Abstract
A family of branched polyrotaxanes (bPRTx+), threaded with multiple cationic α-cyclodextrins (α-CDs) onto a multi-armed poly(ethylene glycol) (PEG) core, were synthesized and studied as gene silencing vectors. These bPRTx+ formed stable, positively charged complexes with diameters of 150–250 nm at N/P ratios as low as 2.5. The bPRTx+ materials were shown to have gene-silencing efficiencies comparable to those of Lipofectamine 2000 (L2k) and bPEI, while displaying similar toxicity profiles. The unique structure of these polyrotaxanes allows them to effectively condense and complex siRNA into nanoparticles at much lower N/P ratios than L2k or bPEI. These findings suggest that bPRTx+ may be useful materials for gene therapy applications.
Introduction
Since its discovery in 1998, RNA interference (RNAi) has gained considerable attention for its potential application in the treatment of diseases such as varied neurological, viral, cancer and heart diseases.1 Small interfering RNAs (siRNAs) are 21–23 nucleotide long fragments that are incorporated into the RNA-interference silencing complex (RISC) in the cytoplasm of the cell.2 A multifunctional protein present inside the RISC known as Argonaut 2 then unwinds the siRNA, leading to cleavage of the sense strand.2 The antisense strand is retained within the activated RISC where it helps to selectively bind and degrade its complementary mRNA target.2 Due to its catalytic nature, appropriately designed siRNA can theoretically silence any gene in the body via cleavage of multiple mRNA strands, thus making it a promising tool for therapy.2 The primary advantage of RNAi over conventional chemotherapy is its high specificity and the action of siRNA “upstream” from most chemotherapeutic agents, thereby conferring the ability to potentially evade drug resistance by targeting virtually any transcript to knock down protein expression of the selected sequence.3
Despite advances in the identification of various gene targets and therapeutic siRNAs, the clinical success of RNAi is greatly impeded by the lack of a robust, safe, efficient, and manufacturable delivery vector.4 Many viral and non-viral vectors have been studied for this purpose, but they all suffer from key limitations. Immunogenicity and safety issues hamper the broad utilization of otherwise efficient and persistent viral vectors.5 Non-viral vectors, on the other hand, are primarily limited by their lack of efficiency; however, they are generally safer, far less immunogenic and scalable. Such features make non-viral gene delivery an attractive option for gene therapy.6
A variety of non-viral vectors have been studied for siRNA delivery, including cyclodextrin-oligomers,7–10 cationic lipids,11–13 lipid/calcium based formulations,14 and gold15 or PLGA16 nanoparticles. All these vehicles are capable of forming nanoparticles that are smaller than 200 nm and can efficiently deliver siRNA into target cells. These particles can also protect the genetic material from degradation and enhance their cell permeability. However, most of these particles face serum stability and acute toxicity challenges. Development of biodegradable, low toxicity materials for use as siRNA vehicles is one means to address these problems.17
Cyclodextrins (CDs) are naturally occurring oligosaccharides that are extensively used in the pharmaceutical industry to improve the bioavailability of hydrophobic drugs, prevent undesired side effects and improve permeability across biological membranes.17 CDs have been used for gene delivery due to their ability to stabilize the nucleic acids in biological media.17 A variety of CD-based systems such as CD polymers,18 CD dendrimers,19 and CD polyrotaxanes have been developed as promising materials for non-viral gene delivery. Davis and coworkers have reported a diverse class of β-CD oligomers coupled via cationic linkers.7–10 One of these derivatives was successfully used as a vector for siRNA delivery in a clinical trial for treatment of melanoma in humans.7 Recently, our group has reported pendant polymer:CD based guest:host systems capable of delivering siRNA with efficiencies comparable to those of Lipofectamine 2000 (L2k) and branched poly(ethylene imine) (bPEI), while being 3–4 orders of magnitude less toxic.20 Polyrotaxanes are supramolecular structures in which cyclic molecules are threaded onto an “axle” and endcapped by bulky groups at the terminal positions of the “axle”. The construction of such supramolecular architectures from FDA approved materials such as α-CD and poly(ethylene glycol) (PEG) makes them extremely attractive for biomedical applications.21 Such CD-based polyrotaxanes have been used as hydrogels for drug delivery,22 biodegradable drug delivery constructs,23 scaffolds for tissue engineering24 and gene delivery.
Cationic CD polyrotaxanes have been studied for their DNA complexation and transfection abilities in cells where cationic substituents have been introduced by post-modification reactions after the macrocycle has been threaded onto the polymer backbone.25–27 Biocleavable cationic α-CD polyrotaxanes have been synthesized where the endcaps are conjugated via disulfide bonds to render them degradable in a reducing environment.28–30 The versatility of polyrotaxanes has further been established in systems where they have been encapsulated within a lipid bilayer to improve their biological performance31 or synthesized with endcaps that can act as targeting moieties to enhance its uptake via receptor mediated endocytosis.32 While these materials have previously been studied for their siRNA complexation ability, their efficiency as siRNA delivery vectors has never been reported.33
Herein, we report for the first time the design and synthesis of delivery vectors based on cationic multi-armed α-cyclodextrin (α-CD):PEG polyrotaxane (bPRTx+) materials that have been synthesized and used as siRNA delivery vehicles (Fig. 1). It was anticipated that the multi-armed structure, combined with the mobility of the cationic CDs on the PEG chain, would result in enhanced complexation with the siRNA, leading to particle formation at lower N/P ratios than the previously reported linear constructs. We have demonstrated that these bPRTx+ materials are capable of forming positively charged nanoparticles less than 200 nm in size at much lower N/P ratios and are able to effectively deliver siRNA.
Fig. 1.
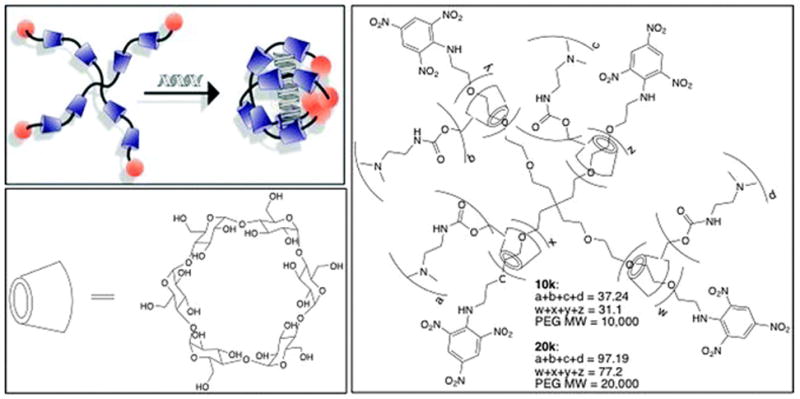
(top left) Conceptual diagram showing formation of bPRTx+:siRNA complex; (bottom left) structure of α-CD; (right) general structure of bPRTx+.
Materials and methods
Materials
All solvents were reagent grade, purchased from commercial sources, and used without further purification, except for DMF and toluene, which were dried over CaH2 under N2, filtered and distilled at reduced pressure. 1,1-Carbonyldiimidazole (CDI), α-CD, and N,N′-dimethylethylenediamine (DMEDA) were obtained from Sigma-Aldrich. The 4-arm 10k and 20k PEG amines were obtained from Creative PEGworks. 2,4,6-Trinitrobenzenesulfonic acid (TNBS) was obtained from Research Organics. Anti-GFP siRNA and All start negative control siRNA were purchased from Qiagen. Lipofectamine 2000 (L2k) was purchased from Invitrogen. Dialysis membranes of MWCO = 6000–8000 were purchased from Fischer Scientific. 1H NMR spectra were recorded on a 300 MHz VARIAN INOVA 300 NMR spectrometer at 30 °C. Chemical shifts were referenced to the residual protonated solvent peak.
Cell culture
Mouse fibroblast (NIH3T3-GFP) cells stably transfected with green fluorescent protein were obtained from Cell Biolabs (CA). The cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum, 1 mM glutamine, and 1% penicillin–streptomycin.
Synthesis of 4 arm 10k bPRTx
Branched 4-arm PEG10k amine (250 mg, 0.0250 mmol) was dissolved in 100 mL H2O containing 10 g α-CD (10.3 mmol) and stirred for 2 d. TNBS (2.93 mL, 0.996 mmol) as a 10% aqueous solution was pre-activated by NaHCO3 addition until pH 8 before addition to the 4-arm PEG10k amine–α-CD mixture and the solution stirred at 20 °C for additional 2 d. The crude solution was then washed with 18 MΩ MilliQ water and the product gathered by centrifugation at 9000 rpm multiple times until the aqueous solution became colorless. The crude bPRTx solid was then dried under vacuum to give an orange powder. Yield: 1.116 g. bPRTx loading: ~26.1 α-CD, 23.6% threading efficiency. MW: 34.18 kDa. 1H NMR (ESI†, Fig. S1) (300 MHz, DMSO-d6, δ): 8.99–8.93 (s, TNBS), 5.84–5.60 (s, C2–OH of CD), 5.61–5.40 (s, CD C3–OH), 4.93–4.65 (s, CD C1H), 4.58–4.32 (s, CD C6–OH), 3.87–3.15 (m, CD C2H, C3H, C4H, C5H, and C6CH2, PEG CH2).
Synthesis of 4 arm 10k DMEDA bPRTx
The 4-arm 10k PRTx (1.116 g, 0.0327 mmol) was dissolved in minimal DMSO under Ar with CDI (4.14 g, 25.5 mmol) and stirred for 1 d at 20 °C. DMEDA (2.79 mL, 25.6 mmol) was then added under Ar and the reaction mixture was stirred for additional 2 d before dialyzing (Fisherbrand, MWCO 6–8 kDa) against DMSO (2× 1 L) and H2O (3× 1 L) and drying under vacuum to give a red solid. Yield: 949 mg. bPRTx loading: ~32 α-CD, 29% threading efficiency; ~1.2 DMEDA/CD, 37 DMEDA/mol. MW: 45.36 kDa. 1H NMR (ESI†, Fig. S2) (300 MHz, D2O, δ): 5.23–4.81 (b, CD C2–OH), 4.80–4.72 (b, CD C1H, D2O), 4.44–3.98 (b, CD C6–OH), 3.98–3.31 (m, CD C2H, C3H, C4H, C5H, and C6CH2, PEG CH2), 3.25–2.87 (b, DMEDA NH-CH2), 2.43–2.20 (b, DMEDA CH2-NMe2), 2.20–1.86 (b, DMEDA N-(CH3)2).
Synthesis of 4 arm 20k bPRTx
The 4-arm PEG20k amine (250 mg, 0.0125 mmol) was dissolved in 100 mL H2O containing 10 g α-CD and stirred for 2 d. A 10% aqueous solution of TNBS (1.47 mL, 0.500 mmol), pre-activated as described above, was added and the solution stirred at 20 °C for additional 2 d. The crude solution was then washed with 18MΩ MilliQ water and the product gathered by centrifugation at 9000 rpm multiple times until the aqueous solution became colorless. The crude bPRTx solid was then dried under vacuum to give an orange powder. Yield: 1.465 g. bPRTx loading: ~77 α-CD, 35% threading efficiency. MW: 91.91 kDa. 1H NMR (ESI†, Fig. S1) (300 MHz, D2O, δ): 5.29–4.89 (b, CD C2–OH), 4.89–4.82 (b, CD C1H, D2O), 4.60–4.25 (b, CD C6–OH), 4.00–3.41 (m, CD C2H, C3H, C4H, C5H, and C6CH2, PEG CH2), 3.36–2.89 (b, DMEDA NH-CH2), 2.57–2.27 (b, DMEDA CH2-NMe2 of DMEDA), 2.28–1.87 (b, DMEDA N-(CH3)2).
Synthesis of 4 arm 20k DMEDA bPRTx
4-arm 20k bPRTx (1.465 g, 0.0189 mmol) was dissolved in minimal DMSO under Ar with CDI (6.01 g, 37.06 mmol) and stirred for 1 d at 20 °C. N,N-Dimethylethylenediamine (DMEDA) (4.05 mL, 37.09 mmol) was then added under Ar, and the reaction mixture was stirred for 2 d. The solution was then dialyzed (Fisherbrand, MWCO 6–8 kDa) against 2× DMSO and 3× H2O, and dried under vacuum to give a red solid. Yield: 1.652 g. Loading: ~77 α-CD, 35% threading efficiency; ~1.3 DMEDA/CD, 97 DMEDA/mol. MW: 104.56 kDa. 1H NMR (ESI†, Fig. S2) (300 MHz, D2O, δ): 5.29–4.89 (b, C2–OH), 4.89–4.82 (b, C1H of CD, D2O), 4.60–4.25 (b, C6–OH of CD), 4.00–3.41 (m, C2H, C3H, C4H, C5H, and C6H2 of CD, CH2 of PEG), 3.36–2.89 (b, NH-CH2 of DMEDA), 2.57–2.27 (b, CH2-NMe2 of DMEDA), 2.28–1.87 (b,N-(CH3)2 of DMEDA).
Gel retardation assay
The complexation ability of bPRTx+ with siRNA was determined by 4% agarose (low melting point) gel electrophoresis. The agarose gels were precast in TBE buffer with GelRed dye at 1: 10 000 dilution. bPRTx+:siRNA complexes containing 0.2 μg of siRNA at different N/P ratios were loaded onto the gels. A 1: 5 dilution of loading dye was added to each well and electrophoresis was carried out at a constant voltage of 55 V for 2 h in TBE buffer. The siRNA bands were then visualized under a UV transilluminator at a wavelength of 365 nm.
Dynamic light scattering (DLS)
The sizes, size distributions and zeta potentials of the materials were evaluated by dynamic light scattering using a particle size analyzer (Zetasizer Nano S, Malvern Instruments Ltd.) at 20 °C at a 90° scattering angle. The bPRTx+ and bPEI were solubilized in nanopure water and the measurements were carried out in HBG buffer (10 mM HEPES, 5% glucose, pH 7.4).
Atomic force microscopy (AFM)
AFM imaging of the nanoparticles was conducted in tapping mode (MultiMode, Veeco, U.S.) using dry samples on mica. The AFM tips (PPP-NCH, Nanoscience Instruments, Inc., U.S.) had a typical radius of 7 nm or less, and the images were recorded at a 0.5 or 1 Hz scan rate. Samples were prepared by dropping 2 mL of solution onto the mica surface, followed by overnight drying at 20 °C.
MTS cell viability assay
The cytotoxicity of the bPRTx+ relative to bPEI (25 kDa) was evaluated as a function of amine density using the MTS assay in NIH3T3-GFP cells. These cells were seeded in 96-well microtiter plates (Nunc, Wiesbaden, Germany) at densities of 7500 cells per well and cultured at 37 °C (5% CO2 and 95% relative humidity) in complete DMEM medium supplemented with 10% FBS. After 24 h, the culture media were replaced with serum-free culture media containing increasing N/P ratios of bPRTx+:siRNA particles and the cells were incubated for 4 h. MTS reagent (20 μL) was then added to each well and incubated for 2 h. Following the incubation period, the absorbance was measured using a microplate reader (Spectra Plus, TECAN) at a wavelength of 492 nm. The cell viability (%) relative to control cells cultured in media without polymers was calculated as [A]test/[A]control × 100%, where [A]test is the absorbance of the wells with polymers and [A]control is the absorbance of the control wells lacking polymers. All experiments were conducted for a minimum of three samples and averaged. Absorbance of MTS with bPEI or bPRTx+ was also recorded as controls.
LDH release assay
LDH is a cytosolic enzyme that is secreted upon damage to cell membranes and thus serves as a cytotoxicity indicator. NIH3T3-GFP cells were seeded in 96-well microtiter plates (Nunc, Wiesbaden, Germany) at densities of 7500 cells per well and cultured at 37 °C (5%CO2 and 95% relative humidity) in complete DMEM medium supplemented with 10% FBS. After 24 h, the culture media were replaced with serum-free culture media containing increasing N/P ratios of bPRTx+:siRNA particles and the cells were incubated for 4 h. CytoTox 96® Non-Radioactive Cytotoxicity Assay from Promega was used for the analysis. After incubation, 50 μL of the solution was transferred to a new 96-well plate and allowed to equilibrate to room temperature. To this, 50 μL of substrate mix was added and incubated for 30 min at 20 °C with protection from light. Stop mix (50 μL) was then added and the absorbance measured at 492 nm. The maximum LDH release was determined by addition of lysis buffer provided by the manufacturer to untreated cells. Cytotoxicity was determined as a percentage of the maximum LDH release caused by the lysis buffer.
In vitro gene knockdown experiments
NIH3T3-GFP cells were seeded in a 24-well plate at a density of 60 000 cells per well and cultured at 37 °C (5% CO2 and 95% relative humidity) in complete DMEM media supplemented with 10%FBS. After 24 h, the culture media were replaced with serum-supplemented media containing the siRNA complexes (90 pmol siRNA per well). The cells were incubated with the complexes for 4 h, after which the media were aspirated and fresh serum-supplemented media were added. At the end of the incubation, the media were aspirated and the cells were washed with PBS, trypsinized and analyzed by FACS using excitation and emission filters of 488 nm and 530 nm, respectively. The % GFP mean fluorescence intensity was calculated relative to untreated samples. For the kinetic study, the complexes were formulated at an N/P ratio of 10 for all materials and the samples analyzed at 24, 36 and 48 h. For the dose response study, the complexes were formulated from 30 pmol, 60 pmol and 90 pmol siRNA.
Results
Synthesis and characterization of bPRTx+
Two bPRTx+ were synthesized by threading α-CDs onto branched PEG amine (MW = 10 000 and 20 000) in a saturated α-CD aqueous solution with stirring at 20 °C, then endcapped with an aqueous solution of TNBS. The crude solution was exhaustively washed with 18 MΩ H2O until the aqueous solution became colorless, indicating removal of unthreaded α-CD and unreacted TNBS. The crude bPRTx solid was then dried under vacuum and the products characterized by 1H NMR to calculate the average number of α-CDs threaded onto the branched PEG scaffold. Post-modification of this bPRTx was achieved via CDI activated coupling with DMEDA in DMSO, giving a bPRTx+ product with random carbamate-linked DMEDA modifications. The reaction mixture was then dialyzed against DMSO and H2O to remove excess DMEDA and other low molecular weight impurities. Finally, the dialyzed product was dried under vacuum and characterized by 1H NMR to calculate the average α-CD threading and average DMEDA density. 1H NMR revealed that the 10k bPRTx+ had a threading efficiency of ~29%, indicating an average of 32 α-CDs threaded onto the PEG with ~37 DMEDA groups attached to it, giving an overall charge density of 1.2 DMEDA/α-CD. Similarly, the 20k bPRTx+ had a threading efficiency of ~35%, indicating an average of 77 α-CDs threaded onto the PEG core with ~97 DMEDA groups attached to it, to produce a DMEDA density of 1.3 DMEDA/α-CD.
Particle characterization of bPRTx+:siRNA complexes
The ability of the bPRTx+ materials to condense siRNA was evaluated by determining its complexation ability, particle size and net surface charge of the complexes formed at different N/P ratios (Fig. 2A and B).
Fig. 2.
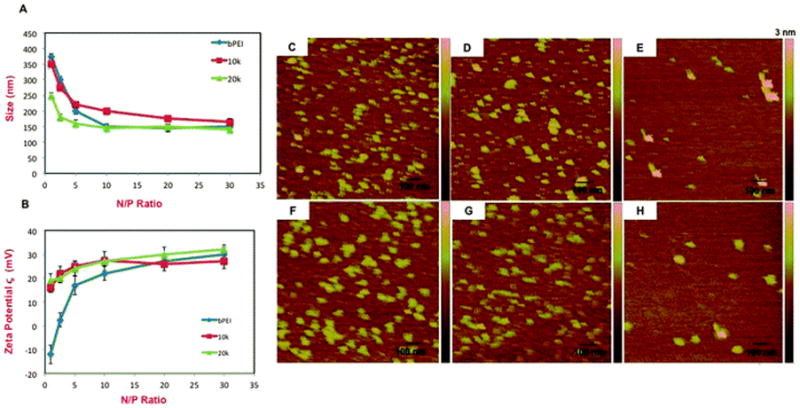
Characterization of siRNA complexes formed by bPEI, 10k and 20k bPRTx+. (A) DLS measurements; (B) ζ potentials. AFM images of (C) 10k; (D) 10k:siRNA at N/P = 2.5; (E) 10k:siRNA at N/P = 10; (F) 20k; (G) 20k:siRNA at N/P = 2.5; (H) 20k:siRNA at N/P = 10.
Gel shift assays of bPRTx+:siRNA complexes indicate that all the bPRTx+ had complexation abilities that were far superior to those of bPEI (ESI†). It was observed that while bPEI can complex siRNA only at higher N/P ratios such as 30, the bPRTx+ materials can condense siRNA at lower N/P ratios such as 2.5, 5 and 10. We attribute this to a combination of the branched multi-armed structure of the bPRTx+ materials and electrostatic adaptation of the DMEDA-α-CD units via rotational and lateral mobility of the CDs to complex siRNA more effectively.
Our DLS data show that bPEI and both of the bPRTx+ formed particles with diameters below 300 nm. While 10k bPRTx+ formed complexes that were slightly larger than bPEI, 20k bPRTx+ complexes were of similar dimensions or slightly smaller than those formed by bPEI. This can be attributed to the longer arms of 20k, which can result in more effective nucleic acid complexation and condensation than the 10k derivative. It also was observed that with increasing N/P ratio, the siRNA complex diameters decreased until an N/P ratio of 5–10, after which the increasing charge did not significantly affect sizes of the complexes. An interesting observation was that even at N/P ratios as low as 0.5–1, 20k bPRTx+ formed complexes that were smaller than 250 nm in diameter, indicating that the longer arms and larger size of 20k bPRTx+ produce effective condensation even at very low N/P ratios.
The ζ potentials were measured for both bPRTx+ to evaluate the surface charge of their siRNA complexes (Fig. 3A, left). We observed that complexes formed by both 10k and 20k bPRTx+ had positive ζ, regardless of the N/P ratios in the complexes and that their ζ potentials were similar to that of bPEI at high N/P ratios. At lower N/P ratios; however, the surface charges on the bPRTx+:siRNA complexes were significantly higher than those on bPEI:siRNA complexes. Interestingly, the bPRTx+:siRNA complexes displayed apparent surface charges of ζ=15–20 mV, even at N/P ratios as low as 0.5–2.5. In contrast, the surface charge of bPEI:siRNA complexes is negative at lower N/P ratios and increases sharply with increasing N/P ratio, eventually increasing more slowly above N/P=10. We infer from these findings that the high charge density and multi-armed structure of the bPRTx+ enable the formation of positively charged particles at much lower N/P ratios than those of bPEI. Since positive surface charges are important for promoting cellular binding and uptake of nanoparticles, bPRTx+:siRNA complexes may be internalized by cells at lower N/P ratios than bPEI complexes.
Fig. 3.
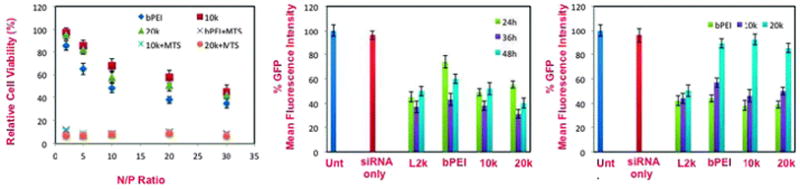
Relative cell viabilities of (left) bPEI:siRNA and bPRTx+:siRNA complexes as a function of N/P ratios; (center) in vitro GFP knockdown efficiencies of 10k and 20k bPRTx+:siRNA complexes at N/P = 10 in 10% serum-supplemented media with L2k and bPEI (@ N/P = 20) as controls (90 pmol siRNA per well, 4 h incubation, flow cytometry analysis of GFP expression 24, 36, and 48 h after removal of siRNA complexes); (right) in vitro GFP knockdown efficiencies of 10k and 20k bPRTx+:siRNA complexes at N/P = 10 in 10% serum-supplemented media with L2k and bPEI (@ N/P = 20) as controls (30 pmol, 60 pmol and 90 pmol siRNA per well, 4 h incubation, flow cytometry analysis of GFP expression 36 h after removal of siRNA complexes).
AFM images of bPRTx+ samples revealed the presence of a mixture of approximately spherical particles with diameters of 35–45 nm (Fig. 2C and F). Upon complexation with siRNA, 80–90 nm diameter spherical particles were observed. At N/P = 2.5, AFM analysis revealed that a mixture of small (10–15 nm) and large (80–90 nm) spherical particles was produced, suggesting that a heterogeneous population of siRNA complexes is formed under these conditions (Fig. 2D and E). Particle homogeneity improved at N/P = 10, with observed particle diameters in the range of 80–100 nm. Both bPRTx+ exhibited the same particle complexation behavior as a function of N/P, although the complexes formed by 20k were smaller than those formed by 10k (Fig. 2G and H), a finding that mirrors the results of the DLS experiments. The discrepancy in sizes determined by DLS and the AFM is attributed to the dry nature of the AFM samples, which may have led to increased electrostatic condensation within the particles. The observation of compact, relatively uniform positively charged bPRTx+:siRNA particles suggests that they are in a size regime that can be efficiently internalized by cells via endocytosis.
Acute cytotoxicity and gene silencing properties of bPRTx+
The cytotoxicity of cationic non-viral siRNA vectors is an important consideration for their successful translation into clinically relevant delivery vehicles. MTS cell viability assays were performed with 10k and 20k bPRTx+ and their bPRTx+:siRNA complexes, using bPEI as a benchmark. The uncomplexed cationic vectors were evaluated as a function of the amine densities on all the polymers (ESI†) in order to perform a direct comparison of the toxicity profiles of the materials based on the nitrogen concentration. The observed toxicity profiles of 10k and 20k bPRTx+ were comparable to bPEI (i.e. the measured LD50 ≅ 1 mM for bPEI and 10k bPRTx+; LD50 ≅ 0.35 mM for 20k bPRTx+) (ESI†).
Interestingly, the 10k bPRTx+:siRNA complexes were less toxic than bPEI or 20k bPRTx+ complexes, with observed cell viabilities of 70%, 50% and 60%, respectively, at N/P = 10 (Fig. 3, left). The LDH assay performed for the bPRTx+:siRNA complexes indicated a similar trend. Despite being more toxic at higher N/P ratios than bPEI complexes, the bPRTx+ complexes had cytotoxicities of 20–30% at an N/P = 5 at which the RNAi performance of these materials was far superior to that of bPEI (ESI†).
The in vitro RNAi performance of these complexes was evaluated using anti-GFP siRNA in NIH3T3-GFP cells in the presence of 10% serum-supplemented media. Upon screening different N/P ratios of complexes formulated from 90 pmol of siRNA, it was observed that the gene silencing efficiencies of both bPTRx+ constructs were found to be comparable to those of bPEI and RNAimax (L2k) (ESI†). Specifically, the RNAi profile of 10k bPRTx+:siRNA complexes was similar or slightly better than those of bPEI:siRNA complexes at all N/P ratios. The most noteworthy difference in performance, however, was found for 20k bPRTx+:siRNA complexes. These vectors generated a more pronounced RNAi effect than bPEI at all N/P ratios (even as low as N/P = 2.5) and silencing efficiencies that were comparable to those of the best commercially available vector, RNAimax. These results are consistent with the particle characterization and cell viability studies wherein it was found that 20k bPRTx+ forms consistently smaller and more highly positively charged siRNA complexes than bPEI or 10k bPRTx+ at N/P≤10. Even though bPEI displays good silencing performance at N/P = 20, the bPRTx+ materials have similar RNAi performance at N/P=5 (20k bPRTx+) or N/P=10 (10k bPRTx+), indicating that the branched cationic polyrotaxane motif is capable of promoting gene silencing at much lower charge densities where improved cell viability occurs (i.e. the observed cell viabilities with siRNA complexes yielding similar RNAi performance were 65%, 70% and 40% for 5/1 20k bPRTx+:siRNA, 10/1 10kbPRTx+:siRNA, and 20/1 bPEI:siRNA, respectively; Fig. 3, left and ESI†).
We then performed a kinetic study to determine the optimal in vitro knockdown of these materials as a function of time. This study was performed at previously determined optimal N/P ratios of the materials and the samples were analyzed at time points of 24, 36 and 48 h. It was observed that while the performance of the L2k remained unaffected over a period of 48 h, bPEI had a significant loss in activity after 36 h and the bPRTx+ materials also had a moderate reduction in activity after 36 h. These studies indicate that maximum gene knockdown was achieved after 36 h for the bPRTx+ complexes.
We further evaluated the dose dependence of the bPRTx+ materials. We studied the in vitro knockdown performance of the complexes at siRNA concentrations of 30 pmol, 60 pmol, and 90 pmol. Based on the previous studies, an N/P ratio of 10 was employed for the bPRTx+ materials and N/P = 20 was employed for bPEI. The samples were analyzed after 36 h, which was the time required for maximum knockdown. These studies revealed that while the performance of L2k is the same regardless of the amount of siRNA used, bPEI and bPRTx+ complexes had a significant loss in activity at an siRNA concentration of 30 pmol. These encouraging findings suggest that bPRTx+ materials may have a more useful RNAi performance window than that of bPEI when cell viability is an important consideration at low N/P ratios.
Conclusions
We have designed two novel multi-armed cationic polyrotaxanes that have been shown to be efficient gene silencing vectors. These materials form positively charged complexes, 100–250 nm in size, even at N/P ratios as low as 2.5. They also display excellent gene silencing properties, especially at lower N/P where good viability of the treated cells is retained. These characteristics suggest that bPRTx+ may be an attractive vehicle for siRNA delivery in cells that are sensitive at high N/P ratios. Although the bPRTx+ materials reported herein are highly efficient toward gene silencing, they still suffer from substantial cytotoxicity at high N/P. It is presumed that this side effect may be obviated by incorporating degradable linkages within the bPRTx+ construct to enable degradation of the material inside the cell, thereby improving the toxicity profile greatly. Efforts in this regard are in progress.
Supplementary Material
Insight, innovation, integration.
The delivery of siRNA using novel cationic multi-armed polyrotaxanes is demonstrated. These unique materials utilize supramolecular chemical principles to produce nucleic acid vectors from low-toxicity precursors. The multi-armed polyrotaxanes synthesized are able to effectively condense siRNA at low N/P ratios and deliver them inside cells. The gene silencing efficiencies and cytotoxicities of these materials are comparable to those of both bPEI and Lipofectamine2000. The ability to form complexes at lower N/P ratios allows us to use much lower concentrations of the polyrotaxanes, thus avoiding possible issues of systemic toxicity due to high doses and off-target effects. These materials can lay the foundation for the construction of safe, degradable materials that may be able to condense siRNA at lower N/P ratios, while producing gene silencing efficiencies comparable to those of bPEI and L2k.
Acknowledgments
We would like to express our special thanks for the support of this work by NIH grant GM087016 and the Purdue Department of Chemistry. 1H NMR data were acquired in the Purdue Interdepartmental NMR Facility supported by NCI CCSG CA23168 to the Purdue University Center for Cancer Research. We would also like to thank Rob Reason for help with the illustrations.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c2ib20107k
Notes and references
- 1.Tamura A, Nagasaki Y. Nanomedicine. 2010;5:1089. doi: 10.2217/nnm.10.76. [DOI] [PubMed] [Google Scholar]
- 2.Kole R, Krainer AR, Altman S. Nat Rev Drug Discovery. 2012;11:125. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitehead KA, Langer R, Anderson DG. Nat Rev Drug Discovery. 2009:8129. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waehler R, Russell SJ, Curiel DT. Nat Rev Genet. 2007;8:573. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mancuso K, Hauswirth WW, Li Q, Connor TB, Kuchenbecker JA, Mauck CM, Neitz J, Neitz M. Nature. 2009;461:784. doi: 10.1038/nature08401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pack DW, Hoffman AS, Pun S, Stayton PS. Nat Rev Drug Discovery. 2005;4:581. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 7.Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Nature. 2010;464:1067. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez H, Hwang SJ, Davis ME. Bioconjugate Chem. 1999;10:1068. doi: 10.1021/bc990072j. [DOI] [PubMed] [Google Scholar]
- 9.Reineke TM, Davis ME. Bioconjugate Chem. 2003;14:247. doi: 10.1021/bc025592k. [DOI] [PubMed] [Google Scholar]
- 10.Reineke TM, Davis ME. Bioconjugate Chem. 2003;14:255. doi: 10.1021/bc025593c. [DOI] [PubMed] [Google Scholar]
- 11.Sparks J, Slobodkin G, Matar M, Congo R, Ulkoski D, Rea-Ramsey A, Pence C, Rice J, McClure D, Polach KJ, Brunhoeber E, Wilkinson L, Wallace K, Anwer K, Fewell JG. J Controlled Release. 2012;158:269. doi: 10.1016/j.jconrel.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Whitehead KA, Sahay G, Li GZ, Love KT, Alabi CA, Ma M, Zurenko C, Querbes W, Langer RS, Anderson DG. Mol Ther. 2011;19:1688. doi: 10.1038/mt.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen DN, Mahon KP, Chikh G, Kim P, Chung H, Vicari AP, Love KT, Goldberg M, Chen S, Krieg AM, Chen J, Langer R, Anderson DG. Proc Natl Acad Sci U S A. 2012;109:797. doi: 10.1073/pnas.1121423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Li SD, Huang LJ. J Controlled Release. 2007;123:181. doi: 10.1016/j.jconrel.2007.09.004. [DOI] [PubMed] [Google Scholar]; (b) Lee SK, Han MS, Asokan S, Tung CH. Small. 2011;7:364. doi: 10.1002/smll.201001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi J, Xiao Z, Votruba AR, Vilos C, Farokhzad OC. Angew Chem, Int Ed. 2011;50:7027. doi: 10.1002/anie.201101554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang H, Tang G, Wang Q, Li D, Shen F, Zhou J, Yu H. Chem Commun. 2006:2382. doi: 10.1039/b601130f. [DOI] [PubMed] [Google Scholar]
- 17.Davis ME, Brewster ME. Nat Rev Drug Discovery. 2004;3:1023. doi: 10.1038/nrd1576. [DOI] [PubMed] [Google Scholar]
- 18.Lee CC, Mackay JA, Frechet JMJ, Szoka FC. Nat Biotechnol. 2005;23:1517. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 19.Helms B, Meijer EW. Science. 2006;313:929. doi: 10.1126/science.1130639. [DOI] [PubMed] [Google Scholar]
- 20.Kulkarni A, DeFrees K, Hyun SH, Thompson DH. J Am Chem Soc. 2012;134:7596. doi: 10.1021/ja300690j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li JJ, Zhao F, Li J. Appl Microbiol Biotechnol. 2011:90427. doi: 10.1007/s00253-010-3037-x. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Li J. J Biomed Mater Res, Part A. 2008;86:1055. doi: 10.1002/jbm.a.31710. [DOI] [PubMed] [Google Scholar]
- 23.Moon C, Kwon YM, Lee WK, Park YJ, Yang VC. J Controlled Release. 2007;124:43. doi: 10.1016/j.jconrel.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ichi T, Watanabe J, Ooya T, Yui N. Biomacromolecules. 2001:2204. doi: 10.1021/bm005617n. [DOI] [PubMed] [Google Scholar]
- 25.Yang C, Wang X, Li H, Goh SH, Li J. Biomacromolecules. 2007;8:3365. doi: 10.1021/bm700472t. [DOI] [PubMed] [Google Scholar]
- 26.Yang C, Li H, Wang X, Li J. J Biomed Mater Res, Part A. 2009;89:13. doi: 10.1002/jbm.a.31976. [DOI] [PubMed] [Google Scholar]
- 27.Yang C, Wang X, Li H, Tan E, Lim CT, Li J. J Phys Chem B. 2009;113:7903. doi: 10.1021/jp901302f. [DOI] [PubMed] [Google Scholar]
- 28.Ooya T, Choi HS, Yamashita A, Yui N, Sugaya Y, Kano A, Maruyama A, Akita H, Ito R, Kogure K, Harashima H. J Am Chem Soc. 2006;128:3852. doi: 10.1021/ja055868+. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita A, Kanda D, Katoono R, Yui N, Ooya T, Maruyama A, Akita H, Kogure K, Harashima H. J Controlled Release. 2008:131137. doi: 10.1016/j.jconrel.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Yamada Y, Nomura T, Harashima H, Yamashita A, Yui N. Biomaterials. 2012;33:3952. doi: 10.1016/j.biomaterials.2012.01.049. [DOI] [PubMed] [Google Scholar]
- 31.Yamada Y, Nomura T, Harashima H, Yamashita A, Katoono R, Yui N. Biol Pharm Bull. 2010;33:1218. doi: 10.1248/bpb.33.1218. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Y, Wang H, Wang C, Li Y, Lu W, Chen S, Luo J, Jiang Y, Chen J. Mol Pharmacol. 2012;9:1067. doi: 10.1021/mp200315c. [DOI] [PubMed] [Google Scholar]
- 33.Yamada Y, Hashida M, Nomura T, Harashima H, Yamasaki Y, Kataoka K, Yamashita A, Katoono R, Yui N. Chem Phys Chem. 2012;13:1161. doi: 10.1002/cphc.201100800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


