Abstract
Aging brings about numerous cellular defects. Amongst the most prominent are elevated levels of persistent DNA damage, changes to chromatin structure and epigenetic modifications, and alterations of global transcription programs. These are not independent events and recent work begins to shed light on the intricate interplay between these aging-related defects.
Introduction
Aging is a complex process. Long thought of as merely an unprogrammed series of largely random deleterious events and hence considered experimentally intractable, the identification of mutations that promote longevity has made it clear that specific cellular pathways underpin the aging process. While damage to almost any biological molecule has been implicated in aing-related deterioration, it is notable that most human premature aging syndromes are caused by defects in genome surveillance, indicating that DNA damage repair is a central pathway in aging (Freitas et al., 2011; Lombard et al., 2005). This notion is further supported by the fact that one of the most prominent hallmarks of aging cells is the accumulation of various types of DNA damage of which DSBs are the most deleterious (Sedelnikova et al., 2008; Sedelnikova et al., 2004).
In addition to DNA damage, aging brings about dramatic changes in the packaging of DNA into higher-order chromatin structure. Perhaps the most significant of these changes are the evolutionarily conserved, global loss of highly condensed, transcriptionally silent chromatin, or heterochromatin, as well as alterations in histone composition during replicative aging (Feser et al., 2010; O’Sullivan et al., 2010; Tsurumi and Li, 2012). Aging-related chromatin defects are pronounced features of cells from patients with premature aging disorders but are also prominent in aging cell populations in humans, worms and flies (Pegoraro et al., 2009; Scaffidi and Misteli, 2006). The physiological relevance of aging-associated chromatin changes is most evident in the brain, where altered chromatin plasticity has been linked to transcriptional deregulation and concomitant age-related memory impairment (Peleg et al., 2010)**. Notably, reversal of some of these changes abolishes neurodegeneration-associated memory impairments in a mouse model (Peleg et al., 2010)** (Graff et al., 2012).
DNA damage, chromatin defects and changes in global gene expression programs associated with aging are not unrelated events (Fig. 1). We discuss here recent findings highlighting the complex interplay between DNA damage, chromatin and transcription as they occur in the context of aging.
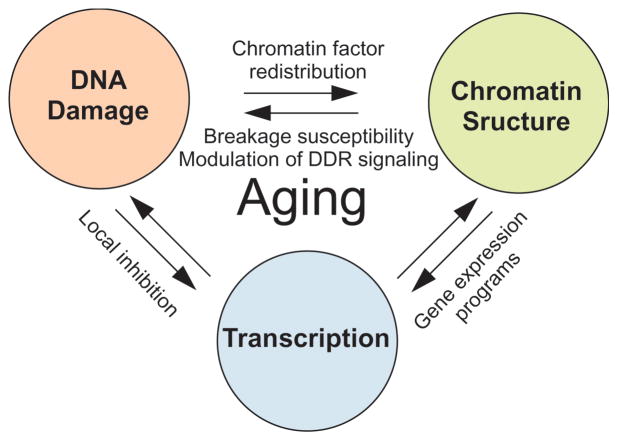
Figure 1. The trinity of DNA damage, chromatin and transcription in aging.
Accumulated DNA damage, altered chromatin structure and changes to local and global gene expression are all hallmarks of aging. These processes are intimately linked. DNA damage leads to local and global changes in gene expression and alterations in chromatin structure, aging-related transcriptional changes affect expression of DNA repair factors and chromatin proteins, and changes in chromatin structure modulate DNA damage responses, breakage susceptibility and influence transcriptional profiles.
Chromatin context affects DNA damage signaling
The sensing of DNA lesions by the DNA damage response (DDR) machinery occurs in the context of the highly complex and heterogeneous chromatin environment (Misteli and Soutoglou, 2009; Shi and Oberdoerffer, 2012). One of the classic hallmarks of the DDR is the phosphorylation of the histone variant H2AX (γ-H2AX), which is important for recruitment and retention of downstream DNA repair factors (Polo and Jackson, 2011). γ-H2AX is primarily generated by the ATM kinase, and subsequent transduction and amplification of the response results in the spreading of this mark to form megabase domains surrounding the damage site (Burma et al., 2001; Rogakou et al., 1999). Recent genome-wide profiling studies have revealed a discontinuous pattern of γ-H2AX spreading, as well as its depletion from actively-transcribed genes after DNA damage, suggesting that precisely controlled γ-H2AX propagation might protect the transcriptional status of genes (Iacovoni et al., 2010). Notably, accumulation of γ-H2AX foci is a characteristic feature of both aged cells and cells from several premature aging disorders (Sedelnikova et al., 2008; Sedelnikova et al., 2004), and may contribute to aging-associated transcriptional deregulation.
The formation of γ-H2AX domains is limited in areas with compact heterochromatin structure, including senescence-associated heterochromatin foci (SAHF) (Di Micco et al., 2011; Goodarzi et al., 2010). The simplest interpretation of the reduced levels of γ-H2AX in heterochromatin is that damage cannot be efficiently recognized in heterochromatin. However, this might be an oversimplification, as damage is efficiently marked by γ-H2AX in highly-condensed mitotic chromosomes, but fails to fully activate the DDR (Giunta et al., 2011)*. An alternative interpretation is that, alterations in chromatin structure, rather than the DSB itself, may be sensed by the DNA damage machinery (Bakkenist and Kastan, 2003; Bencokova et al., 2009; Hunt et al., 2007). It is thus possible that the initial signaling of DNA damage occurs within, and is facilitated by, chromatin structure, and it is instead the amplification of γ-H2AX and the transmission of a full-scale DDR that is restrained by compact chromatin (Fig. 2). This is supported by the finding that experimentally-induced breaks in repetitive heterochromatin move to the periphery of cytologically-detectable heterochromatin compartments prior to recombinational repair factor recruitment (Chiolo et al., 2011; De Piccoli et al., 2006)**. Thus, structural features of heterochromatin may protect, through restraint of the downstream DDR, sequences prone to recombinational rearrangement. Incidentally, heterochromatin domains are reduced in aged cells where recombination and checkpoint signaling appear deregulated (Pegoraro et al., 2009; Sinclair and Guarente, 1997).
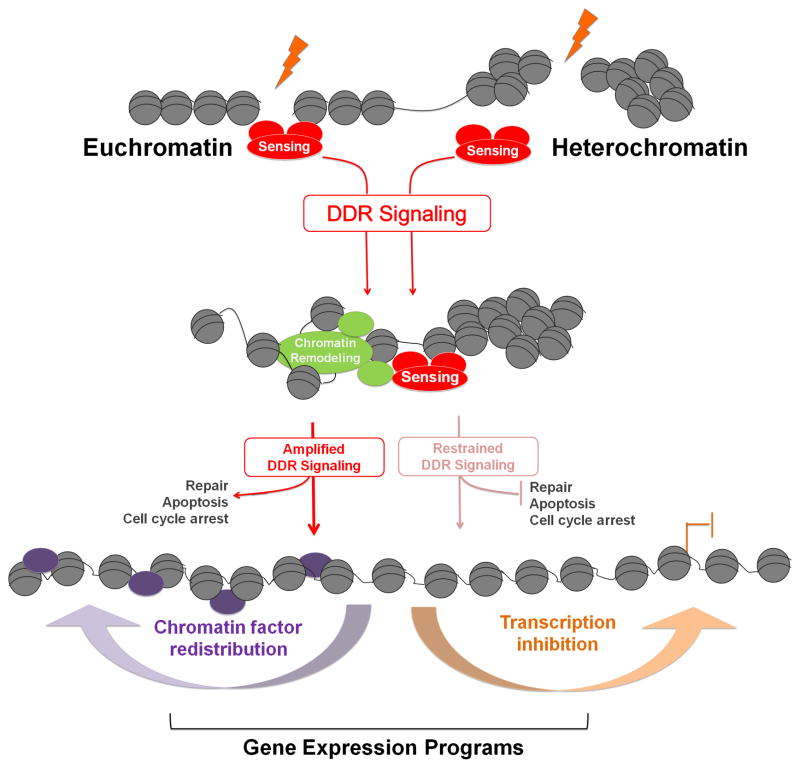
Figure 2. Chromatin context affects DDR signaling.
Double-strand breaks occurring in eu- or hetero-chromatin are sensed by the DDR surveillance machinery and signaling is initiated. Chromatin remodeling complexes are recruited and relax chromatin structure (left pathway). This leads to amplification of DDR signaling and the initiation of downstream effector pathways such as DNA repair, apoptosis, and cell cycle arrest. In the absence of effective chromatin remodeling, and possibly in highly condensed heterochromatin, (right pathway), DDR signaling is restrained and downstream pathways are inhibited. As a result of DDR signaling, global gene expression programs can be altered through chromatin factor redistribution and DDR-induced local and global transcriptional changes.
Intriguingly, the downstream DDR is also restrained in the context of senescence-associated heterochromatin, and shifts the outcome of the DDR from apoptosis to survival and execution of the senescent state (Di Micco et al., 2011), which promotes aging via negative effects on tissue homeostasis (Campisi, 2005). Thus, current evidence suggests that chromatin structure is not refractory to damage detection, but rather that heterochromatin limits and directs the downstream effects of the DDR as a mechanism to protect cells from unwanted consequences of DDR signaling, such as apoptosis or unregulated recombination (Fig. 2).
DNA damage signaling and chromatin changes
Following the sensing of DNA lesions, the site of damage has to be made available for the repair machinery, which requires sequence-level access to the DNA (Heyer et al., 2010; Lieber, 2010). This is achieved via the recruitment of multiple ATP-dependent nucleosome remodeling complexes to DSBs (van Attikum and Gasser, 2009) (Fig. 2). Of particular interest from an aging perspective is the recruitment of the nucleosome remodeling and deacetylase (NuRD) complex, including the CHD4 ATPase subunit, the transcriptional co-repressors MTA1 and MTA2, and the histone deacetylases (HDACs) 1 and 2 (Chou et al., 2010; Larsen et al., 2010; Polo et al., 2010; Smeenk et al., 2010)*. NuRD has previously been associated with the formation of repressive chromatin structures and its loss has been linked to the lack of heterochromatin observed in cells from patients with the premature-aging disease Hutchinson-Gilford Progeroid Syndrome and during normal aging (Pegoraro et al., 2009). On the other hand, CHD4 appears to render chromatin accessible possibly enabling DNA repair factor recruitment (Luijsterburg et al., 2012). Whether these two opposing functions are mediated by the same or different complexes is unknown.
The sirtuin family of NAD+-dependent deacetylases may play a similar role in aging. Mammalian SIRT1, like its yeast ortholog Sir2, is recruited to DNA damage sites where it deacetylates several DNA repair proteins directly and promotes repair (Haigis and Sinclair, 2010). In addition, sirtuins have a role in establishing silent heterochromatin structure in repetitive rDNA and telomeres both in yeast and mammals, where they prevent gene de-repression and aberrant recombination of repeat sequences (Haigis and Sinclair, 2010; Palacios et al., 2010; Tennen et al., 2011). Accordingly, SIRT6 knockout mice show genome instability and premature aging phenotypes (Mostoslavsky et al., 2006). Sirtuins, thus seem to have dual functions as they promote DSB repair but protect from aberrant recombination of repetitive DNA. These properties place NuRD and sirtuins at a potentially central position in preventing aging-associated DNA and chromatin damage.
DDR-induced chromatin alterations and transcription
A plethora of chromatin modifications have been reported to accumulate at DSBs. Emerging evidence suggests that these changes affect gene expression globally and locally (Shi and Oberdoerffer, 2012) (Fig. 2, 3).
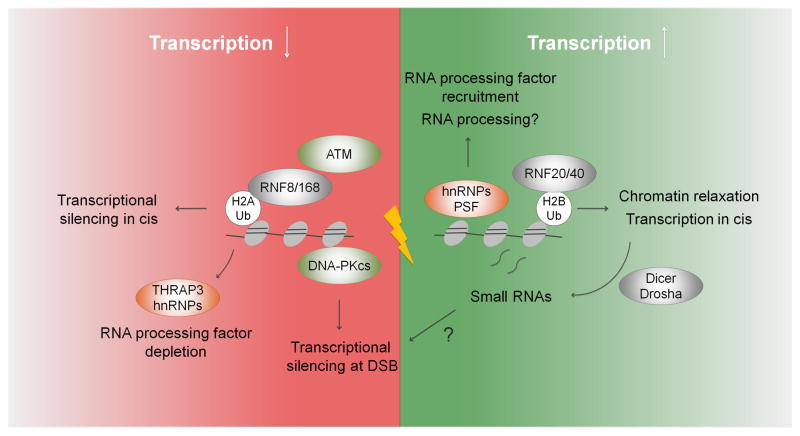
Figure 3. The ups and downs of transcription at DSBs.
DNA damage both promotes and represses transcription. DNA damage stimulates the (transient) recruitment of some RNA processing factors to DSBs and triggers local ubiquitination of H2B by RNF20/40. Both processes have been linked to active transcription, which may aid chromatin relaxation required during the early phases of repair. DSB site-specific RNA transcripts were found to be processed into small RNAs required for DSB repair. On the other hand, downstream DDR signaling causes transcriptional repression at DSBs within genes via DNA-PK and in cis to DSBs over a range of several kb via ATM, RNF8/168 and Ub-H2A. Depletion of a subset of RNA processing factors from DSBs is consistent with loss of transcription near DSBs. Notably, small RNAs have been implicated in gene silencing in several species, suggesting a possible role in DSB-proximal silencing.
Of particular relevance is the ubiquitin landscape in the chromatin regions surrounding the DSB. Ubiquitination of H2A (Ub-H2A) by the RNF8/RNF168 ubiquitin ligase complex is a key chromatin feature involved in many aspects of DDR signaling, and is also strongly associated with transcriptional repression (Bekker-Jensen and Mailand, 2011) (Fig. 3). The spreading of Ub-H2A from sites of damage is correlated with DSB-proximal transcriptional repression in cis, a phenomenon dependent upon ATM signaling as well as RNF8 and RNF168 (Shanbhag et al., 2010)**. Moreover, the ubiquitin ligase subunits of Polycomb repressive complex 1 (PRC1), RING1 and RING2, accumulate at DNA breaks and recruitment of Polycomb components such as EZH2 have been linked to exclusion of transcriptionally active RNA Pol II from sites of damage (Chou et al., 2010; Ginjala et al., 2011; Ismail et al., 2010). While RNF8/168-dependent polyubiquitination of H2A is required for the recruitment of downstream repair factors, the exact role of PRC1 recruitment and concomitant H2A-monoubiquitination in DSB repair is less clear. Given that both modifications have been associated with a DSB-proximal transcriptional silencing, it will be interesting to determine if, and how, DSB-induced repressive chromatin modulates DSB repair.
In addition to Ub-H2A, ubiquitinated H2B also accumulates at DSBs (Moyal et al.; Nakamura et al.). Unlike Ub-H2A, Ub-H2B is normally associated with transcriptional elongation (Weake and Workman, 2008), which appears to be at odds with the observation of DSB-associated RNA PolII depletion from DSBs. One possibility is that Ub-H2B creates a permissive chromatin environment for repair in the immediate vicinity of the break, while the ubiquitin-H2A-mediated transcriptional silencing radiates further from the break (Shiloh et al., 2011). In agreement with the idea of multiple chromatin compartments around a damage site, longer-range silencing of RNAPII transcription appears to be ATM-mediated, while expression of RNAPII-transcribed genes suffering from a single internal strand break appears to be arrested by DNA-PK-dependent mechanisms (Pankotai et al., 2012; Shanbhag et al., 2010)** (Fig. 2).
Many of the DNA damage-related transcriptional changes to the genome are likely transient and swiftly restored upon cessation of DDR signaling. Nevertheless, the inability to restore the original chromatin environment after DDR-enforced chromatin restructuring has the potential to lead to misregulation of large sets of genes, not limited to those at the site of damage. A mechanism for such global misregulation may be the redistribution of the histone deacetylase SIRT1 (Oberdoerffer et al., 2008). Upon DNA damage SIRT1 was found to be recruited to damage sites to facilitate repair, leading in turn to its depletion elsewhere in the genome, which can result in derepression of SIRT1-regulated genes, mimicking transcriptional changes seen in aging cells. This redistribution of a chromatin modifier appears to be an evolutionarily conserved process, as the Sir proteins in yeast also redistribute to DNA damage leading to derepression of undamaged loci and an aging phenotype (Neame, 2009). Similarly, oxidative stress results in the relocalization of members of the SIRT1-containing Polycomb repressive complex 4 and DNA methyltransferase 1 and 3B, which was shown to produce persisting epigenetic changes involving DNA methylation at damaged genes (O’Hagan et al., 2011)**. Notably, DNA damage was also found to promote transcriptional changes that resemble those seen in aged heart tissue, although the underlying mechanisms for this observation are not clear (Bahar et al., 2006).
DNA damage and RNA processing
A novel aspect of DNA break repair has emerged recently that may have significant consequences for how DNA damage affects gene expression: the involvement of RNA processing components in DNA repair (Fig. 3). The first hint at this link was the observation that a member of the SR family of pre-mRNA splicing factors, SRSF1, was required for the maintenance of genomic stability. Loss of SRSF1 resulted in increased DNA break formation, a process that was attributed, at least in part, to increased RNA:DNA hybrid, or R loop, formation (Li and Manley, 2005). Extending these observations, an unbiased, genome-wide siRNA screen for pathways that mediate genome stability revealed a striking overrepresentation of genes involved in mRNA processing and pre-mRNA splicing, and splicing factors were found to be one of the most prominently mis-regulated groups of genes in response to inhibition of clinically used toposiomerase I inhibitors (Paulsen et al., 2009; Solier et al., 2010). In addition, proteomic screens for mediators of the DDR showed enrichment for RNA processing factors and several of them appeared to be targets of DDR-associated post-translational modifications (Adamson et al., 2012; Beli et al., 2012; Matsuoka et al., 2007)**. DNA damage also promotes changes in the expression patterns of RNA processing factors, and several interactions between heterogeneous nuclear ribonucleoproteins (hnRNPs) and effectors of the DDR have been reported (Haley et al., 2009). Circumstantial evidence for a role of RNA processing factors in the DNA break repair process itself came from a recent genome-wide search for mediators of homologous recombination, which identified RNA post-transcriptional modifiers as the most significantly overrepresented gene category (Adamson et al., 2012). Together, these findings link DDR signaling to biochemical, and possibly functional, changes in RNA processing factors.
Consistent with a DSB site-specific function of RNA processing factors in the DDR, their nuclear distribution is affected by DNA damage. For example, the PPM1G phosphatase is recruited to sites of laser-induced DNA damage, whereas the splicing-associated protein THRAP3 is depleted (Beli et al., 2012)**, and several factors including NONO and the hnRNPs RBMX and hnRNPUL-1 and -2 transiently associate with DSBs (Adamson et al., 2012; Polo et al., 2012; Salton et al., 2010). The only current evidence for functional relevance of these factors is the finding that the hnRNPUL1 and -2, which act downstream of the DSB sensor complex MRE11-RAD50-NBS1 (MRN), mediate resection of broken DNA ends, a process essential for repair by homologous recombination (Polo et al., 2012). Notably, recruitment of HNRNPUL proteins to sites of damage appears to be independent of its role in RNA metabolism as it remained unaffected by transcriptional inhibition or RNase treatment. Similarly, transient recruitment of RBMX to DNA breaks was independent of its RNA recognition motif (RRM) (Adamson et al., 2012). Clearly, much remains to be learned about the role of RNA processing factors in DNA repair, but this tentative link has recently been strengthened, by the observation that site-specific RNA transcripts processed by Dicer and Drosha into small RNAs are required for effective DSB repair (Francia et al., 2012)** (Fig. 3).
Conclusions
DNA damage has long been considered a major culprit in aging. However, our view of its effect has been limited to its role as a driver in the accumulation of mutations. It is becoming evident that DNA damage can affect nuclear function and aging in many more ways. We now know that DNA damage can promote global reorganization of chromatin, which in turn affects susceptibility to DNA damage. DNA damage-induced changes in chromatin structure may also affect the integrity of the transcriptome both by altering gene expression patterns and possibly by titrating away RNA binding factors normally needed for accurate pre-mRNA processing. As if the problem were not complex enough, it now appears that understanding aging will not only require identification of the multiple pathways involved in the process, but maybe more importantly, it will be necessary to uncover the intricate interplay between the various pathways.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Mailand N. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 2011;585:2914–2919. doi: 10.1016/j.febslet.2011.05.056. [DOI] [PubMed] [Google Scholar]
- **.Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C. Proteomic Investigations Reveal a Role for RNA Processing Factor THRAP3 in the DNA Damage Response. Mol Cell. 2012;46:212–225. doi: 10.1016/j.molcel.2012.01.026. A parallel quantification of the DDR-regulated acetylome and phospho-proteome identifies a network of RNA processing factors as modified in response to DNA damage. At least some of these factors are recruited to or depleted from sites of damage, suggesting a diverse role for RNA metabolism in the repair process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencokova Z, Kaufmann MR, Pires IM, Lecane PS, Giaccia AJ, Hammond EM. ATM activation and signaling under hypoxic conditions. Mol Cell Biol. 2009;29:526–537. doi: 10.1128/MCB.01301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- **.Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. This report shows, for the first time in metazoans, that breaks induced in heterochromatin relocate to the periphery of the chromatin domain before the initiation of recombinational repair. This suggests that there might be a mechanism, as in yeast, that relocalizes repeats undergoing repair to the outside of their heterochromatin compartment, to control recombinational repair and stabilize repeats. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. This study demonstrates the recruitment of the NuRD complex component CHD4 to DNA damage sites. CHD4 was further shown to be involved in efficient repair of damage, suggesting that downstream chromatin changes facilitate repair factor assembly and subsequent repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piccoli G, Cortes-Ledesma F, Ira G, Torres-Rosell J, Uhle S, Farmer S, Hwang JY, Machin F, Ceschia A, McAleenan A, et al. Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat Cell Biol. 2006;8:1032–1034. doi: 10.1038/ncb1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Sulli G, Dobreva M, Liontos M, Botrugno OA, Gargiulo G, dal Zuffo R, Matti V, d’Ario G, Montani E, et al. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol. 2011;13:292–302. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. Elevated histone expression promotes life span extension. Mol Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **.Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d’Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012 doi: 10.1038/nature11179. In press This is the first description of a role for DSB site-specific small RNAs (DDRNAs) in DSB repair. DDRNAs are generated by Dicer and Drosha and are required for the formation of downstream repair factor recruitment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas AA, Vasieva O, de Magalhaes JP. A data mining approach for classifying DNA repair genes into ageing-related or non-ageing-related. BMC Genomics. 2011;12:27. doi: 10.1186/1471-2164-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginjala V, Nacerddine K, Kulkarni A, Oza J, Hill SJ, Yao M, Citterio E, van Lohuizen M, Ganesan S. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol. 2011;31:1972–1982. doi: 10.1128/MCB.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Giunta S, Belotserkovskaya R, Jackson SP. DNA damage signaling in response to double-strand breaks during mitosis. J Cell Biol. 2011;190:197–207. doi: 10.1083/jcb.200911156. This study shows for the first time that damaged mitotic chromosomes initiate DDR signaling. Interestingly, downstream effects of DDR, such as checkpoint and repair are not engaged in the mitotic chromatin environment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Jeggo P, Lobrich M. The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax. DNA Repair (Amst) 2010;9:1273–1282. doi: 10.1016/j.dnarep.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, Nieland TJ, Fass DM, Kao PF, Kahn M, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Paunesku T, Protic M, Woloschak GE. Response of heterogeneous ribonuclear proteins (hnRNP) to ionising radiation and their involvement in DNA damage repair. Int J Radiat Biol. 2009;85:643–655. doi: 10.1080/09553000903009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt CR, Pandita RK, Laszlo A, Higashikubo R, Agarwal M, Kitamura T, Gupta A, Rief N, Horikoshi N, Baskaran R, et al. Hyperthermia activates a subset of ataxia-telangiectasia mutated effectors independent of DNA strand breaks and heat shock protein 70 status. Cancer Res. 2007;67:3010–3017. doi: 10.1158/0008-5472.CAN-06-4328. [DOI] [PubMed] [Google Scholar]
- Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol. 2010;191:45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Rendtlew Danielsen JM, Menard P, Sand JC, Stucki M, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010;190:731–740. doi: 10.1083/jcb.200912135. This study demonstrates the recruitment of the NuRD complex component CHD4 to DNA damage sites. CHD4 was further shown to be involved in efficient repair of damage, suggesting that downstream chromatin changes facilitate repair factor assembly and subsequent repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Luijsterburg MS, Acs K, Ackermann L, Wiegant WW, Bekker-Jensen S, Larsen DH, Khanna KK, van Attikum H, Mailand N, Dantuma NP. A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO J. 2012;31:2511–2527. doi: 10.1038/emboj.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Misteli T, Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- **.O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. This study documents the relocalization of members of the Polycomb repressive complex 4, including SIRT1 and DNA methyltransferases upon oxidative DNA damage. Mobilization of this silencing complex may lead to aberrant global transcriptional changes and aberrant cancer -specific DNA methylation as a result of DNA damage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, Blasco MA. SIRT1 contributes to telomere maintenance and augments global homologous recombination. J Cell Biol. 2010;191:1299–1313. doi: 10.1083/jcb.201005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **.Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol. 2012;19:276–282. doi: 10.1038/nsmb.2224. This article reports that upon induction of a single double-strand break in a transcriptionally-active gene, RNA Polymerase II elongation is halted by a DNA-PK-dependent mechanism. This suggests that active mechanisms inhibit transcriptional activity at the site of a lesion. Together with Shanbhag et al.,( 2010), this suggests that inhibition of transcriptional activity at the site of a lesion is context dependent. [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro G, Kubben N, Wickert U, Gohler H, Hoffmann K, Misteli T. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **.Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. Using a genome-wide approach, this study demonstrates that a change in chromatin plasticity, and specifically histone H4 K12 acetylation is linked to age-associated memory impairment in mice. Importantly, restoration of physiological H4K12 acetylation reinstates cognitive abilities. [DOI] [PubMed] [Google Scholar]
- Polo SE, Blackford AN, Chapman JR, Baskcomb L, Gravel S, Rusch A, Thomas A, Blundred R, Smith P, Kzhyshkowska J, et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol Cell. 2012;45:505–516. doi: 10.1016/j.molcel.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. This study demonstrates the recruitment of the NuRD complex component CHD4 to DNA damage sites. CHD4 was further shown to be involved in efficient repair of damage, suggesting that downstream chromatin changes facilitate repair factor assembly and subsequent repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton M, Lerenthal Y, Wang SY, Chen DJ, Shiloh Y. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2010;9:1568–1576. doi: 10.4161/cc.9.8.11298. [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova OA, Horikawa I, Redon C, Nakamura A, Zimonjic DB, Popescu NC, Bonner WM. Delayed kinetics of DNA double-strand break processing in normal and pathological aging. Aging Cell. 2008;7:89–100. doi: 10.1111/j.1474-9726.2007.00354.x. [DOI] [PubMed] [Google Scholar]
- Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- **.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. Using a single-cell reporter system, these authors show that ATM kinase and RNF8 and 168 ubiqutin ligases are involved in producing chromatin changes that result in transcriptional silencing in cis to double-strand breaks. This phenomenon, which the authors have dubbed DISC (DNA double-strand break-induced silencing in cis) appears to require repressive ubiquitinated H2A chromatin marks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Oberdoerffer P. Chromatin dynamics in DNA double-strand break repair. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbagrm.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y, Shema E, Moyal L, Oren M. RNF20-RNF40: A ubiquitin-driven link between gene expression and the DNA damage response. FEBS Lett. 2011;585:2795–2802. doi: 10.1016/j.febslet.2011.07.034. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- *.Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190:741–749. doi: 10.1083/jcb.201001048. This study demonstrates the recruitment of the NuRD complex component CHD4 to DNA damage sites. CHD4 was further shown to be involved in efficient repair of damage, suggesting that downstream chromatin changes facilitate repair factor assembly and subsequent repair. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solier S, Barb J, Zeeberg BR, Varma S, Ryan MC, Kohn KW, Weinstein JN, Munson PJ, Pommier Y. Genome-wide analysis of novel splice variants induced by topoisomerase I poisoning shows preferential occurrence in genes encoding splicing factors. Cancer Res. 2010;70:8055–8065. doi: 10.1158/0008-5472.CAN-10-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennen RI, Bua DJ, Wright WE, Chua KF. SIRT6 is required for maintenance of telomere position effect in human cells. Nat Commun. 2011;2:433. doi: 10.1038/ncomms1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi A, Li W. Global heterochromatin loss: A unifying theory of aging? Epigenetics. 2012:7. doi: 10.4161/epi.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]