SUMMARY
The interaction of outer membrane protein A (OmpA) with its receptor, Ecgp96 (a homologue of Hsp90β) is critical for the pathogenesis of E. coli K1 meningitis. Since Hsp90 chaperones Toll-like receptors (TLRs), we examined the role of TLRs in E. coli K1 infection. Herein, we show that newborn TLR2−/− mice are resistant to E. coli K1 meningitis, while TLR4−/− mice succumb to infection sooner. In vitro, OmpA+ E. coli infection selectively upregulates Ecgp96 and TLR2 in human brain microvascular endothelial cells (HBMEC), whereas OmpA− E. coli upregulates TLR4 in these cells. Furthermore, infection with OmpA+ E. coli causes Ecgp96 and TLR2 translocate to the plasma membrane of HBMEC as a complex. Immunoprecipitation studies of the plasma membrane fractions from infected HBMEC reveal that the C-termini of Ecgp96 and TLR2 are critical for OmpA+ E. coli invasion. Knockdown of TLR2 using siRNA results in inefficient membrane translocation of Ecgp96 and significantly reduces invasion. In addition, the interaction of Ecgp96 and TLR2 induces a bipartite signal, one from Ecgp96 through PKC-α while the other from TLR2 through MyD88, ERK1/2 and NF-κB. This bipartite signal ultimately culminates in the efficient production of NO, which in turn promotes E. coli K1 invasion of HBMEC.
Keywords: Outer membrane protein A, Escherichia coli K1, Toll-like receptors, infection, brain, cell signaling
INTRODUCTION
Escherichia coli K1 continues to be one of the leading causes of bacterial meningitis in both developed and developing countries, and this disease is associated with fatality rates of around 40% in infected neonates (Aletayeb et al.,2010, Sunakawa et al., 2010, Houdouin et al., 2008, Zaidi et al., 2009). Survivors of this infection frequently suffer from debilitating neurological sequelae (Kaper et al., 2004). E. coli K1 employs a complex pathogenic mechanism to enter the bloodstream, evade host defenses, and eventually reach the blood brain barrier (BBB). Studies from our group have shown previously that the outer membrane protein A (OmpA) of E. coli K1 interacts with a receptor, Ecgp96, which is a homologue of gp96 or HSP90β that is specifically expressed on human brain microvascular endothelial cells (HBMEC) (Prasadarao, 2002). We have also demonstrated that specific amino acids in the extracellular loops 1 and 2 of OmpA are necessary for E. coli K1 to invade HBMEC (Pascal et al., 2010). Importantly, mutation of these key amino acids in OmpA does not affect the expression of other bacterial surface structures such as S-fimbriae, type 1 fimbriae or total polysaccharide content (includes LPS and capsular polysaccharide), further demonstrating that OmpA expression is critical for HBMEC invasion (Mittal et al., 2011). We have also found that the interaction of OmpA with Ecgp96 triggers recruitment of signal transducer and activator 3 (STAT3) to the C-terminal domain of Ecgp96 (Maruvada et al., 2008). In addition, activation of focal-adhesion kinase (FAK) and rearrangement of actin microfilaments in HBMEC are necessary for these cells to internalize E. coli K1. Furthermore, actin condensation near the bacterial attachment sites requires signaling molecules such as phosphatidyl inositol-3 kinase (PI3K), phospholipase C-γ (PLC-γ) and protein kinase C-α (PKC-α) (Prasadarao et al., 1999, Reddy et al., 2000, Sukumaran et al., 2003a, Sukumaran and Prasadarao., 2002a, Sukumaran et al., 2002b). The bacterium subsequently invades the brain endothelium via caveolae to disseminate into the central nervous system (Sukumaran et al., 2002b). E. coli K1 infection also triggers nuclear factor-κB (NF-κB) to increase intercellular adhesion molecule (ICAM-1) expression (Selvaraj et al., 2007). NF-κB activation in response to infection leads to pro-inflammatory cytokine secretion (Carmody and Chen, 2007) and upregulation of inducible nitric oxide synthase (iNOS), which results in increased nitric oxide (NO) production (Iwashita et al., 1995). It has been shown that NO production upregulates Ecgp96 in HBMEC during E. coli K1 infection and subsequently promotes more invasion (Mittal et al., 2010). However, the signaling mechanism(s) connecting Ecgp96 and NO production is incompletely understood.
Toll-like receptors (TLRs) are pathogen recognition receptors (PRRs) that play an essential role in the innate immune response against microbial pathogens and are considered as indispensable components of the adaptive immune response. They are also one of the most potent activators of NF-κB. These receptors are characterized by an ectodomain composed of leucine-rich repeats, and an intra-cytoplasmic domain that is conserved among members of the interleukin-1 receptor (IL-1R) family (Toll/IL-1R or TIR domain). Upon ligand stimulation, cytoplasmic TIR domains recruit several adaptor proteins to relay the signal. At this point, TLR-mediated signaling bifurcates into MyD88-dependent and MyD88-independent pathways. Activation of the MyD88-dependent pathway leads to the early phase of NF-κB activation, whereas signaling through the MyD88-independent pathway results in late phase of NF-κB activation along with the induction of interferon-responsive genes and the synthesis of TNF-α. Mammalian TLR2 and TLR4 have been shown previously to associate with gp96, which regulates TLR folding and surface expression (Saitoh, 2009, Tsan and Gao, 2009). In mice with gp96-deficient macrophages, there is loss of response to TLR ligands and resistance to Listeria infection, demonstrating that gp96 is crucial for TLR mediated signaling and inflammatory response (Yang et al., 2007). The expression of gp96 on the surface of LPS stimulated B cells has been implicated in the activation of Th2 cells (Banerjee et al., 2002). Moreover, gp96 has been found to physically interaction with TLRs in B cells in a mouse model (Randow and Seed, 2001), suggesting that a possible surface interaction of TLRs and gp96 in immune cells. Because Ecgp96 is a receptor for E. coli K1 OmpA in HBMEC, we investigated whether TLRs might regulate Ecgp96 expression during E. coli K1 invasion of HBMEC. In this study, we discovered that newborn TLR2−/− mice are resistant to E. coli K1 induced meningitis, whereas TLR4−/− mice succumbed to infection earlier than wild type C57BL6 mice. In vitro studies revealed that the expression of Ecgp96 and its association with TLR2 at the plasma membrane of HBMEC increase in response to OmpA+ E. coli infection. Conversely, an OmpA− strain of E. coli K1 induces greater translocation of TLR4 to the cell surface. We also show for the first time that OmpA+ E. coli binding to HBMEC induces nitric oxide (NO) production through a bipartite signaling, one mediated by Ecgp96 and PKC-α and the other by TLR2, MyD88, ERK1/2 and NF-κB.
RESULTS
Newborn TLR2−/− mice are resistant to E. coli K1 induced meningitis
Our studies have demonstrated previously that suppression of gp96 prevents E. coli K1 traversal across the BBB (Mittal et al., 2010). Since, gp96 acts as a chaperone for TLR2 and TLR4, we determined whether lack of either of these molecules would affect the outcome of meningitis. The virulence of E. coli K1 was analyzed in newborn TLR2−/− or TLR4−/− mice. As shown in Fig. 1A, all of the TLR2−/− mice survived until four days post-infection similar to wild type animals that received saline. In contrast, 50% of E. coli K1 infected TLR4−/− newborn mice became extremely sick by 48 h and were euthanized. The remaining 50% became moribund and were subsequently euthanized at 72 h. In contrast, the wild type newborn mice did not become moribund until 72 to 86 h post infection. These observations indicate that lack of TLR2 is protective against E. coli meningitis whereas absence of TLR4 aggravates the disease process.
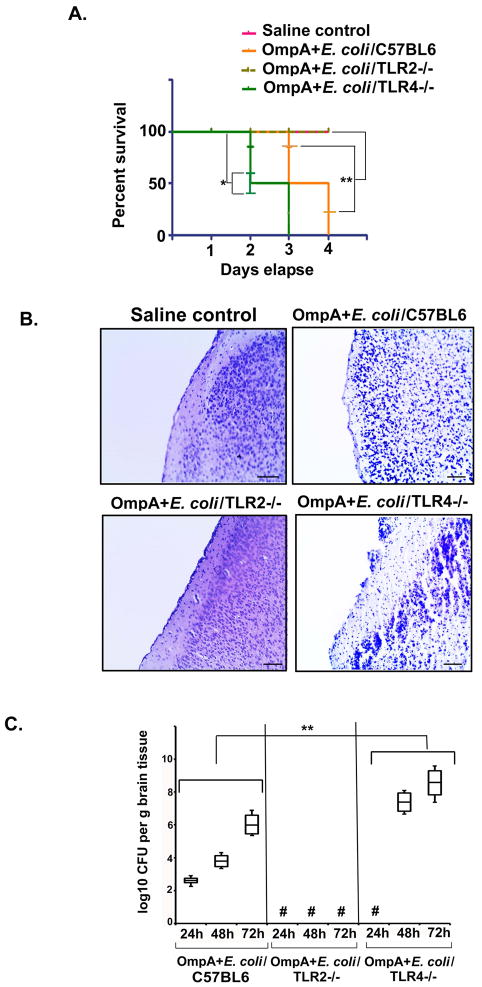
Figure 1. TLR2−/− mice are resistant to E. coli K1 meningitis.
(A) 3 day old C57BL6 (n=12), TLR2−/− (n=15) and TLR4−/− (n=15) pups were infected intranasally with 103 CFU of OmpA+ E. coli. Another set of 3 day old C57BL6 (n=11) received pyrogen free saline and served as controls, where “n” is number animals used per group. The pups were monitored for survival at 24 h, 48 h, 72 h and 96 h post-infection. Percent survival of the pups at the given time points are presented as Kaplan Meier curve. All pups were sacrificed if they were in moribund state due to ethical reasons. Percent survival of C57BL6 and TLR4−/− mice was statistically significant (*P<0.01 and **P<0.02 with respect to saline control by Student’s t test). (B) Control and infected pups were euthanized at 72 h post-infection and half of the brain samples were processed as described in experimental procedures, and stained with H and E. Scale bar = 100 μm. Arrows indicate either loss of meninges or neutrophil infiltration. (C) The other half of the brain was homogenized in saline, serial dilution made and plated on LB agar with rifampicin. Three animals from these experiments were used to determine the bacterial load in the brains at 24, 48 and 72 h post-infection. The results are representative of three different experiments. Asterisks represent lack of bacterial colonies in the brains. Fold increase in log CFU between control and TLR4−/− groups was statistically significant for each time point (**P<0.001 by ANOVA). # indicates no bacteria recovered from the brains.
Histopathologic analysis of brain sections of the infected wild-type mice revealed a significant neutrophil infiltration in the cortex and signs of gliosis (Fig. 1B). However, the brains of the TLR2−/− mice appeared normal, indicating that E. coli K1 did not cause detectable brain injury in these animals. In contrast, the brain histopathology of the TLR4−/− mice was similar to or worse than that of the infected wild-type mice at 48 to 72 h post-infection. By quantitative culture, the brain bacterial load in the TLR4−/− mice was two-fold greater than in the control infected mice at 48 and 72 h post-infection, while no bacteria were cultured from TLR2−/− mice (Fig. 1C). These results indicate that TLR2 expression is necessary for E. coli K1 to cause meningitis, whereas TLR4 is required for the maximal host defense against meningitis.
OmpA expression in E. coli K1 is critical for inducing Ecgp96 and TLR2 upregulation in HBMEC
Traversal of E. coli K1 from the circulation across the BBB, which constitute a single cell layer of endothelial cells, is a critical step in the pathogenesis of meningitis (Prasadarao, 2002). Since our in vivo experiments showed that TLR2 and TLR4 plays important roles in E. coli K1-induced pathology in newborn mice, we used real-time PCR to determine the effects of OmpA+ and OmpA− E. coli K1 infection on HBMEC ecgp96, tlr2 and tlr4 mRNA expression. Infection of HBMEC with OmpA+ E. coli caused ecgp96 transcript levels to increase 3.0 ± 0.75 fold by 60 and 90 min post-infection (Fig. 2A). In contrast, infection of HBMEC with OmpA− E. coli infection actually suppressed ecgp96 mRNA expression below that of uninfected control HBMEC at 30–90 min (**P<0.01). A similar pattern was seen for tlr2, with OmpA+ E. coli inducing tlr2 expression that peaked at 2.3 ± 0.3 fold by 90 min and OmpA− E. coli suppressing tlr2 expression (**P<0.01) (Fig. 2B). In contrast, although infection with OmpA+ E. coli did not induce any apparent increase in tlr4 mRNA levels (Fig. 2C), infection with OmpA− E. coli increased tlr4 expression by 9.8 ± 0.2 fold (*P<0.001). As a control, we determined the effects of OmpA+ and OmpA− stains of E. coli on tlr5 and tlr9 mRNA expression. Neither strain affected the transcript levels of these genes in HBMEC (Fig. 2D and E). These results show that OmpA+ E. coli induces ecgp96 and tlr2 gene expression in HBMEC while OmpA− E. coli induces the expression of tlr4.
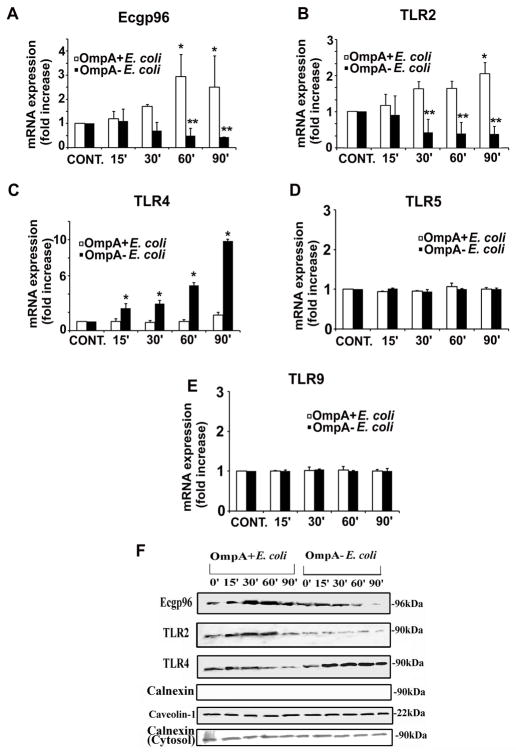
Figure 2. Expression of Ecgp96 and TLRs in HBMEC in response to E. coli K1 infection.
(A) Confluent monolayers of HBMEC in 6-well culture plates were infected with OmpA+ E. coli or OmpA− E. coli for various time points as indicated. Total RNA was isolated, cDNA prepared and subjected to real time qPCR using primers for Ecgp96 (A), TLR2 (B), TLR4 (C), TLR5 (D), TLR9 (E), and GAPDH. Fold increase in mRNA expression was graphed after normalized to GAPDH. The error bars represent means ± SD from three independent experiments performed in duplicate and the fold increase/decrease in transcript levels of Ecgp96, TLR2 and TLR4 was statistically significant, *P<0.01, *P<0.01 and *P<0.0001 respectively by Student’s t test. (F) HBMEC grown in 100 mm dishes were treated with OmpA+ E. coli or OmpA− E. coli for indicated time points and the membrane fractions were prepared. Equal amounts of membrane proteins were subjected to Western blotting with antibodies to Ecgp96, TLR2, TLR4, Calnexin or Caveolin-1. Equal amounts of cytosolic fractions were also subjected to Western blotting using antibodies to Calnexin.
Previously, we determined by flow cytometry that E. coli infection of HBMEC induces the expression of Ecgp96 at the cell surface (Mittal et al., 2010). Therefore, we speculated that increased transcription of ecgp96 and tlr2 genes might translate to an increase in protein levels, with subsequent translocation to the cell surface. To examine this, HBMEC were infected with OmpA+ or OmpA− E. coli at various time points, after which the plasma membranes were isolated and subjected to Western blotting with antibodies to Ecgp96 and TLRs. Consistent with the real-time PCR data, OmpA+ E. coli infection resulted in increased Ecgp96 and TLR2 in the plasma membranes between 30 and 60 min (Fig. 2F). Furthermore, OmpA− E. coli, reduced plasma membrane Ecgp96 and TLR2 content, but increased TLR4 content. Similar results were obtained when lipid raft fractions were subjected to Western blotting with antibodies to Ecgp96, TLR2 or TLR4 (data not shown). To verify that this upregulation of Ecgp96 was an OmpA+ E. coli specific chaperone response, we analyzed the membrane fractions for the presence of another representative endoplasmic reticulum (ER) chaperone, calnexin (Fig. 2F). No calnexin expression in the membrane fraction was detected, whereas the cytosolic fractions contained a significant amount of calnexin, which was unchanged by the presence of either strain of E. coli. Collectively, these results indicate that the plasma membrane preparations were not contaminated with the membranes of other organelles such as the ER and that the increased expression of Ecgp96 at the surface is not due to a general chaperone response. We also examined the expression of additional chaperones such calreticulin, Hsp70 and Erp57 was examined, and all showed patterns similar to that of calnexin (data not shown). These results suggest that OmpA+ E. coli upregulates Ecgp96 and TLR2 mRNA and protein, while OmpA− E. coli upregulates TLR4 both at mRNA and protein levels. Subsequently, Ecgp96 and TLR2 complex translocates to the plasma membrane, providing additional receptors for E. coli K1 for more bacteria to invade.
Absence of TLR2 expression significantly affects the translocation of Ecgp96 and subsequent E. coli K1 invasion
Our observation that lack of TLR2 expression in newborn mice renders the animals resistant to E. coli K1 infection and the fact that Hsp90, which is a homologue of gp96 acts as a chaperone for TLRs (Nicchitta, 2003), suggest that the interaction of TLR2 and Ecgp96 in HBMEC may be important for E. coli K1 invasion. Therefore, to examine the roles of TLR2 and TLR4 during E. coli K1 invasion HBMECs, the expression of these proteins were knocked down using siRNA and the efficiency of invasion was subsequently examined. Transfection of HBMECs with TLR2 or TLR4 siRNA suppressed the respective mRNA transcripts by 74.3% ± 8.7% when compared to control, untransfected HBMEC as analyzed by densitometric analysis (Fig. 3A). Of note, knockdown of one of the TLRs using siRNA did not affect the transcription of the other, and did not affect the change in ecgp96 mRNA in HBMEC induced by infection with OmpA+ or OmpA− E. coli (Fig. 3B). To investigate whether the TLR suppression had any effect on Ecgp96 translocation to the plasma membrane, the surface expression of Ecgp96 in TLR2 or TLR4 siRNA transfected HBMEC was analyzed using flow cytometry. Ecgp96 surface expression was reduced by 48% ± 7.3% in the absence of TLR2 but only by 14.9% ± 3.1% in the absence of TLR4 (*P<0.005, Fig. 3C). Therefore, TLR2 is more important for the translocation of Ecgp96 to the cell surface than TLR4. TLR2 knockdown inhibited the binding of OmpA+ E. coli to HBMEC by 19.4% ± 2.7%, whereas knockdown of TLR4 did not significantly alter this process. OmpA− E. coli exhibited reduced binding to untransfected control cells (51.1% ± 5.6% when compared to OmpA+ E. coli) and this binding pattern was not affected by knockdown of either TLR2 or TLR4 (Fig. 3D). In addition TLR2 knockdown inhibited OmpA+ E. coli invasion by 73.6% ± 4.7% while TLR4 knockdown inhibited invasion only by 18.7% ± 2.4% (Fig. 3E). OmpA− E. coli invasion was not affected by knockdown of TLR2 or TLR4. These result suggest that TLR2 is required for maximal surface expression of Ecgp96 in HBMEC and thereby the invasion of OmpA+ E. coli.
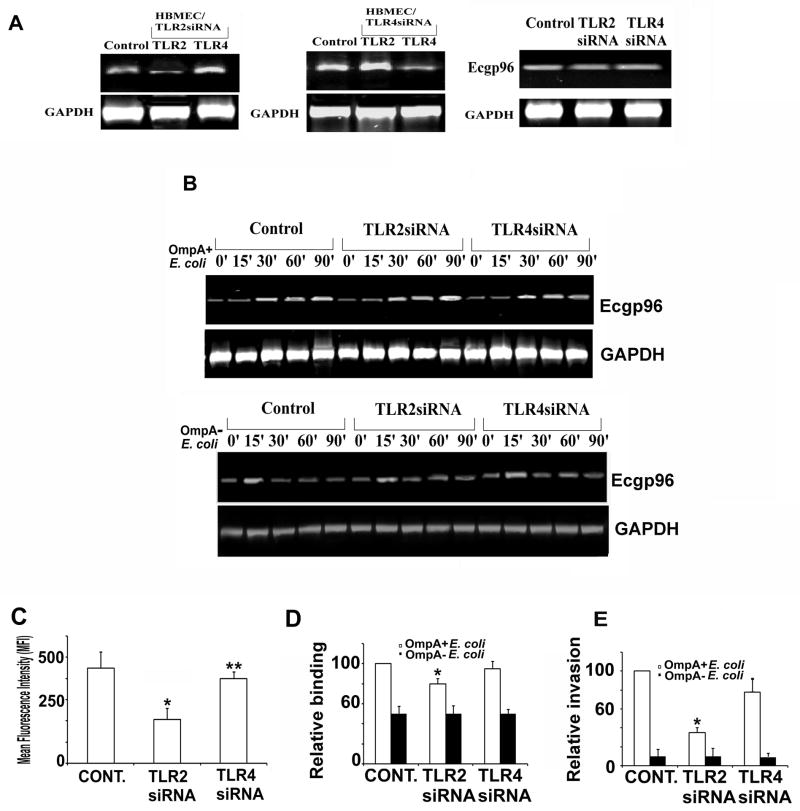
Figure 3. TLR2 knockdown by siRNA affects the surface expression of Ecgp96.
(A) HBMEC in 6-well plates were transfected with either TLR2 or TLR4 siRNA at 90% confluence, allowed to recover for 24 h, total RNA was isolated and subjected to RT-PCR with primers specific to tlr2, tlr4, ecgp96, or gapdh. ‘Control’ indicates untransfected HBMEC. (B) In separate experiments, the transfectants were infected with OmpA+ E. coli or OmpA− E. coli for indicated periods; total RNA was isolated and subjected to RT-PCR with ecgp96 primers. Expression of gapdh was used as a control for normalization. (C) Flow cytometry of HBMEC transfected with TLR2 or TLR4 siRNA was performed with anti-Ecgp96 antibodies. The data represent mean fluorescence intensities (MFI) after subtracting the isotype matched control values. The decrease in the expression of Ecgp96 in TLR2 siRNA transfected HBMEC as compared to untransfected control was statistically significant, *P<0.01 by Student’s t test, whereas the expression of Ecgp96 in TLR4 siRNA transfected HBMEC was similar compared to untransfected HBMEC and not statistically significant, **P=0.15 by Student’s t test. In separate experiments, the transfected HBMEC were subjected to binding (D) or invasion assays (E). The data represent mean ± SD from three different experiments performed in triplicate. Relative binding or invasion was expressed with respect to control cell values taken as 100%. The increase or decrease in binding or invasion is statistically significant compared control cells,*P<0.01 by Student’s t test.
Previously, we found that anti-Ecgp96 antibodies block E. coli K1 invasion of HBMEC (Sukumaran and Prasadarao, 2003b). Therefore, we examined the effect of anti-TLR antibodies on invasion. Both anti-Ecgp96 and anti-TLR2 antibodies reduced the binding of E. coli K1 to HBMEC by 13.5% ± 1.7%, whereas antibodies against TLR4 and TLR5 had no effect (Fig. 4A). In addition, anti-Ecgp96 and anti-TLR2 antibodies inhibited invasion by 78.7% ± 3.6%, but antibodies against TLR4 and TLR5 had no effect on this process (Fig. 4B). Of note, none of the antibodies influenced OmpA− E. coli binding to or invasion of HBMEC. These data not only confirm the role of TLR2 in OmpA+ E. coli invasion but also show that increased TLR4 expression during OmpA− E. coli infection does not affect bacterial binding or invasion.
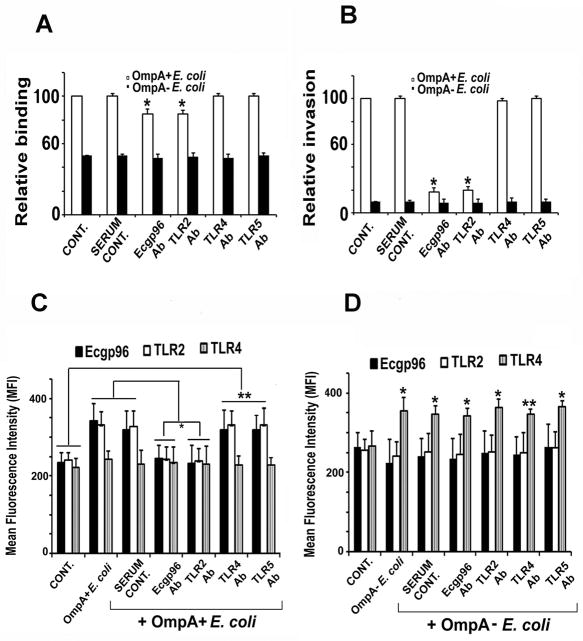
Figure 4. Effect of antibodies to Ecgp6 and TLRs on the binding, invasion and surface expression of Ecgp96.
HBMEC grown in 24 well plates to confluence were pre-incubated with normal serum or antibodies to Ecgp96, TLR2, TLR4 or TLR5 for 1 h and then infected with OmpA+ E. coli K1. Bound (A) or invaded (B) bacteria were determined as described in Methods sections. Data represent mean ± SD from three different experiments performed in triplicate. Relative binding or invasion was expressed with respect to control cell values taken as 100%. The decrease in binding and invasion of E. coli was statistically significant compared with control, *P<0.01 by Student’s t test. In separate experiments, HBMEC were pre-treated with respective antibodies for 1 h prior to infection with OmpA+ E. coli and subjected to flow cytometry analysis using antibodies to Ecgp96, TLR2 or TLR4. Increase or decrease in the expression of the respective proteins in the presence of various antibodies were statistically significant compared to control (*P<0.05 by Student’s t test).
We further determined that blocking the binding of E. coli K1 with either Ecgp96 or TLR2 antibodies prevented the subsequent increase in the expression of Ecgp96 and TLR2 during OmpA+ E. coli infection, while pre-incubation with TLR4 or TLR5 antibodies did not reduce surface expression of Ecgp96 or TLR2 (Fig. 3C). On the other hand, the anti-Ecgp96, anti-TLR2, and anti-TLR5 antibodies did not reduce the capacity of OmpA− E. coli to induce TLR4 surface expression (Fig. 4D). Also, treatment of HBMEC with the anti-TLR4 antibody prior to infection with OmpA− E. coli did not alter TLR4 expression levels. Collectively, these results indicate that initial interaction of OmpA+ E. coli with Ecgp96 and TLR2 results in a positive feedback loop that induces additional Ecgp96 and TLR2 expression on the HBMEC surface.
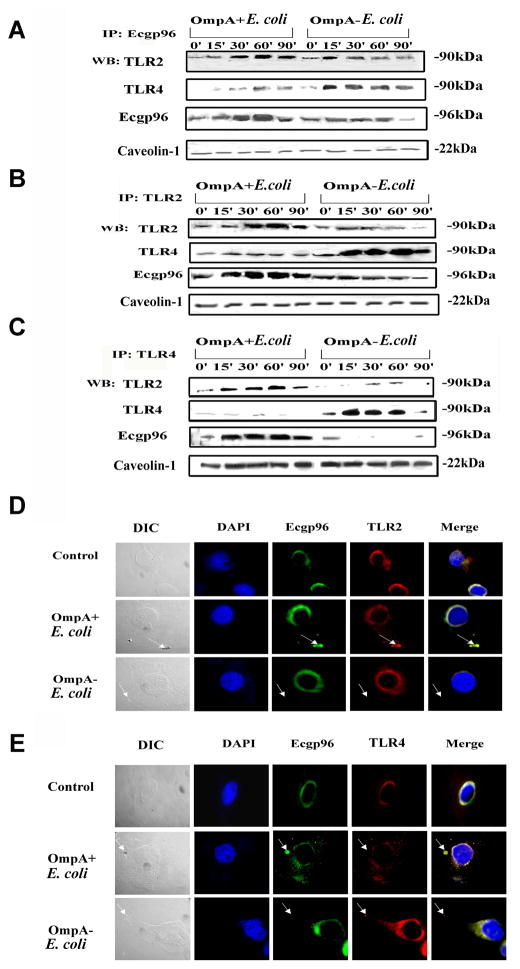
Infection with OmpA+ E. coli increases the association of Ecgp96 with TLR2 at the HMBEC plasma membrane
Since both Ecgp96 and TLR2 antibodies blocked invasion, we next determined whether Ecgp96 physically associates with TLR2 and/or TLR4 at the plasma membrane using immunoprecipitation (IP) studies. Plasma membrane fractions isolated from HBMEC infected with OmpA+ E. coli revealed a significant association of TLR2 with Ecgp96 between 30 and 90 min post-infection after IP with an anti-Ecgp96 antibody (Fig. 5A). IP with a non-specific isotype matched antibody showed no proteins after blotting with the anti-TLR or anti-Ecgp96 antibodies (data not shown). Of note, a slight increase in the levels of TLR4 was also observed when the blots were probed with anti-TLR4 antibody. The expression of Ecgp96 increased between 30 and 90 min post-infection, similar to TLR2 expression. Equal loading of proteins was confirmed by similar levels of caveolin-1 expression. OmpA− E. coli infection resulted in an initial increase in TLR2 association with Ecgp96 at 15 min post-infection, which decreased considerably by 90 min. Importantly, there was increased association of TLR4 with Ecgp96 between 15 to 90 min post-infection, even though Ecgp96 levels did not increase. IP with anti-TLR2 and anti-TLR4 antibodies of plasma membrane fractions confirmed that the association of TLR2 with Ecgp96 increases with OmpA+ E. coli infection, while TLR4 levels increase, albeit with no apparent increase in Ecgp96 expression with OmpA− E. coli infection (Fig. 5B and C).
Figure 5. Association of TLR2 with Ecgp96 at the surface of HBMEC upon infection with OmpA+ E. coli.
60 μg of membrane fractions of HBMEC, infected with OmpA+ E. coli and OmpA− E. coli for indicated time points, were immunoprecipitated with anti-Ecgp96 antibodies (A), anti-TLR2 antibodies (B) or anti-TLR4 antibodies (C) and the resulting immune complexes were subjected to Western blotting using antibodies to TLR2, TLR4 and Ecgp96. To evaluate equality of loading, 10 μg of membrane proteins were subjected to Western blotting with anti-Caveolin-1 antibodies. (D) HBMEC were infected with OmpA+ E. coli or OmpA− E. coli for 30 min in 8-well chamber slides, fixed and stained with anti-Ecgp96 antibodies followed by Alexa 488 coupled secondary antibody. The cells were also stained with anti-TLR2 antibody followed by Alexa 568 coupled secondary antibody. (E) Similarly, in separate experiments, HBMEC infected with the bacteria were stained with anti-Ecgp96 antibodies and anti-TLR4 antibodies. Arrows indicate the position of bacteria and respective accumulation of Ecgp96, TLR2 or TLR4. (Magnification: 63X).
To confirm Ecgp96/TLR2/TLR4 localization patterns during infection, immunocytochemistry of infected HBMEC was performed. As expected, because Ecgp96 acts as a chaperone for both TLR2 and TLR4, Ecgp96 co-localized with both proteins in uninfected control cells (Fig. 5D). Incubation of HBMEC with secondary antibodies alone did not reveal such localization (data not shown). However, when HBMEC were infected with OmpA+ E. coli, both Ecgp96 with TLR2 accumulated beneath the bacterial binding sites. Such an association was not observed in OmpA− E. coli infected cells. In contrast, Ecgp96 was associated with TLR4 in a random distribution along the membranes of HMBEC infected with OmpA+ E. coli, which we speculate is due to the normal chaperone activity of Ecgp96. Nonetheless, TLR4 was never observed to accumulate beneath invading OmpA+ E. coli, indicating that TLR4 association with Ecgp96 was minimal in these cells and does no contribution to invasion. Furthermore, OmpA− E. coli infected HBMEC showed no or little TLR4 association with Ecgp96 (Fig. 5E). Taken together, these results indicate that TLR2, Ecgp96 and TLR4 might form a complex at the basal level in the plasma membrane, which upon interaction with OmpA+ E. coli induces a strengthened interaction between Ecgp96 and TLR2, accompanied by upregulation of TLR2/Ecgp96 levels in the plasma membrane. During this process, the levels of TLR4 do not change. In contrast, when HBMEC were infected with OmpA− E. coli, only TLR4 expression is upregulated in the infected cells, while the levels of Ecgp96 and TLR2 in the same complex remain at basal levels.
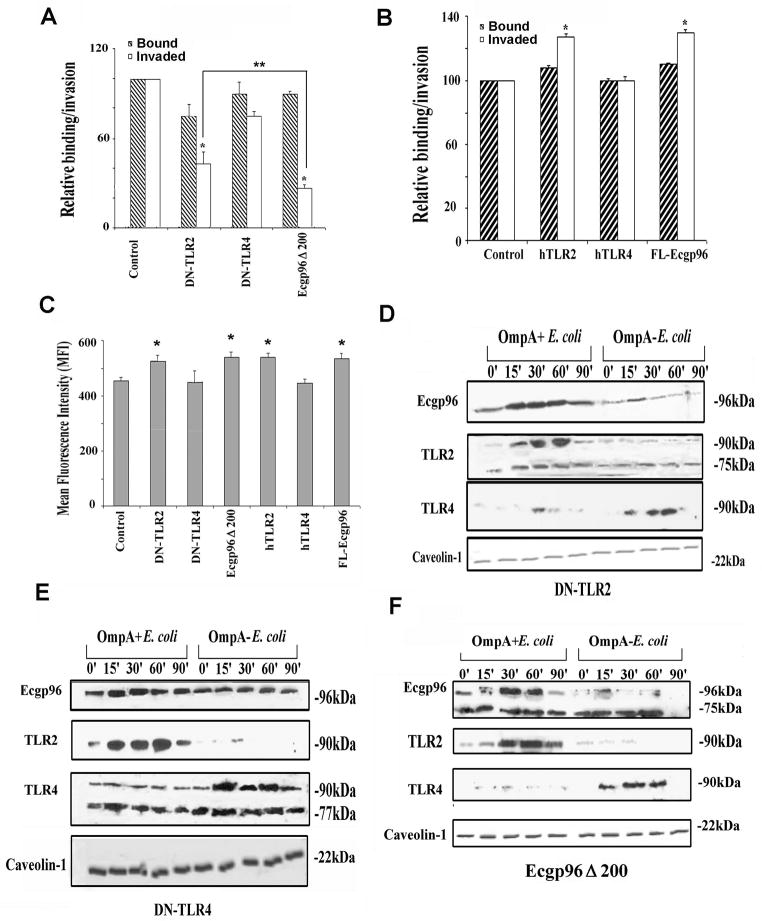
The TLR2 C-terminal domain is required for maximal invasion of HBMEC by OmpA+ E. coli
The increased association of TLR2 and Ecgp96 upon infection suggested that TLR2 may play a role in the signaling events required for the invasion of E. coli. To examine this possibility, we overexpressed dominant negative (DN) forms of TLR2 or TLR4 in HBMEC and determined there effects on OmpA+ E. coli invasion. DN-TLR2 and DN-TLR4 plasmids express truncated TLRs that lack the TIR domain necessary for downstream signaling (Smith et al., 2003). These plasmids were compared with a plasmid containing truncated Ecgp96 that lacks the C-terminal 214 amino acids of the protein (Ecgp96Δ200), which we have shown in our previous studies to inhibit invasion (Maruvada et al., 2008). For each transfection experiment, overexpression of the truncated proteins was analyzed by Western blotting using antibodies to TLRs or Ecgp96 (data not shown). Overexpression of DN-TLR2 in HBMEC inhibited the invasion of OmpA+ E. coli by 60% ± 5.2%, while Ecgp96Δ200 expression caused a significantly greater inhibition of invasion by 81% ± 4.8% (Fig. 6A). However, DN-TLR4 inhibited the invasion by only 23.6% ± 1.9%. The total cell-associated bacteria decreased by 25% ± 4.9% in DN-TLR2 and 18% ± 1.2% in Ecgp96Δ200 transfected HBMEC compared to cells transfected with the control plasmid or DN-TLR4. It should be noted that total cell associated bacteria includes both extracellular and intracellular bacteria. Since E. coli invades HBMEC at a frequency of ~0.1%, changes in the total cell associated bacteria may not appear significant. However, change in total cell associated bacteria by ~10% is sufficient to reduce or increase the invasion significantly.
Figure 6. Effect of overexpression of dominant negative constructs of Ecgp96, TLR2 or TLR4 on OmpA+ E. coli invasion and on the surface expression of Ecgp6.
(A) HBMEC transfected with DN-TLR2, DN-TLR4, or Ecgp96Δ200 were grown to confluence, and used for binding and invasion assays. The data presented are mean ± SD from three different experiments performed in triplicate. Relative bound/invaded values were expressed with respect to control values taken as 100%. The decrease in the invasion was statistically significant, *P<0.03 by Student’s t test. The difference in invasion between DN-TLR2 and DN-TLR4 transfected HBMEC was also statistically significant, **P<0.05 by Student’s t test. (B) HBMEC transfected with hTLR2, hTLR4 or FL-Ecgp96 plasmids were grown to confluence, and used for total cell associated and invasion assays. The data presented are mean ± SD from four different experiments performed in triplicate. The increase in the invasion was statistically significant in comparison to control cells, *P<0.02 by Student’s t test. (C) HBMEC were subjected to flow cytometry 24 h after transfection with the respective plasmids and probed with anti-Ecgp96 antibody followed by Alexa 488 coupled secondary antibody to analyze the surface expression of Ecgp96. Values represent mean fluorescence intensities (MFI) after subtracting values obtained from isotype-matched control antibody. The data represent mean ± SD from three different experiments. The increase or decrease in Ecgp96 expression is statistically significant compared to the expression in control cells, *P<0.05 by Student’s t test. DN-TLR2 (D), DN-TLR4 (E) or Ecgp96Δ200 (F) transfected HBMEC were infected with OmpA+ E. coli or OmpA− E. coli for indicated time points. HBMEC membranes were prepared and 60 μg of the proteins were immunoprecipitated with anti-Ecgp96 antibody. The immune complexes were then subjected to Western blot analysis with antibodies to TLR2 and TLR4. In a separate gel, equal amounts of plasma membrane proteins used for immunoprecipitation studies were subjected to Western blotting with anti-caveolin-1 antibody.
We also found that overexpression of a full length Ecgp96 (FL-Ecgp96) or TLR2 (hTLR2) increased bacterial binding by 10% ± 1.1% and increased bacterial invasion by 30% ± 2.7% compared to control HBMEC, while overexpression of full- length TLR4 (hTLR4) had no effect on binding or invasion (Fig. 6B). Flow cytometry analysis of Ecgp96 in HBMEC transfected with various dominant-negative and full length constructs of Ecgp96, TLR2 and TLR4 revealed that overexpression of either DN-TLR4 or hTLR4 did not influence the surface expression of Ecgp96, but that expression of either dominant-negative or full length Ecgp96 or TLR2 notably increased Ecgp96 on the membrane (Fig. 6C).
Although the surface expression of Ecgp96 increased upon infection with E. coli in HBMEC overexpressing DN-TLR2, invasion of these cells was significantly reduced. One possibility for this outcome is that Ecgp96 may not associate with the truncated TLR proteins at the plasma membrane. Therefore, IP studies were performed using plasma membrane preparations to examine whether the dominant negative constructs could still associate with Ecgp96 in HBMEC. IP of DN-TLR2/HBMEC membranes with anti-Ecgp96 antibody revealed that truncated TLR2 protein (75kDa) was transported to the plasma membrane (Fig. 6D). Native TLR2 (90kDa) was also observed in the membranes, indicating that Ecgp96 was associated with both transfected and endogenous forms of TLR2. The association of Ecgp96 with TLR2 was also observed in DN-TLR4/HBMEC membranes of HBMC infected with OmpA+ E. coli. OmpA− E. coli infection of DN-TLR4/HBMEC resulted in increased levels of both native and truncated TLR4 in the plasma membranes, although the level of Ecgp96 was unchanged (Fig. 6E). We also observed that Ecgp96Δ200 (74kDa) was expressed in the membranes of Ecgp96Δ200/HBMEC during infection with OmpA+ E. coli (Fig. 6F). Overexpression of Ecgp96Δ200 did not affect the expression of TLR2 or TLR4 in the membranes. These data demonstrate that the N-terminal interaction of TLR2 with Ecgp96 is sufficient for recruitment of these proteins to the plasma membrane and their association with each other. However, lack of invasion of OmpA+ E. coli into DN-TLR2/HBMEC suggests that the C-terminal portions of TLR2 and Ecgp96 are ncessary for efficient bacterial invasion.
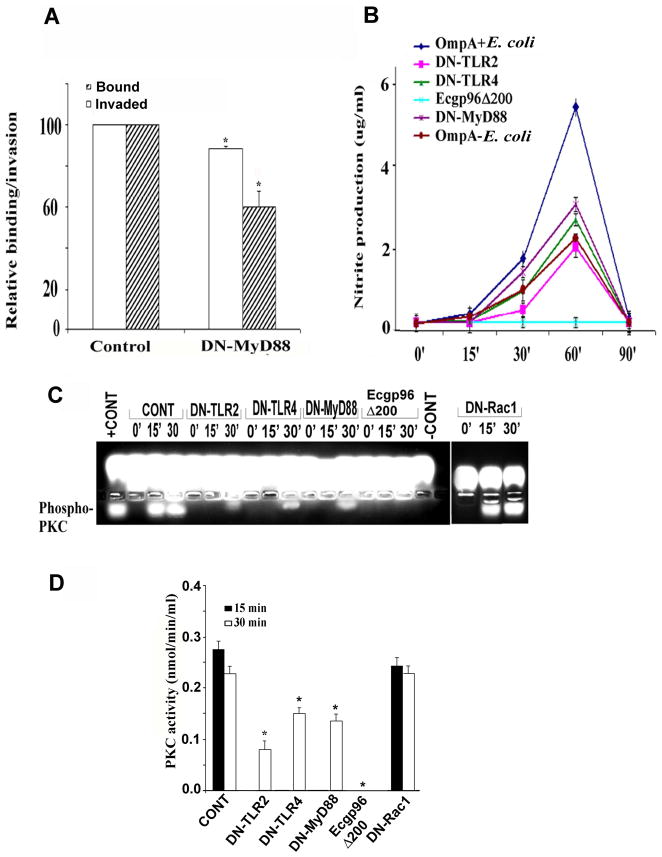
Overexpression of a dominant negative MyD88 construct in HBMEC partially inhibits invasion of E. coli
Our experimental observations have highlighted the critical role of Ecgp96 and TLR2 in OmpA+ E. coli invasion of HBMEC. Previous studies demonstrated that induction of NO enhances Ecgp96 expression in HBMEC upon infection with OmpA+ E. coli and overexpression of Ecgp96Δ200 prevents NO production and OmpA+ E. coli invasion (Mittal et al., 2010). To map the possible mechanisms by which Ecgp96 and/or TLR2 facilitate invasion, the role of MyD88, a downstream adapter protein of TLRs, was examined. Overexpression of a dominant negative MyD88 construct (DN-MyD88) in HBMEC prevented the invasion of E. coli K1 by 38.3% ± 2.3% (Fig. 7A). In addition, the number of bacteria that were cell-association with DN-MyD88/HBMEC was reduced by 14.7% ± 0.8% compared to control cells, suggesting that MyD88 might be playing a partial role in the invasion of E. coli, perhaps due to its interaction with TLR2. OmpA+ E. coli infection of HBMEC transfected with DN-TLR2 induced 2.5 ± 0.3 -fold less NO production compared to infected control cells, whereas infection of DN-TLR4 transfected HBMEC resulted in only a 1.5 ± 0.09-fold decrease in NO production (Fig. 7B). DN-MyD88/HBMEC infected with OmpA+ E. coli also produced 1.3 ± 0.07 fold less NO than infected, control HBMEC, indicating that the binding of E. coli K1 to Ecgp96 may be responsible for the production of NO partially through the TLR2/MyD88 pathway. In agreement with the essential role of Ecgp96 function in NO production, overexpression of Ecgp96Δ200 in HBMEC completely reduced NO production to basal levels.
Figure 7. Overexpression of dominant negative-TLR2, -TLR4 or -MyD88 inhibits the production of nitric oxide and activation of PKC-α.
(A) HBMEC transfected with DN-MyD88 or plasmid alone (control) were grown to confluence and used for binding and invasion assays. The data represent means ± SD from three different experiments performed in triplicate. Relative bound/invaded values were expressed with respect to control cell values taken as 100%. The reduction in the cell association and invasion of E. coli in DN-MyD88/HBMEC was statistically significant compared with plasmid-alone transfected cells, *P<0.02 by Student’s t test. (B) HBMEC transfected with various plasmid constructs were infected with OmpA+ E. coli for varying time points, supernatants were collected and analyzed for NO production as mentioned in Experimental Procedures. The data represent mean ± SD from three different experiments performed in triplicate. (C) HBMEC transfected with various plasmids were infected with OmpA+ E. coli for 0, 15 or 30 min, total cell lysates were prepared, and subjected for PKC substrate phosphorylation assay using PepTag non-radioactive kit. +ve represents the positive standard provided by the manufacturer. −ve represents a reaction performed without cell lysates. (D) Spectrophotometric analysis of phosphorylated PKC substrate bands was determined as described in Experimental Procedures. The reduction in the phosphorylation of PKC substrate in DN-TLR2, DN-TLR4, DN-MyD88 and Ecgp96Δ200 transfected and E. coli K1 infected HBMEC was statistically significant, *P<0.05 by Student’s t test.
One of the major roles of PKC-α is to govern the signaling network that induces actin polymerization by invading microbial pathogens. Previous studies showed that PKC-α phosphorylation increases between 15 and 30 min in HBMEC after infection with OmpA+ E. coli, and this phosphorylation was significantly inhibited by pre-incubating the cells with anti-Ecgp96 antibodies (Sukumaran and Prasadarao, 2002a). Therefore, the activity of PKC was evaluated in HBMEC overexpressing DN-TLR2, DN-TLR4 or DN-MyD88 using a non-radioactive PepTag assay. Although the PepTag assay determines the activity of various PKC isoforms, we have previously demonstrated that PKC-α is the only isoform activated in HBMEC upon E. coli K1 infection (Sukumaran and Prasadarao, 2002a). We found that DN-TLR2 reduced PKC activity by 81.1% ± 6.9%, whereas overexpression of DN-MyD88 and DN-TLR4 reduced it by 48.7% ± 2.1% at 30 min post-infection as determined by spectrophotometric analysis of phosphorylated protein from agarose gel slices (Fig. 7C and D). Of note, PKC activity was observed only at 30 min post infection in HBMEC overexpressing DN-TLR2, DN-TLR4 and DN-MyD88. Overexpression of Ecgp96Δ200 completely prevented PKC activity. In addition, overexpression of a dominant-negative form of Rac1 (DN-Rac1), which was previously shown to have no effect on E. coli K1 invasion in HBMEC (Rudrabhatla et al., 2006), did not block PKC activity, suggesting that the observed effect on PKC activity by the other dominant-negative constructs was not an artifact. These observations parallel the effects of the dominant negative constructs on invasion and suggest a model in which Ecgp96 associated with TLR2 to mediate OmpA+ E. coli invasion. The above results also indicate that OmpA+ E. coli induces NO production by two distinct signals, via Ecgp96 and TLR2/MyD88, but the induction predominantly occurs through Ecgp96.
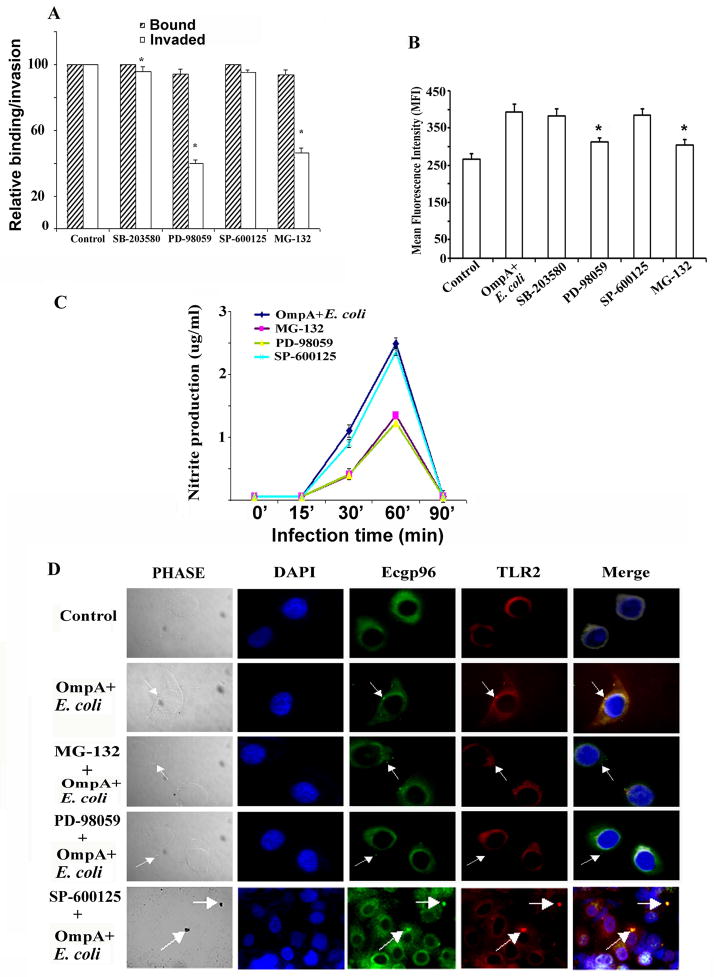
ERK1/2 and NF-κB signaling mediates TLR2 and Ecgp96 recruitment to HBMEC membranes
TLR2/MyD88 dependent signaling commonly leads to NF-κB activation through MAP kinases and thereby triggers the activation of iNOS (Iwashita et al., 1995). Therefore, we analyzed whether the inhibition of NF-κB or MAPK activation affects the invasion of OmpA+ E. coli. Cells were pretreated with the NF-κB inhibitor, MG-132, the ERK1/2 inhibitor, PD-98059, the p38 inhibitor, SB-203580, or the JNK inhibitor, SP-600125 for 30 min prior to performing the invasion assays. Cells with DMSO alone were used as controls. MG-132 blocked OmpA+ E. coli invasion by 50% ± 3.2% while PD-98059 reduced invasion by 60% ± 2.1% (Fig. 8A). Neither SB-203580 nor SP-600125 reduced invasion. Of note, the total cell associated bacteria in HBMEC treated with PD-98059 or MG-132 was slightly lower than that of control cells.
Figure 8. ERK1/2 and NF-κB inhibitors prevent the invasion of OmpA+ E. coli in HBMEC by reducing the production of NO.
(A) HBMEC were pre-treated with various inhibitors for 30 min before performing invasion assays with OmpA+ E. coli. The data presented are mean ± SD from three different experiments performed in triplicate. Relative bound/invaded values were expressed with respect to control values taken as 100%. Inhibitors of ERK1/2 and NF-κB significantly reduced the invasion compared with control cells, *P<0.01 by t test. (B) HBMEC were pre-treated with various inhibitors for 30 min, infected with OmpA+ E. coli and subjected to flow cytometry. The experiment was performed three times and reduction in Ecgp96 levels in PD-98059 and MG-132 treated HBMEC was statistically significant (*P<0.05 by Student’s t test). (C) HBMEC were pre-treated with various inhibitors for 30 min and infected with OmpA+ E. coli for different time points. Supernatants were collected and analyzed for NO production as nitrite by Griess method. The values represent mean ± SD from three different experiments performed in triplicate. (D) In separate experiments, pre-treated and infected HBMEC in 8-well chamber slides were subjected to immunocytochemistry with anti-Ecgp96 and anti-TLR2 antibodies. (Magnification: 63X). Arrows indicate the position of the bacteria or the accumulation of Ecgp96 or TLR2.
Finding that the ERK1/2 and NF-κB inhibitors inhibited invasion to a similar extent to DN-TLR2, suggested that inhibition of downstream effectors partly influences TLR2 or Ecgp96 membrane expression. To test this hypothesis were performed flow cytometric analysis of HBMEC treated with ERK1/2 and NF-κB inhibitors. We found that both inhibitors caused a 48.9% ± 2.6% reduction in the surface expression of Ecgp96 in HBMEC infected with OmpA+ E. coli, whereas the JNK inhibitor had no effect (Fig. 8B). Since our studies demonstrated that NO production enhances the expression of Ecgp96 after infection with E. coli, we postulated that ERK1/2 and NF-κB inhibitors may interfere with NO production. Therefore, the production of NO was analyzed in HBMEC that were pre-treated with ERK1/2 and NF-κB inhibitors and infected with OmpA+ E. coli for various time points. NO production was reduced by 1.5 ± 0.04 fold by inhibition of ERK1/2 or NF-κB, but not by inhibition of JNK (Fig. 8C).
To examine the effects of NF-κB and ERK1/2 inhibition on the localization of Ecgp96 and TLR2, immunocytochemistry analysis was performed. We found that in HBMEC pre-treated with NF-κB or ERK1/2 inhibitors, Ecgp96 and TLR2 failed to co-localized beneath OmpA+ E. coli (Fig. 8D). In contrast, HBMEC treated with JNK inhibitor showed normal co-localization of TLR2 and Ecgp96 similar to OmpA+ E. coli infected cells. These results suggest that ERK1/2 and NF-κB pathways are involved in Ecgp96/TLR2 mediated production of NO, which in turn induces increased Ecgp96/TLR2 localization beneath bacteria in the membrane and finally leads to invasion of more bacteria.
DISCUSSION
Breaching the BBB and dissemination into the central nervous system precedes the neurological sequelae caused by invasive bacterial pathogens. Endothelial cells express various receptors that can act as microbial sensors and transduce signals to activate innate and adaptive immune responses upon stimuli with pathogen associated molecular patterns (PAMPs). PAMPs are sensed by pathogen recognition receptors (PRRs), of which TLRs are the best characterized. However, the role of TLRs in E. coli K1 induced pathogenesis is still unclear. Despite the conserved nature of OmpA on many gram-negative bacteria, the three-dimensional structure acquired by OmpA due to the presence of other structures such as K1 capsular polysaccharide may contribute to its ability to interact with various receptors on mammalian cells (Kisiela and Czuprynski, 2009, Smith et al., 2007). Our previous studies established that OmpA of E. coli K1 interacts with Ecgp96 to invade HBMEC (Prasadarao, 2002). In addition, mutation of three residues to alanines in loop 1 and loop 2 of the extracellular domains of OmpA prevented the invasion by 95% (Pascal et al., 2010). Since Hsp90 (a homologue of Ecgp96), which has been shown to act as a chaperone for TLRs, we hypothesize that Ecgp96/TLR complex plays an important role in E. coli K1 induced pathogenesis. In this study, we demonstrate that E. coli K1 does not cause brain infection in TLR2−/− newborn mice, whereas TLR4−/− pups were more vulnerable to infection and succumbed earlier than the wild type, infected animals.
OmpA of Klebsiella pneumoniae (KpOmpA) has been previously shown to induce TLR2, but not TLR4 expression in dendritic cells (Jeannin et al., 2000). However, KpOmpA does not bind directly to TLR2, but instead binds to the scavenger receptors LOX-1 and SREC-1 (Jeannin et al., 2005). This shows that OmpA, though highly conserved among the members of Enterobacteriaceae, employs specific responses in phagocytic versus non-phagocytic cells. In agreement, infection of HBMEC with OmpA+ E. coli specifically upregulated the transcription of ecgp96 and tlr2 genes, but not tlr4, while OmpA− E. coli induced only tlr4 transcription. Similar upregulation of Ecgp96 and TLR2 at the protein level was also observed with OmpA+ E. coli. Suppression of TLR4 expression could possibly be a survival mechanism of the bacteria since it has been reported that TLR4 induction by LPS or E. coli in endothelial cells causes the recruitment of neutrophils, which effectively clear bacteria from the infection site (Andonegui et al., 2009).
There has been increasing evidence that gp96 is a receptor for bacteria and bacterial toxins. Vip surface protein of Listeria monocytogenes and toxin A of Clostridium difficile have been shown to bind gp96 (Cabanes et al., 2005, Na et al., 2008). Neisseria meningitidis porin PorB1A is responsible for binding to epithelial gp96 under low phosphate conditions but requires SREC for invasion and there appears to be no role for TLR2 or TLR4 in this process (Rechner et al., 2007). However, PorB also binds TLR2 in transfected HEK293 cells, although the caveat remains that TLR2 in these experiments was not a native molecule (Massari et al., 2006). In addition, gp96 also acts as a receptor for adherent-invasive E. coli on ileal epithelial cells (Rolhion et al., 2010). It is interesting to note that both KpOmpA and Neisseria PorB1A require SREC for invasion and to some extent gp96 expression or TLR2 activation.
Interaction of TLR2 and gp96 on the surface of B-cells requires both N-terminal and C-termini of gp96. Deletions in either of the termini of gp96 prevent gp96-mediated recruitment of TLR2 to the membrane (Randow and Seed, 2001). A recent report also showed that lack of C-terminal but not N-terminal of gp96 results in failure to chaperone TLRs (Wu et al., 2012). On the contrary, our current study shows that TLR2 is recruited to the plasma membranes in HBMEC overexpressing a C-terminal truncated form of Ecgp96. A possible reason for this contradiction could be due to the difference in cell types and also due to the fact that the previous studies used soluble TLR ligands and not whole bacteria. Our results show that knockdown of TLR2 using siRNA reduces surface expression of Ecgp96 by ~50%. This is indeed in agreement with the concept that gp96 chaperones a very limited set of proteins to the cell surface, namely the TLR family and the integrin family (Randow and Seed, 2001). Since siRNA knockdown of Ecgp96 expression in HBMEC also prevents E. coli K1 invasion (Mittal et al., 2010), it is possible that TLR2 and Ecgp96 act together for efficient invasion of the bacteria in HBMEC. Thus, enhanced expression of Ecgp96 upon infection with OmpA+ E. coli could be due to the recruitment of TLR2 to the plasma membrane.
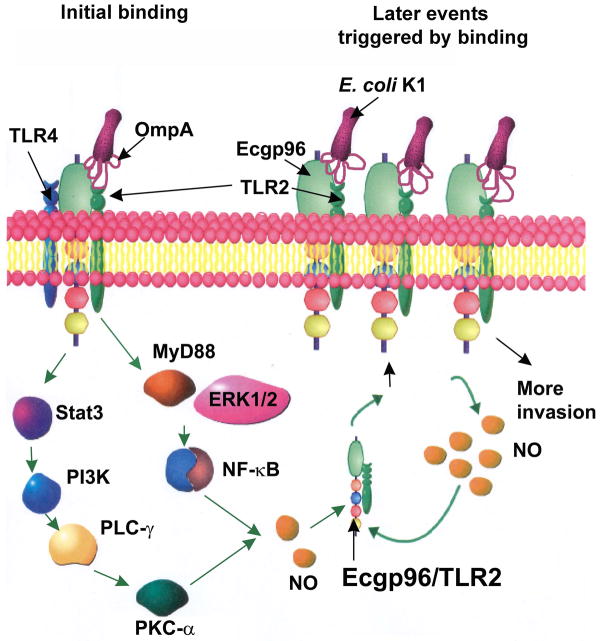
We previously demonstrated that STAT3 interaction with Ecgp96, activation of phosphatidyl inositol 3-kinase (PI3K)/Akt pathway and recruitment of PKC-α are all critical steps for E. coli K1 invasion in HBMEC (Maruvada et al., 2008, Reddy et al., 2000, Sukumaran et al., 2002b). A recent study reported that Neisseria gonorrhoeae infection in Pex cells is mediated by increased Akt levels, which in turn induces NO, which is important for bacterial survival in these cells (Edwards, 2010). E. coli K1 also requires NO for efficient invasion, and inhibitors of iNOS block invasion. In addition, we found previously that a C-terminal truncated form of Ecgp96 (Ecgp96Δ200) prevented STAT3 binding, NO production, and PKC-α activation (Mittal et al., 2010, Maruvada et al., 2008). These data confirm that Ecgp96 mediated NO production is through STAT3, PI3K and PKC-α, which in turn promotes bacterial invasion. Here, we found that blocking of TLR2 associated signaling by overexpression of a DN-TLR2 or DN-MyD88 and inhibitors of ERK1/2 or NF-κB also partially inhibits NO production. A recent report showed that TLR2/MyD88 interaction required for PKC-α to bind to this complex and induce MyD88 dependent activation of NF-κB (Langlet et al., 2010). This is in agreement with our observation that PKC activity was delayed in HBMEC overexpressing DN-TLR2 and DN-TLR4, which could be due to the lack of MyD88 binding domains in these dominant negative constructs. Of note, PKC-α has been shown to interact with Ecgp96 at the membranes of HBMEC during OmpA+ E. coli infection (Krishnan et al., 2012). Hence, it is likely that PKC-α might interact with Ecgp96/TLR2 complex to promote invasion. Based on these results, we propose that resting HBMEC express basal levels of Ecgp96/TLR2/TLR4 complex in the plasma membrane (Fig. 9). Initial interaction of OmpA+ E. coli with Ecgp96/TLR2 induces specific upregulation of Ecgp96/TLR2 complexes in the plasma membrane. OmpA binding to Ecgp96 also recruits PKC-α to the Ecgp96/TLR2 complex. Interaction of Ecgp96 with TLR2 recruits MyD88 to the complex, which in turn induce more PKC-α binding. PKC-α bound to Ecgp96 and MyD88 via ERK1/2 and NF-κB transduces signals to induce higher levels of NO production. This increased production of NO promotes the translocation of additional Ecgp96/TLR2 complexes to the plasma membrane where they act as receptors for additional bacteria to bind and invade.
Figure 9. A proposed model for E. coli K1 invasion of HBMEC mediated by OmpA and Ecgp96/TLR2 interaction.
At the initial stages of infection, HBMEC express Ecgp96/TLR2/ TLR4 complex on the cell surface. E. coli K1 interaction with Ecgp96 on HBMEC mediated by OmpA induced activation of PKC-α via Stat3, PLC-γ, PI3K, which results in the production of NO. Similarly, OmpA interaction with Ecgp96/TLR2 complex activates MyD88 and ERK1/2 followed by NF-κB, which also produces NO. Recruitment of MyD88 induces more association of PKC-α to the complex and triggers more NO production, which induces the translocation of additional Ecgp96/TLR2 complexes to the membrane. These additional Ecgp96/TLR2 complexes act as receptors for the binding of more bacteria and thereby promote increased invasion of E. coli K1.
In summary, the present study has, for the first time dissected the critical roles played by OmpA and Ecgp96/TLR2 in E. coli K1 invasion of HBMEC. TLR2 expression on the plasma membrane appears to be imperative for efficient Ecgp96 expression and optimal invasion. Given the role of OmpA as an important virulence factor for Gram-negative bacteria and the role of gp96 as a bacterial receptor, targeting either of these molecules could form the basis for efficient therapeutic options.
EXPERIMENTAL PROCEDURES
Bacteria, antibodies and other reagents
OmpA+ E. coli, a rifampin-resistant mutant of RS 218 (serotype O18:K1:H7) was isolated from the cerebrospinal fluid of a neonate with meningitis. OmpA− E. coli, a derivative of OmpA+ E. coli in which the ompA gene was disrupted, has been described earlier (Prasadarao et al., 1996). All bacteria were grown in Luria-Bertani (LB) broth with appropriate antibiotics. Bacterial media were purchased from Difco laboratories (Detroit, MI). Antibodies to TLR2, TLR4 and TLR5 were purchased from Imgenex (San Diego, CA) and Cell signaling technology, Inc., (Danvers, MA). Primers and probes for real time PCR were purchased from Roche (Indianapolis, IN). Anti-Caveolin-1 and Calnexin antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to Ecgp96 were raised as described previously (Prasadarao, 2002). Fluorescent-tagged secondary antibodies, TLR2 and TLR4 siRNA were purchased from Invitrogen (Carlsbad, CA). For plasmid transfection, FuGENE HD reagent from Roche (Indianapolis, IN) was used while Lipofectamine from Invitrogen (Carlsbad, CA) was used for siRNA transfection. Griess reagent to determine NO production was purchased from Sigma. Inhibitors of MAP kinases [PD-98059 (ERK inhibitor), SB-203580 (p38 inhibitor) and SP-600125 (JNK inhibitor)] and NF-κB (MG-132) were purchased from Calbiochem (San Diego, CA). RNeasy kit was purchased from Qiagen (Valencia, CA). Zyppy plasmid miniprep kit was obtained from Zymo Research (Orange, CA). All other chemicals were purchased from Sigma.
Cell culture, plasmid transfections and invasion assays
Cerebral cortex was obtained from surgical resections of 4- to 7-year-old children with seizure disorders at Children’s Hospital Los Angeles for capillary isolation. Human brain microvascular endothelial cells were isolated from these capillaries and stored in liquid nitrogen until they were cultured as described previously (Stins et al., 1999). 97% of the the cells were positive for Factor VIII-rag and 100% negative for GFAP as assessed by flow cytometry and 99% were positive for Ac-LDL uptake as determined by immunocytochemistry. HBMEC were maintained at 37°C in a humidified atmosphere of 5% CO2 in medium containing M-199/Ham F-12 (1:1 v/v) supplemented with 10% fetal bovine serum, 10% nu-serum, sodium pyruvate and 2 mM glutamine. Binding and invasion assays using OmpA+ E. coli and OmpA− E. coli in HBMEC were performed as described previously (Prasadarao et al., 1996). For invasion assays, HBMEC grown in 24-well cell culture plates to 95% confluence were infected with 107 cfu of E. coli strains in experimental medium (1:1 mixture of Ham’s F-12 and M-199 containing 5% heat inactivated fetal bovine serum) and incubated for 90 min at 37°C in an atmosphere containing 5% CO2. The monolayers were washed three times with RPMI-1640 medium followed by the addition of gentamicin (100 μg/ml) and further incubated for 1 h at 37°C. The cells were then washed three times with RPMI 1640 and lysed with 0.5% of Triton X-100. The released bacteria were diluted with saline and enumerated by plating on blood agar. The total cell-associated bacteria were determined as described for the invasion, except that the gentamicin step was omitted. Dominant negative TLR2 and TLR4 were a kind gift of Michael F. Smith, University of Virginia. Dominant negative MyD88, full-length human TLR2 (hTLR2) and human TLR4 (hTLR4) were purchased from Addgene (Cambridge, MA). The plasmids were transfected into HBMEC using FuGENE HD reagent according to the manufacturer’s instructions. The cells were allowed to recover for 24 h before performing invasion assays. For inhibition assays, confluent HBMEC monolayers were pre-incubated for 30 min with 50 μM of PD-98059, 1 μM of SP-600125, 0.6 μM of SB-203580 or 10 μM of MG-132. For siRNA studies, 20 pmols each of TLR2 or TLR4 siRNA was transfected into HBMEC using Lipofectamine according to the manufacturer’s instructions and allowed to recover for 24 h before performing invasion assays.
Plasma membrane isolation, immunoprecipitation and Western blotting
HBMEC grown to confluence in 100 mm Petri-dishes were infected with either OmpA+ E. coli or OmpA− E. coli for 0, 15, 30, 60 and 90 min. Subsequently, plasma membrane fractions of control and infected HBMEC were isolated using plasma membrane protein extraction kit from Biovision (Mountain View, CA). Lipid rafts were prepared from the total lysates of HBMEC using the caveolae/raft isolation kit (Sigma, St Louis, MO) as described earlier (Maruvada et al., 2008). The protein content was estimated using BCA protein assay kit (Pierce, Rockford, IL). HBMEC plasma membrane proteins (60 μg) were immunoprecipitated with respective antibodies overnight at 4°C, washed, and further incubated for 2 h at 4°C with protein A/G agarose beads. The beads were collected by centrifugation, washed twice with sterile PBS, suspended in a minimal volume of SDS sample buffer, boiled for 10 min and separated by 10% SDS-PAGE. The proteins were then transferred to a nitrocellulose membrane. The blot was blocked with 5% milk in Tris-buffered saline and 0.1% Tween-20 (TBST) for 1 h, washed four times with TBST for 5 min each and then incubated with 1:1000 anti-Ecgp96 antibody for 2 h. The membrane was washed extensively for 15 min with TBST and incubated with anti-rabbit secondary antibody conjugated to HRP for 1 h. The membrane was finally washed for 15 min with TBST and developed with Super Signal chemiluminescence substrate (Pierce, Rockford, IL) and exposed to X-ray film for protein visualization. For analyzing TLR2 and TLR4, the blot was stripped using Restore PLUS Western blot stripping buffer (Pierce, Rockford, IL) and probed with the respective antibodies according to the manufacturers’ instructions.
Real time PCR
HBMEC monolayers were infected with OmpA+ E. coli or OmpA− E. coli for varying periods, washed and total RNA was isolated using RNA isolation kit (Qiagen) and 1μg of RNA was reverse transcribed to cDNA using QuantiTect kit (Qiagen). Quantitative real-time PCR was carried out with specific primers and probes (Table 1) using the Roche Light Cycler 480 and the Light Cycler TaqMan Master Mix. Real Time PCR conditions were as follows: 95°C for 10 minutes, 45 cycles: 95°C for 10 seconds, 60°C for 30 seconds. Fold increase in expression of mRNA was normalized using GAPDH expression. Real time experiments were run three times in duplicate.
Table 1.
List of primers and probes for qPCR analysis.
| Target mRNA | Primer sequence | TaqMan™ Probe |
|---|---|---|
| gapdh | F- agccacatcgctcagacac R- gcccaatacgaccaaatcc |
60 |
| gp96 | F- ctggaaatgaggaactaacagtca R- tcttctctggtcattcctacacc |
62 |
| tlr2 | F- cgttctctcaggtgactgctc R-cctttggatcctgcttgc |
14 |
| tlr4 | F- ggtttagaagtccatcgtttgg R- cacaggccctctagagcaga |
14 |
| tlr5 | F- attgagaatgttggcgctgt R- tggtgaatcaggagaaaggact |
80 |
| tlr9 | F- tgtgaagcatccttccctgta R- gagagacagcgggtgcag |
56 |
siRNA transfection
The siRNA sequences are as follows: TLR2, primer 1: GGCAGUCUUG AACAU UAGACUUA; primer 2: AUAAGUCUAAAUGUUCAAGACUGC. TLR4: primer 1: GGGUAAGGAAUGAGCUAGUAAAGA; primer 2: UUCUUUACUAGCUCAUUCCUUACC. 20 pmols of TLR2 and TLR4 siRNA were transfected into 90% confluent HBMEC in 60 mm dishes using Lipofectamine. The cells were then allowed to recover for 24 h, RNA was isolated from these cells as mentioned above, and RT-PCR was performed to evaluate the extent of gene silencing.
RT-PCR
HBMEC were grown to 80% confluence in 60 mm dishes, transfected with TLR2 or TLR4 siRNA and analyzed for silencing. In a separate experiment, siRNA transfected HBMEC were infected with OmpA+ or OmpA− E. coli for 0, 15, 30, 60 and 90 min, washed, total RNA was isolated using RNeasy kit and RT-PCR was performed using one-step PCR kit (Qiagen). Primers for Ecgp96, TLR2, TLR4 and GAPDH were synthesized at the DNA sequencing core, USC Norris Cancer Center, University of Southern California. The primer sequences are as follows: Ecgp96 F- 5′ ATGAGGGCCCTGTGGGTG 3′; Ecgp96 R- 5′ ATACACATGACAA GATTT 3′; TLR2 F-5′ TGAGAGTGGGAAATATGGAC 3′; TLR2 R- 5′ CCTGGATCTATAA CTCTGTC 3′; TLR4 F-5′ TGGATACGTTTCCTTATAAG 3′; TLR4 R-5′ GAAATGGAG GCACCCC TTC 3′; GAPDH F-5′ CACAGTCCATGCCATCACTG 3′; GAPDH R-5′ TACTCCTTGGAG GCCATGTG 3′.
Flow cytometry
To detect the expression of Ecgp96, HBMEC were infected with E. coli for various periods. The cells were washed three times with PBS and then detached with 0.05% trypsin-EDTA from the plates. The cells were fixed using BD cytofix and pre-incubated for 30 min with blocking/wash buffer (PBS+5% normal goat serum) to mask non-specific binding sites. Cells were then incubated with anti-Ecgp96 antibody or an isotype-matched control antibody for 60 min at 4°C and washed with buffer. Then FITC-conjugated secondary antibody was added, incubated for 30 min at 4°C and washed with buffer. The stained cells were then analyzed by four-color flow cytometry using FACS Calibur Cell Quest Pro software (BD Biosciences, San Diego, CA) and at least 10,000 events were collected for analysis. Results are expressed as mean fluorescence intensity (MFI) after subtracting the values of isotype matched antibody controls.
Immunofluorescence microscopy
HBMEC grown in the 8-well chamber slides were infected with either OmpA+ E. coli or OmpA− E. coli, washed with RPMI-1640, and fixed with 2% paraformaldehyde in PBS for 15 min at room temperature. In some experiments, the inhibitors of JNK, ERK1/2 or NF-κB were incubated with HBMEC for 30 min prior to addition of the bacteria. The cells were washed and permeabilized with 0.5% Triton X-100 in PBS containing 5% normal goat serum for 30 min at room temperature. The cells were then incubated with anti-Ecgp96 antibodies (1:1000 dilution) for 1 h, washed and further incubated with FITC-coupled secondary antibodies (1:1000 dilution). The cells were further incubated with anti-TLR2 or anti-TLR4 antibodies (1:1000 dilution) for 1 h, washed and incubated with Phycoerythrin (PE)-coupled secondary antibodies (1:1000 dilution). Finally the slides were washed and mounted with Vectashield containing DAPI (Vector Laboratories, Burlingame, CA). Cells were viewed with a Leica (Wetzlar, Germany) DMRA microscope with Plan-apochromat ×63/1.65 NA and ×63/1.40 NA oil immersion objective lenses. The images were processed using Adobe Photoshop 7.0.
PepTag assay to determine PKC activity
The PepTag (Promega) assay to detect PKC activity was performed as described previously (Sukumaran and Prasadarao, 2002a). The PepTag assay uses brightly colored, fluorescent peptide substrates that are highly specific for PKC. Phosphorylation of the peptide alters the net charge from +1 to −1. This change in the net charge allows the phosphorylated and non-phosphorylated versions of the substrate to be rapidly separated on an agarose gel at neutral pH. For quantitative determination of PepTag assay results, the negatively charged phosphorylated bands were excised from the gel with a scalpel, and added to 250μl water in a 1.5ml graduated microcentrifuge tube and heated at 95°C until the gel slice is melted. 175μl of hot agarose was transferred to a tube containing 75μl of gel solubilization solution (that had been warmed to room temperature and mixed well), 100μl of glacial acetic acid and 150μl of distilled water. The mixture was vortexed and the 500μl of the solution was transferred to a 0.5ml cuvette and the absorbance was read at 570 nm.
Estimation of NO production as nitrite
To determine the production of NO by HBMEC following interaction with bacteria, OmpA+ E. coli or OmpA− E. coli was added to the wells and incubated for varying periods. At the completion of the experiment, supernatants were collected, NO produced was converted to nitrite and analyzed by Greiss method at 550 nm as previously described (Mittal et al., 2010). A standard curve was also developed in the same assay using various quantities of sodium nitrite.
Newborn mouse model of meningitis
The animal studies were approved by the Institutional Animal Care and Use Committee of the Saban Research Institute at Children’s Hospital Los Angeles (CHLA) and followed National Institutes of Health guidelines for the performance of animal experiments. Breeding pairs of C57BL/6 wild-type mice were obtained from Jackson Laboratories while TLR2−/− and TLR4−/− knockout mice were bred in Cedars-Sinai and have transferred to CHLA. Three-day-old mouse pups were randomly divided into various groups and infected intranasally with 103 cfu of OmpA+ E. coli or pyrogen free saline. Blood was collected from facial vein at different time periods post-infection, diluted in saline and plated on rifampicin LB agar plates. CSF samples were collected aseptically by cisternal puncture under anesthesia without traumatic tap and directly inoculated into broth containing appropriate antibiotics. Bacterial counts in brain tissue homogenates were determined by plating tenfold serial dilution on rifampicin LB agar plates. Growth of OmpA+ E. coli in rifampicin-containing LB broth from CSF samples was considered positive for meningitis. For histopathological analysis, one half of the brain was fixed in 10% buffered formalin, routinely processed, and embedded in paraffin. 4–5-μm sections were cut using a Leica microtome and stained with hematoxylin and eosin (H&E).
Statistical analysis
Results were analyzed using Student’s t-test and ANOVA, and P values <0.05 were considered statistically significant.
Acknowledgments
Our sincere thanks to Esteban Fernandez, Cellular Imaging Core, Children’s Hospital Los Angeles for assistance with fluorescence imaging. We also thank Denise Al Alam and members of the Bellusci lab, Children’s Hospital Los Angeles, for assistance with brain sectioning and immunohistochemistry and Scott Filler and Barbara Driscoll for critical reading of the manuscript. This work was supported by NIH grants AI40567 and NS73115 (N.V.P).
Abbreviations used
- HBMEC
human brain microvascular endothelial cells
- Ecgp96
Endothelial cell glycoprotein 96
- OmpA
outer membrane protein A
- DN
dominant negative
- PKC-α
Protein kinase C-alpha
- PI3-kinase
Phosphatidyl inositol 3-kinase
- TLRs
Toll-like receptors
Footnotes
All the authors have no conflict of interest.
References
- Aletayeb MH, Ahmad FS, D Masood. An 11-year study of causes of neonatal bacterial meningitis in Ahvaz, Iran. Pediatr Int. 2010;52:463–466. doi: 10.1111/j.1442-200X.2010.03107.x. [DOI] [PubMed] [Google Scholar]
- Andonegui G, Zhou H, Bullard D, Kelly MM, Mullaly SC, McDonald B, et al. Mice that exclusively express TLR4 on endothelial cells can efficiently clear a lethal systemic Gram-negative bacterial infection. J Clin Invest. 2009;119:1921–1930. doi: 10.1172/JCI36411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee PP, Vinay DS, Mathew A, Raje M, Parekh V, Prasad DV, et al. Evidence that glycoprotein 96 (B2), a stress protein, functions as a Th2-specific costimulatory molecule. J Immunol. 2002;169:3507–3518. doi: 10.4049/jimmunol.169.7.3507. [DOI] [PubMed] [Google Scholar]
- Cabanes D, Sousa S, Cebria A, Lecuit M, Garcia-del Portillo F, Cossart P. Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. Embo J. 2005;24:2827–2838. doi: 10.1038/sj.emboj.7600750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RJ, Chen YH. Nuclear factor-kappaB: activation and regulation during toll-like receptor signaling. Cell Mol Immunol. 2007;4:31–41. [PubMed] [Google Scholar]
- Edwards JL. Neisseria gonorrhoeae survival during primary human cervical epithelial cell infection requires nitric oxide and is augmented by progesterone. Infect Immun. 2010;78:1202–1213. doi: 10.1128/IAI.01085-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houdouin V, Bonacorsi S, Bidet P, Blanco J, De La Rocque F, Cohen R, et al. Association between mortality of Escherichia coli meningitis in young infants and non-virulent clonal groups of strains. Clin Microbiol Infect. 2008;14:685–690. doi: 10.1111/j.1469-0691.2008.02019.x. [DOI] [PubMed] [Google Scholar]
- Iwashita E, Miyahara T, Hino K, Tokunaga T, Wakisaka H, Sawazaki Y. High nitric oxide synthase activity in endothelial cells in ulcerative colitis. J Gastroenterol. 1995;30:551–554. doi: 10.1007/BF02347578. [DOI] [PubMed] [Google Scholar]
- Jeannin P, Bottazzi B, Sironi M, Doni A, Rusnati M, Presta M, et al. Complexity and complementarity of outer membrane protein A recognition by cellular and humoral innate immunity receptors. Immunity. 2005;22:551–560. doi: 10.1016/j.immuni.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Jeannin P, Renno T, Goetsch L, Miconnet I, Aubry JP, Delneste Y, et al. OmpA targets dendritic cells, induces their maturation and delivers antigen into the MHC class I presentation pathway. Nat Immunol. 2000;1:502–509. doi: 10.1038/82751. [DOI] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Kisiela DI, Czuprynski CJ. Identification of Mannheimia haemolytica adhesins involved in binding to bovine bronchial epithelial cells. Infect Immun. 2009;77:446–455. doi: 10.1128/IAI.00312-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S, Fernandez GE, Sacks DB, Prasadarao NV. IQGAP1 mediates the disruption of adherens junctions to promote Escherichia coli K1 invasion of brain endothelial cells. Cell Microbiol. 2012 doi: 10.1111/j.1462–5822.2012.01805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlet C, Springael C, Johnson J, Thomas S, Flamand V, Leitges M, et al. PKC-alpha controls MYD88-dependent TLR/IL-1R signaling and cytokine production in mouse and human dendritic cells. Eur J Immunol. 2010;40:505–515. doi: 10.1002/eji.200939391. [DOI] [PubMed] [Google Scholar]
- Maruvada R, Argon Y, Prasadarao NV. Escherichia coli interaction with human brain microvascular endothelial cells induces signal transducer and activator of transcription 3 association with the C-terminal domain of Ec-gp96, the outer membrane protein A receptor for invasion. Cell Microbiol. 2008;10:2326–2338. doi: 10.1111/j.1462-5822.2008.01214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari P, Visintin A, Gunawardana J, Halmen KA, King CA, Golenbock DT, Wetzler LM. Meningococcal porin PorB binds to TLR2 and requires TLR1 for signaling. J Immunol. 2006;176:2373–2380. doi: 10.4049/jimmunol.176.4.2373. [DOI] [PubMed] [Google Scholar]
- Mittal R, Krishnan S, Gonzalez-Gomez I, Prasadarao NV. Deciphering the roles of outer membrane protein A extracellular loops in the pathogenesis of Escherichia coli K1 meningitis. J Biol Chem. 2011;286:2183–2193. doi: 10.1074/jbc.M110.178236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R, Prasadarao NV. Nitric oxide/cGMP signalling induces Escherichia coli K1 receptor expression and modulates the permeability in human brain endothelial cell monolayers during invasion. Cell Microbiol. 2010;12:67–83. doi: 10.1111/j.1462-5822.2009.01379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na X, Kim H, Moyer MP, Pothoulakis C, LaMont JT. gp96 is a human colonocyte plasma membrane binding protein for Clostridium difficile toxin A. Infect Immun. 2008;76:2862–2871. doi: 10.1128/IAI.00326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta CV. Re-evaluating the role of heat-shock protein-peptide interactions in tumour immunity. Nat Rev Immunol. 2003;3:427–432. doi: 10.1038/nri1089. [DOI] [PubMed] [Google Scholar]
- Pascal TA, Abrol R, Mittal R, Wang Y, Prasadarao NV, Goddard WA., 3rd Experimental validation of the predicted binding site of Escherichia coli K1 outer membrane protein A to human brain microvascular endothelial cells: identification of critical mutations that prevent E. coli meningitis. J Biol Chem. 2010;285:37753–37761. doi: 10.1074/jbc.M110.122804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasadarao NV. Identification of Escherichia coli outer membrane protein A receptor on human brain microvascular endothelial cells. Infect Immun. 2002;70:4556–4563. doi: 10.1128/IAI.70.8.4556-4563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasadarao NV, Wass CA, Stins MF, Shimada H, Kim KS. Outer membrane protein A-promoted actin condensation of brain microvascular endothelial cells is required for Escherichia coli invasion. Infect Immun. 1999;67:5775–5783. doi: 10.1128/iai.67.11.5775-5783.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasadarao NV, Wass CA, Weiser JN, Stins MF, Huang SH, Kim KS. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect Immun. 1996;64:146–153. doi: 10.1128/iai.64.1.146-153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F, Seed B. Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat Cell Biol. 2001;3:891–896. doi: 10.1038/ncb1001-891. [DOI] [PubMed] [Google Scholar]
- Rechner C, Kuhlewein C, Muller A, Schild H, Rudel T. Host glycoprotein Gp96 and scavenger receptor SREC interact with PorB of disseminating Neisseria gonorrhoeae in an epithelial invasion pathway. Cell Host Microbe. 2007;2:393–403. doi: 10.1016/j.chom.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Reddy MA, Prasadarao NV, Wass CA, Kim KS. Phosphatidylinositol 3-kinase activation and interaction with focal adhesion kinase in Escherichia coli K1 invasion of human brain microvascular endothelial cells. J Biol Chem. 2000;275:36769–36774. doi: 10.1074/jbc.M007382200. [DOI] [PubMed] [Google Scholar]
- Rolhion N, Barnich N, Bringer MA, Glasser AL, Ranc J, Hebuterne X, et al. Abnormally expressed ER stress response chaperone Gp96 in CD favours adherent-invasive Escherichia coli invasion. Gut. 2010;59:1355–1362. doi: 10.1136/gut.2010.207456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrabhatla RS, Selvaraj SK, Prasadarao NV. Role of Rac1 in Escherichia coli K1 invasion of human brain microvascular endothelial cells. Microbes Infect. 2006;8:460–469. doi: 10.1016/j.micinf.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh SI. Chaperones and transport proteins regulate TLR4 trafficking and activation. Immunobiology. 2009;214:594–600. doi: 10.1016/j.imbio.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Selvaraj SK, Periandythevar P, Prasadarao NV. Outer membrane protein A of Escherichia coli K1 selectively enhances the expression of intercellular adhesion molecule-1 in brain microvascular endothelial cells. Microbes Infect. 2007;9:547–557. doi: 10.1016/j.micinf.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MF, Jr, Mitchell A, Li G, Ding S, Fitzmaurice AM, Ryan K, et al. Toll-like receptor (TLR) 2 and TLR5, but not TLR4, are required for Helicobacter pylori-induced NF-kappa B activation and chemokine expression by epithelial cells. J Biol Chem. 2003;278:32552–32560. doi: 10.1074/jbc.M305536200. [DOI] [PubMed] [Google Scholar]
- Smith SG, Mahon V, Lambert MA, Fagan RP. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol Lett. 2007;273:1–11. doi: 10.1111/j.1574-6968.2007.00778.x. [DOI] [PubMed] [Google Scholar]
- Stins MF, Nemani PV, Wass C, Kim KS. Escherichia coli binding to and invasion of brain microvascular endothelial cells derived from humans and rats of different ages. Infect Immun. 1999;67:5522–5525. doi: 10.1128/iai.67.10.5522-5525.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran SK, McNamara G, Prasadarao NV. Escherichia coli K-1 interaction with human brain micro-vascular endothelial cells triggers phospholipase C-gamma1 activation downstream of phosphatidylinositol 3-kinase. J Biol Chem. 2003a;278:45753–45762. doi: 10.1074/jbc.M307374200. [DOI] [PubMed] [Google Scholar]
- Sukumaran SK, Prasadarao NV. Regulation of protein kinase C in Escherichia coli K1 invasion of human brain microvascular endothelial cells. J Biol Chem. 2002a;277:12253–12262. doi: 10.1074/jbc.M110740200. [DOI] [PubMed] [Google Scholar]
- Sukumaran SK, Prasadarao NV. Escherichia coli K1 invasion increases human brain microvascular endothelial cell monolayer permeability by disassembling vascular-endothelial cadherins at tight junctions. J Infect Dis. 2003b;188:1295–1309. doi: 10.1086/379042. [DOI] [PubMed] [Google Scholar]
- Sukumaran SK, Quon MJ, Prasadarao NV. Escherichia coli K1 internalization via caveolae requires caveolin-1 and protein kinase Calpha interaction in human brain microvascular endothelial cells. J Biol Chem. 2002b;277:50716–50724. doi: 10.1074/jbc.M208830200. [DOI] [PubMed] [Google Scholar]
- Sunakawa K, Sakai F, Hirao Y, Hanaki H, Nonoyama M, Iwata S, et al. Childhood bacterial meningitis trends in Japan from 2007 to 2008. Kansenshogaku Zasshi. 2010;84:33–41. doi: 10.11150/kansenshogakuzasshi.84.33. [DOI] [PubMed] [Google Scholar]
- Tsan MF, Gao B. Heat shock proteins and immune system. J Leukoc Biol. 2009;85:905–910. doi: 10.1189/jlb.0109005. [DOI] [PubMed] [Google Scholar]
- Wu S, Hong F, Gewirth D, Guo B, Liu B, Li Z. The Molecular Chaperone gp96/GRP94 Interacts With Toll-Like Receptors And Integrins Via Its C-Terminal Hydrophobic Domain. J Biol Chem. 2012;287:6735–6742. doi: 10.1074/jbc.M111.309526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu B, Dai J, Srivastava PK, Zammit DJ, Lefrancois L, Li Z. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity. 2007;26:215–226. doi: 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi AK, Thaver D, Ali SA, Khan TA. Pathogens associated with sepsis in newborns and young infants in developing countries. Pediatr Infect Dis J. 2009;28:S10–18. doi: 10.1097/INF.0b013e3181958769. [DOI] [PubMed] [Google Scholar]