel was probed for PDK1 in a paired experiment. As shown in Figure 5B, a specific protein localized to the PDH complex and matching the molecular weight of PDK1 (~48 kDa) is detected. Subsequent probing for PDK2, 3 and 4 with commercial antibodies was attempted, but not found to be as specific as the PDK1 antibody (results not shown). Targeted mass spectrometry revealed the presence of PDH phosphatase 1 and confirmed the identification of PDK2 (Figure 5C), with false discovery rates of less than 1%. This screen also confirmed the presence of PDK1 and PDK3, although the false discovery rates exceeded our 1% threshold. Collectively, these PDH complex results demonstrate the feasibility of our 2D BN/SDS-PAGE approach in screening for active mitochondrial kinases and phosphatases though no kinases other than those previously assigned to the mitochondrial matrix were detected.
Figure 5.
Validation that 2D BN/SDS-PAGE can be used to screen for active mitochondrial kinases. PDH kinase 1 activity is demonstrated by the intense 32P-incorproation of the PDHE1α subunit (A), as highlighted by the red circle. A representative Western blot following 2D BN/SDS-PAGE reveals a specific positive hit for PDH kinase 1, which is of the appropriate molecular weight (~48 kDa) and localized to the region of the BN-PAGE gel that contains the PDH complex (B). Following 1D BN-PAGE, proteins are separated in the vertical direction by molecular weight, from ~150 to 10 kDa. Panel C presents a list of kinases and phosphatases that were detected in Complexes I–V and PDH complex using a combination of mass spectrometry and Western blot analysis. (●) denotes proteins identified by targeted mass spectrometry, and (○) denotes proteins identified by Western blot analysis and mass spectrometry, but at an FDR of >1%.
Screen for additional mitochondrial kinases
Several studies in the literature report a role for cytosolic kinases that enter the mitochondria (despite no evidence for a mitochondrial leader sequence) and phosphorylate matrix proteins. Thus, using the same approaches that detected the PDH kinase and phosphatase system, 2D BN/SDS-PAGE gels were probed for a variety of kinases using Western blot analysis (Figure 5 and Supplemental Figure 3) and targeted mass spectrometry (Figure 5C). No evidence in the Western screens or in the targeted mass spectroscopy for classical cytosolic kinases were found in the pig mitochondrial oxidative phosphorylation complexes.
We also repeated the mass spectrometry studies in the rat heart mitochondria blue native gels where the protein data base is better characterized. The rat heart mitochondria BN-PAGE gels also demonstrated 32P incorporation in a similar pattern as the pig in 2D BN/SDS-PAGE gels (supplemental Figure S5). Protein identifications were made in the 2D BN/SDS-PAGE of the rat mitochondrial proteins (Supplemental Figure S6 and Table S2) as well as the intact 1D- BN-PAGE bands (supplemental Figure S7 and Tables S3–S12). Protein identifications were made with trypsin as well as chymotrypsin digestion with many more peptides being detected with trypsin digestion. As found in the porcine data, no protein kinase reached significance within a mitochondria Complex in the rat. Even though the mitochondrial enzyme complexes demonstrate kinase activity in the rat and pig, these multiple screens did not reveal specific associations of cytosolic kinases with mitochondrial complexes. However, the PDH kinases were detected. This result led us to hypothesize that the kinase activity observed in the current study may be due to 32P metabolite associations, mimicking as protein phosphorylations, or previously unknown autophosphorylation mechanisms within the Complexes.
Demonstration of 32P-metabolite association
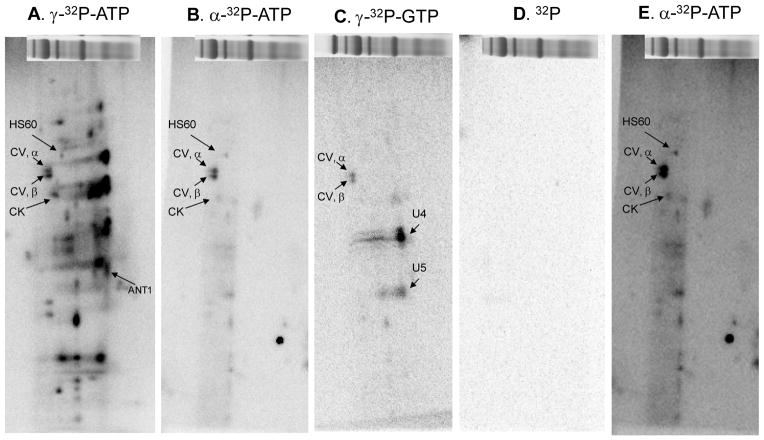
We have previously shown that in crude mitochondrial extracts free inorganic phosphate can bind to succinyl CoA synthetase, α subunit (SCSα) and remain associated throughout the 2D electrophoresis process 25. To test for the contribution of metabolite-associations in these studies, BN-PAGE lanes were incubated with 350 μCi of α-32P-ATP, γ-32P-GTP or 32P for 4.5 hours at room temperature. Incubating with α-32P-ATP can result in ATP-, ADP- and/or AMP-association, whereas incubating with γ-32P-ATP can result in kinase mediated phosphorylation, autophosphorylation and/or ATP-association. Similarly, incubating with γ-32P-GTP can result in kinase dependent phosphorylation, autophosphorylation and/or GTP-association. As shown in Figure 5B, several proteins labeled with α-32P-ATP, including the α- and β-subunits of Complex V as well as heat shock protein 60 and CK (see increased contrast image, Figure 5E). CK and several other proteins showed lower 32P-incorproation when labeled with α-32P-ATP relative to γ-32P-ATP, which implies that their 32P-incorporation results mainly from protein phosphorylation, and not ATP/ADP-binding. In contrast, the extent of 32P-labeling for the α- and β-subunits of Complex V and heat shock protein 60 is approximately equal between α-32P-ATP and γ-32P-ATP incubations (Figures 5A and 5B), which suggests that these proteins label as result of ATP-association and not protein phosphorylation. These findings are consistent with the apparent molecular weight shift associated with 32P incorporation for Complex V’s β-subunit detected Figure 6 and our earlier studies 3,26 as well as reports of an ATP-dependence for heat shock protein 60 27. To screen for additional 32P-metabolite associations, experiments were conducted in the presence of γ-32P-GTP (Figure 5C). As with α- and γ-32P-ATP, the α- and β-subunits of Complex V incorporated γ-32P-GTP. Whether this 32P-label results from phosphorylation or γ-32P-ATP-association is unclear, as Complex V can hydrolyze both ATP and GTP with similar affinity (Supplemental Figure S8). A few additional proteins labeled with γ-32P-GTP, notably two unidentified proteins (U4 and U5) that labeled intensely with γ-32P-GTP. Finally, incubating BN-PAGE lanes with 32P alone did not result in detectable radio-labeling (Figure 5D). This is important because, as mentioned above, incubating BN-PAGE lanes with γ-32P-ATP or -GTP generates free 32P and ADP or GDP at the level of Complex V. The lack of free 32P-labeling confirms that the vast majority of 32P-incorporations observed in these studies result from ATP-dependent covalent processes, rather than artifacts from metabolite associations and the high sensitivity of 32P. These studies did not reveal the previously described 32P-binding to SCSα 25, since this protein was not retained as a complex in this preparation, according to mass spectrometry screens. Overall, these studies demonstrate that a majority of the 32P-labeling observed in these 2D BN/SDS-PAGE gels is dependent on γ-32P-ATP and likely the result of protein phosphorylation. The only tight metabolite incorporations seem to be associated with the α– and β–subunits of Complex V, heat shock protein 60, CK, and a few unidentified proteins.
Figure 6.
Comparison of 2D BN/SDS-PAGE 32P-labeling after 1D BN-PAGE Incubation with Different 32P labeled Metabolites. 2D BN/SDS-PAGE gels following incubation of 1D BN-PAGE porcine heart gels with γ-32P-ATP (A), α-32P-ATP (B), γ-32P-GTP (C), 32P (D), or α-32P-ATP with increased contrast (E). Figure abbreviations correspond to Table 1 as follows: HSP60, protein #5; CV, α, protein #6; CV, β, protein #7; CK, protein #12; and ANT1, protein #22. Additionally, proteins marked U4 and U5 represent unidentified proteins 4 and 5. Following 1D BN-PAGE, proteins are separated in the vertical direction by molecular weight, from ~150 to 10 kDa.
In vitro demonstration of mitochondrial auto-catalytic processes
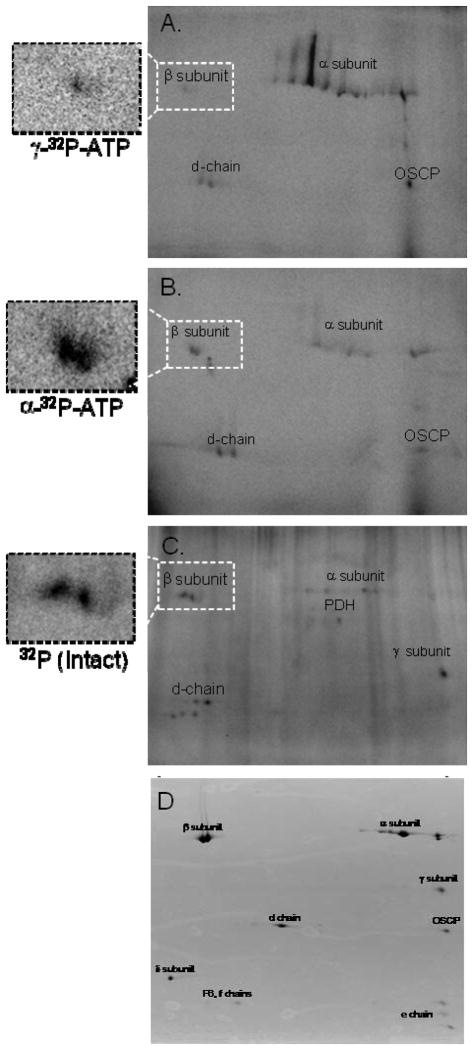
Since exogenous mitochondrial kinases have been difficult to identify, we hypothesized that ATP-dependent autophosphorylation events may be responsible for a majority of the protein kinase activity detected in this study. To evaluate this further, we compared the 32P-labeling patterns of purified Complex V incubated in vitro—with γ-32P-ATP (Figure 7A) or α-32P-ATP (Figure 7B)—with that of Complex V isolated from intact mitochondria that were labeled with 32P under energized conditions (Figure 7C). In vitro 32P-ATP association was shown for the α-, β-, d-, and OSCP subunits, whereas intact 32P-incorporation showed labeling for the α-, β-, γ-, and d-chain subunits, as we previously demonstrated 3,26. For the 32P-ATP studies, the α- and OSCP-subunits were much more intensely labeled with γ-32P-ATP than α-32P-ATP, consistent with a rapid autophosphorylation for these subunits. With regards to Complex V’s β-subunit, incubation with γ-32P-ATP showed weak 32P-incorporation, whereas α-32P-ATP revealed strong labeling and increasing molecular weight for the 32P-labeled component of the β-subunit, similar to Figure 6B and our previous observations in intact mitochondria 3,26. The α-32P-ATP labeling pattern for the β-subunit, coupled with our findings in Figure 7B, suggest that labeling of the β-subunit primarily results from 32P-metabolite associations such as ADP and/or ATP binding. It is important to point out that unlike the β-subunit, which has catalytic metabolite-binding sites, the α-, d- and OSCP subunits do not, and thus it is unlikely that these interactions are related to metabolite binding sites. As described above, incubating purified Complex V with γ-32P-ATP results in free 32P. To ensure that these auto-catalytic studies were not influenced by Complex V’s hydrolytic process, we also incubated purified Complex V with free 32P, but detected no labeling (results not shown). Comparing the in vitro 32P-labeling pattern of purified Complex V (Figures 7A and B) with the pattern obtained from intact mitochondria (Figure 7C), revealed many similarities, with the notable exceptions of OSCP, which only labeled in vitro with 32P-ATP, and the γ-subunit, which only labeled in the intact mitochondria. These data suggest that a majority of Complex V’s 32P-labeling is due to autocatalytic processes that occur within the enzyme complex, whereas the γ-subunit is likely phosphorylated by a matrix kinase that is not tightly associated with Complex V or the configuration of the Complex in the gels inhibits autophosphorylation of this subunit.
Figure 7.
Purified Complex V Radio-labeled in vitro with ATP and in Intact Mitochondria. Panels A and B show Complex V labeled in vitro with γ-32P-ATP and α-32P-ATP, respectively. Panel C shows the labeling pattern of Complex V purified from intact mitochondria following incubation with 32P. Proteins are separated in the horizontal direction by isoelectric focusing point, from pH 3 to 10, and vertically by molecular weight, from ~150 to 10 kDa. Panel D representative protein stain of immuno-captured porcine Complex V as previously described24. All assignments were made by using fiduciary markers on the 32P and protein stained gel as previously described24.
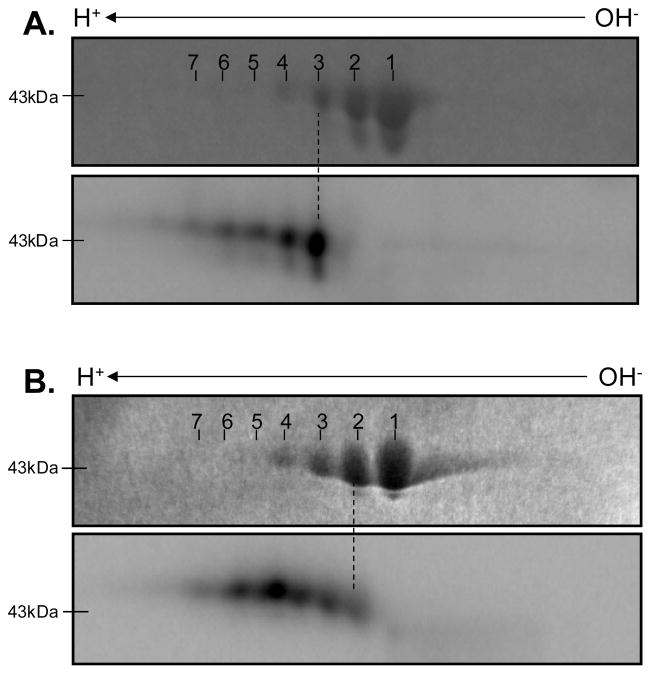
To demonstrate that auto-catalytic processes were not unique to Complex V, we also screened purified creatine kinase (CK). CK was selected for several reasons: 1) it plays an important role in intracellular energy transfer, 2) it labels strongly with γ-32P-ATP and weakly with α-32P-ATP in our 2D BN/SDS-PAGE studies (Figure 6), and 3) there is a debate in the literature as to whether CK is autophosphorylated 20,28,29 or nucleotidylated (i.e., auto-incorporation of the entire adenosine-molecule) 30. For these studies purified CK from the cytosolic MM-fraction was used instead of the mitochondrial-fraction because it was commercially available at very high purity. Incubating CK in vitro with γ-32P-ATP (Figure 8A) revealed that the most intense 32P-incorpation was into the third isoelectric variant, whereas α-32P-ATP-labeling (Figure 8B) was most intense in the fifth isoelectric variant. The differential 32P-labeling of CK’s isoelectric variants show that a greater mole fraction of the protein is labeled with γ-32P-ATP than α-32P-ATP, consistent with the data presented in Figures 7A and 7B above. It should be stressed that extreme care was taken in aligning these gels using four fiduciary points on each gel. This finding suggests that two mechanisms are contributing to CK’s 32P-labeling; we propose that autophosphorylation is dominating the γ–32P-ATP labeling, while the weaker α–32P-ATP labeling reflects nucleotidylation of either AMP or ADP. Thus, both mechanisms debated in the literature seem to be in play for this enzyme, albeit to different extents.
Figure 8.
Purified Creatine Kinase Radio-labeled in vitro with ATP. Panels A and B show γ-32P-ATP and α-32P-ATP incorporation into creatine kinase, respectively. For both panels, the Coomassie blue stained image is above the radio-labeled image. Proteins are separated in the horizontal direction by isoelectric focusing point, from pH 3 to 10, and vertically by molecular weight, from ~150 to 10 kDa.
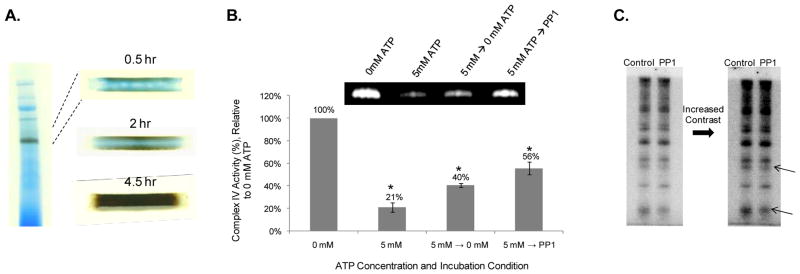
Functional effects of ATP on Complex IV Activity in BN-PAGE gels
To determine if direct incubation of the BN-PAGE lanes with ATP altered enzyme activity, we monitored Complex IV activity in the presence of various ATP concentrations. Complex IV was selected because previous studies have shown that it is inhibited by ATP 31–33. Furthermore, since ATP is not involved in the catalytic process of Complex IV, as is the case for Complex V, any allosteric effect of ATP on this complex could be directly determined in the BN-PAGE gels. Toward this aim, individual BN-PAGE lanes were incubated with ATP contents, ranging from 0 to 5 mM. Complex IV activity was shown to decrease with increasing concentrations of ATP, relative to the 0 mM ATP condition (Figure 9). Whether this ATP-induced inhibition is due to phosphorylation by a kinase associated with CIV, auto-phosphorylation or other allosteric mechanism has yet to be determined. Figure 10 reveals that Complex IV activity recovers in the absence of ATP or with protein phosphatase 1 (PP1), consistent with previous reports 34, further demonstrating that the mitochondrial enzyme complexes are dynamically regulated by ATP.
Figure 9.
Inhibition of Complex IV Activity by ATP. Panel A provides a representative example of the colormetric Complex IV activity assay in the presence of 0 mM and 5 mM ATP. Complex IV activity over an [ATP]-dose response, ranging from 0 mM ATP to 5 mM ATP, is shown in Panel B. The graph in Panel B represents four different biological samples. Data are displayed as a percentage of Complex IV activity, relative to the 0 mM ATP condition.
Figure 10.
Diffusion Limitation of In-Gel Activity Assays. Panel A shows that several hours are required for the small molecules used in the colormetric Complex IV activity assay to penetrate the 1 mm thick BN-PAGE gels. Complex IV activity measurements in response to the absence of ATP and presence of PP1 are shown in Panel B. The graph in Panel B represents four different biological samples, and data are displayed as a percentage of Complex IV activity, relative to the 0 mM ATP condition. Panel C shows a low and high contrast image of the Complex IV subunits, following incubation with γ-32P-ATP for 4.5 hr and a 4.5 hr wash (Control) or a 4.5 hr incubation in PP1 (PP1). The arrows on Panel C point to two potential sites of regulation.
Discussion
The current study garners further support for the notion that mitochondrial protein phosphorylation is extensive and ubiquitous within the enzymes associated with energy metabolism. Widespread 32P-labeling was directly observed in BN-PAGE gels in vitro, with the addition of γ-32P-ATP. ATP is not a metabolic substrate for Complexes I–IV thus 32P incorporation from γ-32P-ATP is not associated with with normal metabolic process. Thus in these Complexes the extensive 32P incorporation demonstrated that, aside from the well recognized energetic functions, they have protein kinase activity. Surprisingly, with the exception of the PDH system, our attempt to detect protein kinases within isolated mitochondrial complexes, using targeted mass spectrometry and Western blot analysis, failed to detect any previously recognized protein kinase. This result suggests that conventional cytosolic kinases may not exist within the mitochondrial complexes, but rather, that these protein complexes have intrinsic kinase activity. Further evidence for autophosphorylation within the mitochondrial complexes was obtained by incubating purified Complex V and CK with α- and γ-32P-ATP in vitro and comparing their labeling pattern to proteins purified following 32P-incorporation in intact mitochondria. With the exception of Complex V’s γ-subunit, the 32P labeling patterns for Complex V in the in vitro and intact mitochondria studies were very similar. Finally, this study revealed that the addition of ATP to the BN-PAGE gels decreased Complex IV activity that was partially reversed by phosphatase, consistent with an inhibitory protein phosphorylation in this complex35.
Detection of Protein Kinases in the Mitochondrial Complexes
The potentially low concentration of mitochondrial protein kinases in total protein mixtures and the uncertainty of isolation artifacts make the detection of matrix protein kinases a major challenge. Take for instance PDH kinase 1 (PDK1). Previous studies have shown that only one or two copies of PDK dimers exist per PDH complex 36. This low mole fraction, among complex mitochondrial protein mixtures, generally places PDK1 below the detection level for most gross mass spectrometry or gel electrophoresis staining efforts. To this point, the most comprehensive mitochondrial proteome study to date by Pagliarini et al 15, did not detect the PDH kinases or virtually any other mitochondrial protein kinases. The 2D BN/SDS-PAGE strategy described here was specifically designed to circumvent these difficulties, by directly assaying for mitochondrial protein kinase activity and screening for protein kinase identifications within the individual complexes. Thus, this approach would enhanced the mole fraction of the kinase molecules in complex mitochondrial protein mixtures. This strategy was validated by detecting several low abundance protein kinases and phosphatases within the well-studied PDH complex. However, even with this enhanced sensitivity, we detected no additional mitochondrial-localized protein kinases, including those that have been previously shown to associate with intact mitochondria.
The fact that each Complex revealed specific protein kinase activity in the gels suggests that the kinase activity is intrinsic to the Complex’s proteins. That is, the protein kinase should be roughly equal in mole fraction to the proteins within the Complex. Since we detected a majority of the subunits for the oxidative phosphorylation complexes, with the exception of the very low molecular weight elements or very hydrophobic, it is unlikely that the concentration is limiting. As further support for this notion, the PDH complex exists at a much lower concentration (>100-fold less) than the oxidative phosphorylation complexes (See Table 2, calculated according to 37,38). The 2D BN/SDS-PAGE approach detected PDH kinases 1, 2 and 3 as well as PDH phosphatases. Thus, the concentration of the oxidative phosphorylation complexes protein kinases/phosphatases should far exceed the sensitivity limit of this approach if the stoichiometry of kinases/phosphatases to target proteins is similar in the PDH and oxidative phosphorylation protein complexes. It is possible that due to the extreme sensitivity of 32P, this assay is able to detect protein kinase activity that affects only a small fraction of the total protein. In other words, protein phosphorylation may only be exhibited by a small percentage of each protein subunit. However, numerous mitochondrial protein phosphorylations have been detected using less sensitive techniques, such as gross mass spectrometry, isoelectric shifts, and phosphorylation sensitive dyes 5,6,8,39,40, implying that a substantial fraction of the proteins are phosphorylated. Additionally, the considerable inhibition of Complex IV by ATP implies that phosphorylation is able to modulate a large percentage of Complex IV’s protein subunit(s) in the gel environment. However, the mole fraction of proteins that are phosphorylated under the conditions used in this study and the specific amino acid sites of the 32P association have yet to be determined.
Table 2.
Protein complex molecules per mitochondria, for PDH Complex and Complexes I–V of oxidative phosphrylation. These values were calculated from 37,38, and 5000 mitochondria per heart cell was assumed.
| Complex molecules/cell | Complex molecules/mitochondrion | |
|---|---|---|
| PDH Complex | 93,372 | 19 |
| Complex I | 6,287,030 | 1,257 |
| Complex II | 9,430,545 | 1,886 |
| Complex III | 18,861,089 | 3,772 |
| Complex IV | 40,865,693 | 8,173 |
| Complex V | 22,004,604 | 4,401 |
What is the nature of 32P association with the mitochondrial protein complexes?
The absence of classical protein kinases the mitochondrial Complexes suggests that the protein phosphorylation processes might be very different from the kinase systems well characterized in the mammalian cytosol. An obvious question becomes what mechanism(s) are responsible for the widespread association of 32P to mitochondrial proteins, as observed in this study and many others 3–10? One suggestion has been that the protein phosphorylations detected by mass spectrometry screen or dyes are the result of phosphorylations that occurred in the cytosol before the protein was transported into the matrix. Though this might be the case for some matrix protein phosphorylations, the fact that we are detecting 32P incorporations in isolated mitochondria and isolated complexes demonstrate that this pool of protein phosphorylations we are evaluating are generated within the mitochondria consistent with protein kinase activity in the matrix. It is very difficult to correlate specific phosphorylation sites detected with protein mass-spectrometry with 32P association. Experimental approaches might include simultaneous studies on isolated peptides using 32P detection and mass spectroscopy or amino acid substitution effects on 32P incorporation
Is the 32P labeling protein phosphorylation or tight metabolite association? We recently demonstrated that non-covalent Pi-binding to SCSα survives the denaturing conditions of SDS gel electrophoresis 25. Thus, to resolve the contribution of protein phosphorylation to the 32P-labeled proteins observed in this study, several controls were performed, including incubation with free 32P to control for inorganic phosphate (Pi) binding, α-32P-ATP to control for ADP and/or ATP association, and γ-32P-GTP to control for GTP metabolites. No Pi-binding was detected in the 2D-BN-PAGE study (Figure 6) or in the purified Complex V studies (Figure 8) upon incubation with 32P. However, strong α-32P-ATP labeling was observed for the α- and β-subunits of Complex V and heat shock protein 60, along with weak association to other mitochondrial proteins, including CK. We confirmed the existence of these reactions by labeling purified CK with α– and γ-32P-ATP (Figure 9). Additionally, a few proteins labeled with γ-32P-GTP, including the α- and β-subunits of Complex V. Therefore, these experiments show that a relatively small degree of metabolite association contributes to the overall 32P-labeling pattern observed in this study. Furthermore, a majority of the 32P-labeled proteins only labeled in the presence of γ-32P-ATP, implying that they were ATP dependent protein phosphorylation. A striking example of this involved ANT1 (Figure 7). ANT1 functions as a gated pore through which ADP and ATP are exchanged between the mitochondrial matrix and the cytoplasm. Thus, while ANT1 contains ADP and ATP binding sites, it did not label with α-32P-ATP, but did label with γ-32P-ATP, providing strong evidence for protein phosphorylation and not metabolite binding in this important transporter. The possible role of metabolite associations is further discussed in the limitations section.
Since mitochondria originated from bacteria and the PDH and branch chain dehydrogenase kinase system shares more sequence homology with bacterial histidine kinases than eukaryotic cytosolic kinases 41, it was logical to search for bacterial characteristics, rather than the conventional eukaryotic kinase targets and pathways within the mitochondrial protein phosphorylations observed here. Bacterial proteins are phosphorylated using a two-component system, which generally involves histidine autophosphorylation and subsequent phospho-transfer to aspartate residues 42. It is important to point out that several conventional serine, threonine and tyrosine protein phosphorylations have now been found in bacteria (for review see 43) thus protein phosphorylations are not limited to 2 component systems in bacteria. Although eukaryotic proteins have also been shown to autophosphorylate 44 to a variety of sites, recently regulatory phosphohistidines have also been described in the eukaryotic cell, such as the mitochondrial branch chain kinase 45 as well as G-proteins and membrane channels46. Interestingly, one of the only other protein kinases detected in this study—nucleoside diphosphate kinase (NDPK) (Figure 5C)—undergoes autophosphorylation on its active histidine residue 47,48. Furthermore, studies have suggested that this phosphate is transferred from the unstable phospho-histidine to a nearby serine residue 48, while others suggest that direct serine autophosphorylation can occur, depending on the concentration of metabolites in the cellular environment 49.
Due to the extensive wide-spread autophosphorylation detected in this study, the bacterial origins of the mitochondria and the phosphohistidine kinases homology of the protein kinases undisputedly in the matrix along with NDPK, makes the possibility that some of these sites could be phospho-histidine or phospho-asparate or other bacterial protein phosphorylation sites is a reasonable consideration. However, the detection of phospho-histidine or aspartate is extremely difficult due to the labiality of these bonds46 especially to the acid conditions normally used in proteomic procedures and the lack of specific antibodies. In early studies on 32P association to mitochondrial proteins it was found that the majority (>90%) of the 32P associations were acid labile 50. Boyer et al described one of these sites as an enzyme catalysis phosphohistidine in succinate thiokinase51,52. Consistent with this notion that many mitochondrial phosphorylation sites are acid labile was our own observations in BN-PAGE or Ghost native gel electrophoresis gels from 32P labeled mitochondria24 that demonstrated the vast majority of the 32P association was acid labile in these minimally solubilized systems consistent with the bacterial protein phosphorylation system (i.e. phosphohistidine, phosphoaspartate etcs) along with possible metabolite associations. Thus, it is possible that some of the 32P protein associations detected in this current study could be small remaining fractions of a phosphor-hisidine or aspartate that survived the electrophoresis process. To test this hypothesis further will require the development of significantly different protein isolation techniques and protein phosphorylation detection schemes46.
Collectively, these findings suggest that mitochondrial protein phosphorylation may be governed by widespread Complex autophosphorylation and/or phospho-transfer mechanisms. As this implies that mitochondrial protein phosphorylation may be more similar to the bacterial two component system than traditional diffusing eukaryotic cytosolic kinases. It is apparent that the protein phosphorylation decreases significantly after the isolation of mitochondria or creation of the BN-PAGE gels based on the observation that initial and nearly irreversible phosphor-protein pools build up upon reintroduction of ATP in the mitochondria matrix 24 or within the BN-PAGE gels (Figure 4). This is also consistent with the original studies on PDHK by Randle53 that also showed a de-phosphorylation after mitochondria isolation that was rapidly reversed with re-energization and warming. The simplest mechanism to explain this behavior is that the ratio of kinase to phosphatase activity is increased with ATP concentration. At high ATP, the kinase activity far exceeds the phosphatase activity explaining the rapid net phosphorylation of protein during rewarming or reintroduction of ATP as well as the fact that once phosphorylated the turnover is extremely slow, revealed in the cold ATP chase experiments, suggesting a very low phosphatase activity at the steady state. These data also suggest that great care must be taken in assuming that freshly isolated mitochondria, for functional studies, reflect the post-translational modification state of the mitochondria, in vivo.
Study limitations: Metabolite Associations, Diffusion Limitations, Mole Fraction of Protein Phosphorylated
As mentioned above, it is important to recognize that metabolite associations involving Pi-binding 25, nucleotidylation 30, and ADP-ribosylation 54 coupled to NAD+ synthesis via NMNAT-3 55 have all been shown to occur in the mitochondrial matrix. It is therefore necessary to take great care when assigning 32P-associations as phosphorylation events. We focused further attention on this topic by comparing the in vitro 32P-labeling patterns of purified Complex V (Figure 7) with the 32P-labeling profile obtained from intact mitochondrial studies 3,5,26. Unlike the α-, β-, and d-subunits, which labeled in vitro with α-32P-ATP and γ-32P-ATP (Figure 6, 7) and with 32P in intact mitochondria 3, the γ-subunit only labels in intact mitochondria. This finding implies that 32P-labeling of the γ-subunit is not “auto-catalytic” within Complex V in the gel environment. As the 2D BN/SDS-PAGE studies described here did not reveal 32P-labeling for Complex V’s γ-subunit, it provides further evidence that γ-subunit phosphorylation is either dependent on a mitochondrial kinase that is not tightly bundled with Complex V, co-factors for the reaction are missing or the conformation of the Complex V in the native gel inhibits this process.
Another important consideration for the current study is the diffusion limitation of assaying directly in BN-PAGE gels. As shown in Figure 10A, several hours are required for small molecules, such as ATP, GTP, Pi, or Ca2+, to penetrate the 1 mm-thick gels. Since this study demonstrated that Complex IV activity was inhibited by ATP (Figure 8) and recovered in the absence of ATP and PP1 (Figure 10B), we next attempted to correlate these changes in activity with changes in phosphorylation (Figure 10C). Unfortunately, due to the large size of PP1 (~38 kDa), this phosphatase was unable to penetrate the BN-PAGE gel and act on the 32P-labeled Complex IV subunits in their entirety. Instead, PP1 only accessed the surface of the BN-PAGE gel, an estimated 15% of Complex IV. This explains the discrepancy between our activity measurement and 32P-data: that is, why a large change in Complex IV activity was observed, with very small changes in the 32P-labeling profile. Since the Complex IV activity assay is run for 30 min, only the protein molecules on the surface are being assayed. Hence, these surface molecules are the same Complex IV molecules that are acted on by PP1, and the activity assay reflects the presumably large mole fraction of Complex IV that is modified by PP1. However, in the 32P-labeling study, ~100% of the Complex IV molecules are exposed to 32P, but only ~15% of them are exposed to PP1. This finding points out that the diffusion limitation of large protein molecules, such as phosphatases, must be considered when conducting this type of analysis in the gel environment. It is also important to point out that these in gel BN-PAGE assays are only evaluating partial reactions of the Complexes outside of the normal membrane environment as well as without a membrane potential. Thus, these assays can only be used as extrapolations to what might be happening in the inner mitochondrial membrane.
Finally, using 32P labeling it is difficult to extrapolate these data to the total mole fraction of an enzyme or complex that is phosphorylated. For signaling molecules, small fractions of a given protein phosphorylation can result in an amplified signal. However, the majority of the phosphorylation sites detected in this study were metabolic enzymes where the only way to significantly influence the activity of these enzymes is to impact a significant fraction of the enzymes 56. While 32P provides insight on protein phosphorylation turnover, it is not very useful in determining the mole fraction of a given protein that is phosphorylated. In addition, the dynamic nature of protein phosphorylation could also complicate this interpretation. 32P labeling, though very sensitive, is a poor marker of the fraction of a protein phosphorylated at any given time. The degree of 32P protein labeling is dependent on turnover of the sites, as well as the total amount of protein phosphorylated. Thus, in the two extremes, a protein could be highly phosphorylated and not turnover providing no labeling at all, while a partially phosphorylated protein with a high turnover could generate a large 32P labeling signal. These complications are why we are interpreting our results as the detection of protein kinase activity, not the degree of phosphorylation of the mitochondria protein complexes. Further studies are required to establish the mole fraction and specific amino acids of the labile protein phosphorylation sites detected in this study,as well as their impact on enzyme function and mechanistic details of the intrinsic protein kinase/phosphatase system.
SUMMARY
The current study demonstrated that the mitochondrial complexes of oxidative phosphorylation have intrinsic protein kinase activity, with virtually no evidence of conventional cytosolic kinases within these Complexes. With the exception of the γ-subunit of Complex V, which may require a matrix dependent kinase reaction, most of the 32P-incorporations detected in intact mitochondria were detected in BN-PAGE resolved Complexes exposed to γ-32P-ATP. This protein kinase activity may represent unrecognized protein kinase motifs in these protein complexes of bacterial origin or highly efficient coupling of a few copies of kinases not detected in this system. It was also demonstrated that the addition of ATP to isolated Complex IV inhibited its enzyme activity, consistent with an inhibitory action of an autophosphorylation reactions or allosteric effects. Further work is required to determine the nature and position of these phosphorylation sites, especially in Complexes I, III and IV which are not classically believed to interact or require ATP for their electron transport function, as well as the kinase and phosphatase system regulating this processes.
Supplementary Material
Acknowledgments
FUNDING INFORMATION: These studies were funded by the NIH Division of Intramural Research.
The authors gratefully acknowledge Ilsa Rovira and Stephanie French for their oversight of the radioisotope studies. We thank Dr Toren Finkel for the laboratory space and Dr Robert Harris for his insightful discussions on the PDH kinases. We are also grateful to Mike Nauman for providing the gel-lane cutting tool.
ABBREVIATIONS
- BN-PAGE
blue native polyacrylamide gel electrophoresis
- 1D BN-PAGE
one-dimensional blue native polyacrylamide gel electrophoresis
- 2D BN/SDS-PAGE
two-dimensional blue native polyacrylamide gel electrophoresis
- PDH
pyruvate dehydrogenase
- BCKDH
branched chain α-ketoacid dehydrogenase
- PDK1
pyruvate dehydrogenase kinase 1
- CK
creatine kinase
- RCR
respiratory control ratio
- ANT1
ADP/ATP translocase 1
- OSCP
oligomycin sensitivity conferring protein
- PP1
protein phosphatase 1
Footnotes
A supplemental file containing supporting data is available. These data include Supplemental FigureS1: Linearity of Complex IV In-Gel Activity Assay. Supplemental Figure S2: 1D BN-PAGE of porcine heart mitochondria. Supplemental Table S1: Mass Spectrometry identification of proteins in the Complex Bands of Figure S2. Supplemental Figure S3. Demonstration that Abgent’s pyruvate dehydrogenase kinase 4 antibody primarily detects the α-subunit of Complex V, not PDK4. Supplemental Figure S4. Negative 2D Western blots of MEK6, PKCδ, and PKCε. Supplemental Figure S5. Rat mitochondria association of 32P in 1D BN/2D SDS PAGE gels after incubation with γ 32P ATP in the 1D BN_PAGE. Supplemental Figure S6. 1D BN/2D SDS PAGE of rate heart mitochondria proteins. Supplemental Table S2. Protein identifications of protein in rat 1DBN/2D SDS PAGE gel. Supplemental Figure S7. 1D BN PAGE gel of rat heart mitochondrial proteins. Supplemental Table S3. Complex 1 band chymotrypsin digestion. Supplemental Table S4. Complex 1 band trypsin digestion. Supplemental Table S5. Complex 2 band chymotrypsin digestion. Supplemental Table S6. Complex 2 band trypsin digestion Supplemental Table S7. Complex 3 band chymotrypsin digestion. Supplemental Table S8. Complex 3 band trypsin digestion. Supplemental Table S9. Complex 4 band chymotrypsin digestion. Supplemental Table S10. Complex 4 band trypsin digestion. Supplemental Table S11. Complex 5 band trypsin digestion. Supplemental Table S12. Complex 5 band chymotrypsin digestion. Supplemental Figure S8. Hydrolysis of ATP and GTP by Porcine Complex V. This material is available free of charge via the Internet at http://pubs.acs.org. All mass spectroscopy data including spectra are available at www.ncbi.nlm.nih.gov/peptidome (accession pending).
Reference List
- 1.Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112(4):481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 2.Pagliarini DJ, Dixon JE. Mitochondrial modulation: reversible phosphorylation takes center stage? Trends Biochem Sci. 2006;31(1):26–34. doi: 10.1016/j.tibs.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Aponte AM, Phillips D, Hopper RK, Johnson DT, Harris RA, Blinova K, Boja ES, French S, Balaban RS. Use of (32)P to study dynamics of the mitochondrial phosphoproteome. J Proteome Res. 2009;8(6):2679–2695. doi: 10.1021/pr800913j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bykova NV, Egsgaard H, Moller IM. Identification of 14 new phosphoproteins involved in important plant mitochondrial processes. FEBS Lett. 2003;540(1–3):141–146. doi: 10.1016/s0014-5793(03)00250-3. [DOI] [PubMed] [Google Scholar]
- 5.Hopper RK, Carroll S, Aponte AM, Johnson DT, French S, Shen RF, Witzmann FA, Harris RA, Balaban RS. Mitochondrial matrix phosphoproteome: effect of extra mitochondrial calcium. Biochemistry. 2006;45(8):2524–2536. doi: 10.1021/bi052475e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinders J, Wagner K, Zahedi RP, Stojanovski D, Eyrich B, van der LM, Rehling P, Sickmann A, Pfanner N, Meisinger C. Profiling phosphoproteins of yeast mitochondria reveals a role of phosphorylation in assembly of the ATP synthase. Mol Cell Proteomics. 2007;6(11):1896–1906. doi: 10.1074/mcp.M700098-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Schieke SM, Phillips D, McCoy JP, Jr, Aponte AM, Shen RF, Balaban RS, Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281(37):27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- 8.Schulenberg B, Aggeler R, Beechem JM, Capaldi RA, Patton WF. Analysis of steady-state protein phosphorylation in mitochondria using a novel fluorescent phosphosensor dye. J Biol Chem. 2003;278(29):27251–27255. doi: 10.1074/jbc.C300189200. [DOI] [PubMed] [Google Scholar]
- 9.Struglics A, Fredlund KM, Konstantinov YM, Allen JF, Moller IM. Protein phosphorylation/dephosphorylation in the inner membrane of potato tuber mitochondria. FEBS Letters. 2000;475(3):213–217. doi: 10.1016/s0014-5793(00)01680-x. [DOI] [PubMed] [Google Scholar]
- 10.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007;104(5):1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee I, Salomon AR, Ficarro S, Mathes I, Lottspeich F, Grossman LI, Huttemann M. cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity. J Biol Chem. 2005;280(7):6094–6100. doi: 10.1074/jbc.M411335200. [DOI] [PubMed] [Google Scholar]
- 12.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87(4):619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 13.Harris RA, Hawes JW, Popov KM, Zhao Y, Shimomura Y, Sato J, Jaskiewicz J, Hurley TD. Studies on the regulation of the mitochondrial alpha-ketoacid dehydrogenase complexes and their kinases. Adv Enzyme Regul. 1997;37:271–293. doi: 10.1016/s0065-2571(96)00009-x. [DOI] [PubMed] [Google Scholar]
- 14.Roche TE, Baker JC, Yan X, Hiromasa Y, Gong X, Peng T, Dong J, Turkan A, Kasten SA. Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog Nucleic Acid Res Mol Biol. 2001;70:33–75. doi: 10.1016/s0079-6603(01)70013-x. [DOI] [PubMed] [Google Scholar]
- 15.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134(1):112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prokisch H, Andreoli C, Ahting U, Heiss K, Ruepp A, Scharfe C, Meitinger T. MitoP2: the mitochondrial proteome database--now including mouse data. Nucleic Acids Res. 2006;34(Database issue):D705–D711. doi: 10.1093/nar/gkj127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schagger H, von JG. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199(2):223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 18.Blinova K, Levine RL, Boja ES, Griffiths GL, Shi ZD, Ruddy B, Balaban RS. Mitochondrial NADH fluorescence is enhanced by complex I binding. Biochemistry. 2008;47(36):9636–9645. doi: 10.1021/bi800307y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boja ES, Hoodbhoy T, Fales HM, Dean J. Structural characterization of native mouse zona pellucida proteins using mass spectrometry. J Biol Chem. 2003;278(36):34189–34202. doi: 10.1074/jbc.M304026200. [DOI] [PubMed] [Google Scholar]
- 20.Stolz M, Hornemann T, Schlattner U, Wallimann T. Mutation of conserved active-site threonine residues in creatine kinase affects autophosphorylation and enzyme kinetics. Biochem J. 2002;363(Pt 3):785–792. doi: 10.1042/0264-6021:3630785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittig I, Karas M, Schagger H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics. 2007;6(7):1215–1225. doi: 10.1074/mcp.M700076-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Reisinger V, Eichacker LA. How to analyze protein complexes by 2D blue native SDS-PAGE. Proteomics. 2007;7(Suppl 1):6–16. doi: 10.1002/pmic.200700205. [DOI] [PubMed] [Google Scholar]
- 23.Henderson NS, Nijtmans LG, Lindsay JG, Lamantea E, Zeviani M, Holt IJ. Separation of intact pyruvate dehydrogenase complex using blue native agarose gel electrophoresis. Electrophoresis. 2000;21(14):2925–2931. doi: 10.1002/1522-2683(20000801)21:14<2925::AID-ELPS2925>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Aponte AM, Phillips D, Hopper RK, Johnson DT, Harris RA, Blinova K, Boja ES, French S, Balaban RS. Use of (32)P to study dynamics of the mitochondrial phosphoproteome. J Proteome Res. 2009;8(6):2679–2695. doi: 10.1021/pr800913j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips D, Aponte AM, French SA, Chess DJ, Balaban RS. Succinyl-CoA synthetase is a phosphate target for the activation of mitochondrial metabolism. Biochemistry. 2009;48(30):7140–7149. doi: 10.1021/bi900725c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aponte AM, Phillips D, Harris RA, Blinova K, French S, Johnson DT, Balaban RS. 32P labeling of protein phosphorylation and metabolite association in the mitochondria matrix. Methods Enzymol. 2009;457:63–80. doi: 10.1016/S0076-6879(09)05004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostermann J, Horwich AL, Neupert W, Hartl FU. Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature. 1989;341(6238):125–130. doi: 10.1038/341125a0. [DOI] [PubMed] [Google Scholar]
- 28.Hemmer W, Furter-Graves EM, Frank G, Wallimann T, Furter R. Autophosphorylation of creatine kinase: characterization and identification of a specifically phosphorylated peptide. Biochim Biophys Acta. 1995;1251(2):81–90. doi: 10.1016/0167-4838(95)00083-7. [DOI] [PubMed] [Google Scholar]
- 29.Quest AF, Soldati T, Hemmer W, Perriard JC, Eppenberger HM, Wallimann T. Phosphorylation of chicken brain-type creatine kinase affects a physiologically important kinetic parameter and gives rise to protein microheterogeneity in vivo. FEBS Lett. 1990;269(2):457–464. doi: 10.1016/0014-5793(90)81215-a. [DOI] [PubMed] [Google Scholar]
- 30.David SS, Haley BE. ATP nucleotidylation of creatine kinase. Biochemistry. 1999;38(26):8492–8500. doi: 10.1021/bi990041b. [DOI] [PubMed] [Google Scholar]
- 31.Arnold S, Kadenbach B. Cell respiration is controlled by ATP, an allosteric inhibitor of cytochrome-c oxidase. Eur J Biochem. 1997;249(1):350–354. doi: 10.1111/j.1432-1033.1997.t01-1-00350.x. [DOI] [PubMed] [Google Scholar]
- 32.Arnold S, Kadenbach B. The intramitochondrial ATP/ADP-ratio controls cytochrome c oxidase activity allosterically. FEBS Lett. 1999;443(2):105–108. doi: 10.1016/s0014-5793(98)01694-9. [DOI] [PubMed] [Google Scholar]
- 33.Helling S, Vogt S, Rhiel A, Ramzan R, Wen L, Marcus K, Kadenbach B. Phosphorylation and kinetics of mammalian cytochrome c oxidase. Mol Cell Proteomics. 2008;7(9):1714–1724. doi: 10.1074/mcp.M800137-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bender E, Kadenbach B. The allosteric ATP-inhibition of cytochrome c oxidase activity is reversibly switched on by cAMP-dependent phosphorylation. FEBS Lett. 2000;466(1):130–134. doi: 10.1016/s0014-5793(99)01773-1. [DOI] [PubMed] [Google Scholar]
- 35.Lee I, Salomon AR, Ficarro S, Mathes I, Lottspeich F, Grossman LI, Huttemann M. cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity. J Biol Chem. 2005;280(7):6094–6100. doi: 10.1074/jbc.M411335200. [DOI] [PubMed] [Google Scholar]
- 36.Yu X, Hiromasa Y, Tsen H, Stoops JK, Roche TE, Zhou ZH. Structures of the human pyruvate dehydrogenase complex cores: a highly conserved catalytic center with flexible N-terminal domains. Structure. 2008;16(1):104–114. doi: 10.1016/j.str.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray J, Gilkerson R, Capaldi RA. Quantitative proteomics: the copy number of pyruvate dehydrogenase is more than 10(2)-fold lower than that of complex III in human mitochondria. FEBS Lett. 2002;529(2–3):173–178. doi: 10.1016/s0014-5793(02)03278-7. [DOI] [PubMed] [Google Scholar]
- 38.Phillips D, Reilley MJ, Aponte AM, Wang G, Boja E, Gucek M, Balaban RS. Stoichiometry of STAT3 and mitochondrial proteins: Implications for the regulation of oxidative phosphorylation by protein-protein interactions. J Biol Chem. 2010;285(31):23532–23536. doi: 10.1074/jbc.C110.152652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19(3):1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulenberg B, Goodman TN, Aggeler R, Capaldi RA, Patton WF. Characterization of dynamic and steady-state protein phosphorylation using a fluorescent phosphoprotein gel stain and mass spectrometry. Electrophoresis. 2004;25(15):2526–2532. doi: 10.1002/elps.200406007. [DOI] [PubMed] [Google Scholar]
- 41.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 42.Umeyama T, Lee PC, Horinouchi S. Protein serine/threonine kinases in signal transduction for secondary metabolism and morphogenesis in Streptomyces. Appl Microbiol Biotechnol. 2002;59(4–5):419–425. doi: 10.1007/s00253-002-1045-1. [DOI] [PubMed] [Google Scholar]
- 43.Leonard CJ, Aravind L, Koonin EV. Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res. 1998;8(10):1038–1047. doi: 10.1101/gr.8.10.1038. [DOI] [PubMed] [Google Scholar]
- 44.Smith JA, Francis SH, Corbin JD. Autophosphorylation: a salient feature of protein kinases. Mol Cell Biochem. 1993;127–128:51–70. doi: 10.1007/BF01076757. [DOI] [PubMed] [Google Scholar]
- 45.Lasker MV, Thai P, Besant PG, Bui CD, Naidu S, Turck CW. Branched-chain 6-ketoacid dehydrogenase kinase: A mammalian enzyme with histidine kinase activity. J Biomol Tech. 2002;13(4):238–245. [PMC free article] [PubMed] [Google Scholar]
- 46.Klumpp S, Krieglstein J. Reversible phosphorylation of histidine residues in proteins from vertebrates. Sci Signal. 2009;2(61):e13. doi: 10.1126/scisignal.261pe13. [DOI] [PubMed] [Google Scholar]
- 47.Morera S, Chiadmi M, LeBras G, Lascu I, Janin J. Mechanism of phosphate transfer by nucleoside diphosphate kinase: X-ray structures of the phosphohistidine intermediate of the enzymes from Drosophila and Dictyostelium. Biochemistry. 1995;34(35):11062–11070. [PubMed] [Google Scholar]
- 48.Shen Y, Kim JI, Song PS. Autophosphorylation of Arabidopsis nucleoside diphosphate kinase 2 occurs only on its active histidine residue. Biochemistry. 2006;45(6):1946–1949. doi: 10.1021/bi051868a. [DOI] [PubMed] [Google Scholar]
- 49.Dorion S, Dumas F, Rivoal J. Autophosphorylation of Solanum chacoense cytosolic nucleoside diphosphate kinase on Ser117. J Exp Bot. 2006;57(15):4079–4088. doi: 10.1093/jxb/erl175. [DOI] [PubMed] [Google Scholar]
- 50.Pressman BC. Metabolic function of phosphohistidine. Biochem Biophys Res Commun. 1964;15(4):556–561. [Google Scholar]
- 51.Peter JB, Boyer PD. The formation of bound phosphohistidine from adenosine triphosphate-P32 in mitochondria. J Biol Chem. 1963;238:1180–1182. [PubMed] [Google Scholar]
- 52.Lindberg O, Duffy JJ, Norman AW, Boyer PD. Characteristics of bound phosphohistidine labeling in mitochondria. J Biol Chem. 1965;240:2850–2854. [PubMed] [Google Scholar]
- 53.Kerbey AL, Randle PJ, Cooper RH, Whitehouse S, Pask HT, Denton RM. Regulation of pyruvate dehydrogenase in rat heart. Mechanism of regulation of proportions of dephosphorylated and phosphorylated enzyme by oxidation of fatty acids and ketone bodies and of effects of diabetes: role of coenzyme A, acetyl-coenzyme A and reduced and oxidized nicotinamide-adenine dinucleotide. Biochem J. 1976;154(2):327–348. doi: 10.1042/bj1540327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frei B, Richter C. Mono(ADP-ribosylation) in rat liver mitochondria. Biochemistry. 1988;27(2):529–535. doi: 10.1021/bi00402a004. [DOI] [PubMed] [Google Scholar]
- 55.Berger F, Lau C, Dahlmann M, Ziegler M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem. 2005;280(43):36334–36341. doi: 10.1074/jbc.M508660200. [DOI] [PubMed] [Google Scholar]
- 56.Phillips D, Reilley MJ, Aponte AM, Wang G, Boja E, Gucek M, Balaban RS. Stoichiometry of STAT3 and mitochondrial proteins: Implications for the regulation of oxidative phosphorylation by protein-protein interactions. J Biol Chem. 2010;285(31):23532–23536. doi: 10.1074/jbc.C110.152652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.