Abstract
Imputation in admixed populations is an important problem but challenging due to the complex linkage disequilibrium (LD) pattern. The emergence of large reference panels such as that from the 1,000 Genomes Project enables more accurate imputation in general, and in particular for admixed populations and for uncommon variants. To efficiently benefit from these large reference panels, one key issue to consider in modern genotype imputation framework is the selection of effective reference panels. In this work, we consider a number of methods for effective reference panel construction inside a hidden Markov model and specific to each target individual. These methods fall into two categories: identity-by-state (IBS) based and ancestry-weighted approach. We evaluated the performance on individuals from recently admixed populations. Our target samples include 8,421 African Americans and 3,587 Hispanic Americans from the Women’s Health Initiative, which allow assessment of imputation quality for uncommon variants. Our experiments include both large and small reference panels; large, medium, and small target samples; and in genome regions of varying levels of LD. We also include BEAGLE and IMPUTE2 for comparison. Experiment results with large reference panel suggest that our novel piecewise IBS method yields consistently higher imputation quality than other methods/software. The advantage is particularly noteworthy among uncommon variants where we observe up to 5.1% information gain with the difference being highly significant (Wilcoxon signed rank test P-value < 0.0001). Our work is the first that considers various sensible approaches for imputation in admixed populations and presents a comprehensive comparison.
Keywords: genotype imputation, admixed populations, large reference panel, uncommon variants, MaCH-Admix
INTRODUCTION
Imputation of untyped genetic markers has been routinely performed in genome-wide association studies (GWAS) [Sanna et al., 2010; Scott et al., 2007; WTCCC, 2007] and meta-analysis [Dupuis et al., 2010; Smith et al., 2010; Willer et al., 2008], and will continue to play an important role in sequencing-based studies [Fridley et al., 2010; The 1000 Genomes Project Consortium, 2010]. We have previously developed a hidden Markov model (HMM) based method for imputation [Li et al., 2010] and shown that it achieves high imputation accuracy in a number of populations [Huang et al., 2009], particularly those with high level of linkage disequilibrium (LD) or having closely matched reference population(s) from the HapMap [The International HapMap Consortium, 2010] or the 1000 Genomes Projects (1000G) [The 1000 Genomes Project Consortium, 2010]. However, little methodological work exists for imputation in admixed populations, such as African Americans and Hispanic Americans, which comprise more than 20% of the US population (see Web Resources).
Admixed populations offer a unique opportunity for gene mapping because one could utilize admixture LD to search for genes underlying diseases that differ strikingly in prevalence across populations [Reich and Patterson, 2005; Rosenberg et al., 2010; Tang et al., 2006;Winkler et al., 2010; Zhu et al., 2004]. Although useful for admixture mapping, admixture LD also imposes challenges for imputation. Because an admixed individual’s genome is a mosaic of ancestral chromosomal segments, to appropriately impute the genotypes, it is imperative to incorporate the underlying ancestry information. Practically, this is equivalent to selecting an appropriate reference panel that matches the corresponding ancestral population(s).
We and others have evaluated a wide range of choices on the construction of a reference panel prior to running the imputation engine. The recommendation is to use a predefined panel that either combines all reference populations (a cosmopolitan panel) [Hao et al., 2009; Li et al., 2009; Shriner et al., 2010] or a weighted combination panel [Egyud et al., 2009; Huang et al., 2009; Pasaniuc et al., 2010; Pemberton et al., 2008]. The cosmopolitan panel may include haplotypes from populations that are irrelevant, and fails to reflect the underlying ancestry proportions and consequently the LD pattern for the target population. The weighted combination panel is generated by duplicating haplotypes according to certain weights, which substantially and unnecessarily increases computational costs [Egyud et al., 2009].
An alternative approach, based on identity-by-state (IBS) sharing between the target individual and haplotypes in the reference populations, can be embedded within existing imputation models. This approach constructs individual-specific effective reference panels, by selecting the most closely related haplotypes (according to IBS score) from the entire reference pool. The IBS-based selection is intuitive and useful for reducing the size of the effective reference panel and is tailored separately for each target individual. The selection is usually conducted by finding pairwise Hamming distances, which is computationally very appealing. A simple IBS-based method, which selects a subset of haplotypes into the effective reference panel according to their Hamming distance with the haplotypes to be inferred across the entire genomic region to be imputed (hereafter referred to as whole haplotype), has been adopted by IMPUTE2 [Howie et al., 2009]. Although some promising results have been shown when compared with random selection, no work has examined alternatives to this simple whole-haplotype based matching, partly due to the heavy computational burden posed.
In this work, we evaluated two classes of reference selection methods: IBS-based and ancestry-weighted approaches. Among the IBS-based approaches, we propose a novel method based on IBS matching in a piecewise manner. The method breaks genomic region under investigation into small pieces and finds reference haplotypes that best represent every small piece, for each target individual separately. The method can be incorporated directly into existing imputation algorithms and has identical computational complexity to that of the existing whole-haplotype IBS-based method. Results from all real datasets evaluated suggest that our piecewise IBS method is highly robust and stable even when a small number of reference haplotypes are selected. Importantly, for uncommon variants, our piecewise IBS selection method manifests more pronounced advantage with large reference panels.
We have implemented all methods evaluated, including our piecewise IBS selection method, in our software package MaCH-Admix. Besides the new reference selection functionality, our software also retains high flexibility in two major aspects. First, both regional and whole-chromosome imputation can be accommodated. Second, both data-independent and data-dependent model parameter estimation are supported. Thus, besides standard reference panel with precalibrated parameters, we can elegantly handle study-specific reference panels and target samples with unknown ethnic origin.
The rest of the paper is organized as follows. We first present the general framework of our imputation algorithm, followed by the intuition and formulation of our piecewise IBS and various other effective reference selection methods. Then we evaluate all these methods implemented in MaCH-Admix, the whole-haplotype IBS method implemented in IMPUTE2 [Howie et al., 2009], and BEAGLE[Browning and Browning, 2009] using the following datasets:
3,587 Hispanic American individuals from the Women’s Health Initiative (WHI),
8,421 African American individuals from the WHI,
49 HapMap III African American individuals,
50 HapMap III Mexican individuals.
All datasets are imputed with reference from the 1000 Genomes Project (2,188 haplotypes). We also explored the performance with small/medium reference set from HapMap II/III. Finally, we provide practical guidelines for imputation in admixed populations in the Discussion section.
MATERIAL ANDMETHODS
Assume that we have n individuals in the target population that are genotyped at a set of markers denoted by Mg. In addition, we have an independent set of H reference haplotypes, for example, those from the International HapMap or the 1000 Genomes Projects, encompassing a set of markers denoted by Mr. Without loss of generality, we assume that the set of markers assayed in the target population, Mg, is a subset of Mr, the markers in the reference population. The goal of genotype imputation is to fill in missing genotypes including those missing by design (e.g., genotypes at markers in Mr but not Mg, commonly referred to as untyped markers). As described earlier [Li et al., 2010], our HMM as implemented in MaCH fulfills the goal by inferring the haplotypes encompassing Mr markers for each target individual, from unphased genotypes at the directly assayed markers in Mg. Haplotype reconstruction is accomplished by building imperfect mosaics using some of the H reference haplotypes.
GENERAL FRAMEWORK
Because admixed individuals have inherited genetic information from more than one ancestral population, we start with a pooled panel: a panel with haplotypes from all relevant populations, for example, CEU+YRI for African Americans and CEU+YRI+JPT+CHB for Hispanic Americans, where CEU is an abbreviation for Utah residents (CEPH) with Northern and Western European ancestry; YRI for Yoruba in Ibadan, Nigeria; JPT for Japanese in Toyko, Japan; and CHB for Han Chinese in Beijing, China. Let 𝒢 = (g1, g2, g3, …, gMr) denote the unphased genotypes at Mr markers for a target individual. Furthermore, we define a series of variables Sm, m = 1, 2, …, Mr to denote the hidden state underlying each unphased genotype gm. The hidden state Sm consists of an ordered pair of indices (xm, ym) indicating that, at marker m, the first chromosome of this particular target individual uses reference haplotype xm as the template and the second chromosome uses reference haplotype yM as the template, where xm and ym both take values from {1, 2, …, H}.
We seek to infer the posterior probabilities of the sequence of hidden states 𝒮 = (S1, S2, …, SMr) for each individual as the knowledge of 𝒮 will determine genotype at each of the Mr markers. Define P(Sm |ℋ, 𝒢) as the posterior probability for Sm, the hidden state at marker m with ℋ denoting the pool of reference haplotypes and 𝒢 denoting the genotype vector of the target individual. To infer these posterior probabilities, we run multiple Markov iterations. Within each iteration, we calculate the conditional joint probabilities P(Sm, 𝒢|ℋ) at each marker m via an adapted Baum’s forward and backward algorithm as previously described [Li et al., 2010].
For admixed populations, as we tend to include more reference haplotypes in the pool under the philosophy of erring on the safe side, and as we attempt not to duplicate haplotypes, one key aspect of the modeling is on how to traverse the sample space harboring the most probability mass with minimum computational efforts.
PIECEWISE IBS-BASED REFERENCE SELECTION
In piecewise IBS selection, we seek to construct a set of t effective reference haplotypes from the pool of H haplotypes within each HMM iteration for each target individual separately. Selected reference panels are therefore tailored for each target individual. For presentation clarity, we consider a single target individual. Specifically, we calculate the genetic similarity (measured by IBS, the Hamming distance between two haplotypes) in a piecewise manner between the individual and each haplotype in the reference pool, ignoring the subpopulations (e.g., CEU or YRI) within the reference.
Denote as the current haplotype guess for the target individual. We break haplotype into a maximum of pieces so that the typed markers are evenly placed across pieces. Each piece has a minimum length of ν typed markers to ensure that the calculated Hamming distance is informative. Denote the number of pieces by p. For each haplotype piece, we calculated the piece-specific IBS score between and each reference haplotype and selects the top reference haplotypes, resulting in a total of selected for across all p regions. We repeat the same procedure for and select a second set of reference haplotypes. In our implementation, we set ν = 32, which corresponds to an average length of <200 kb for commonly used genome-wide genotyping platforms. To avoid creating spurious recombinations at piece boundary, we apply a random offset to the first piece in each sampling so that the boundaries differ across iterations. In the case where is not an integer, we selects (the ceiling integer) reference haplotypes in each piece for each target haplotype. Then we sample randomly from the selected reference haplotypes. Note that the piecewise selection is repeated for each individual in each sampling iteration. Thus, the selection will change along with the intermediate sampling results.
We have also implemented two whole-haplotype IBS-based methods, IBS Single Queue (IBS-SQ) and IBS Double Queue (IBS-DQ). The former defines IBS score with any reference haplotype as the minimum Hamming distance to , thus ordering the H reference haplotypes in a single queue. The top t reference haplotypes will be selected accordingly. The latter defines two separate IBS scores for , thus ordering the H reference haplotypes in two queues. The top t/2 reference haplotypes will be selected for according to IBS scores for . Similarly, another t/2 reference haplotypes will be selected for .
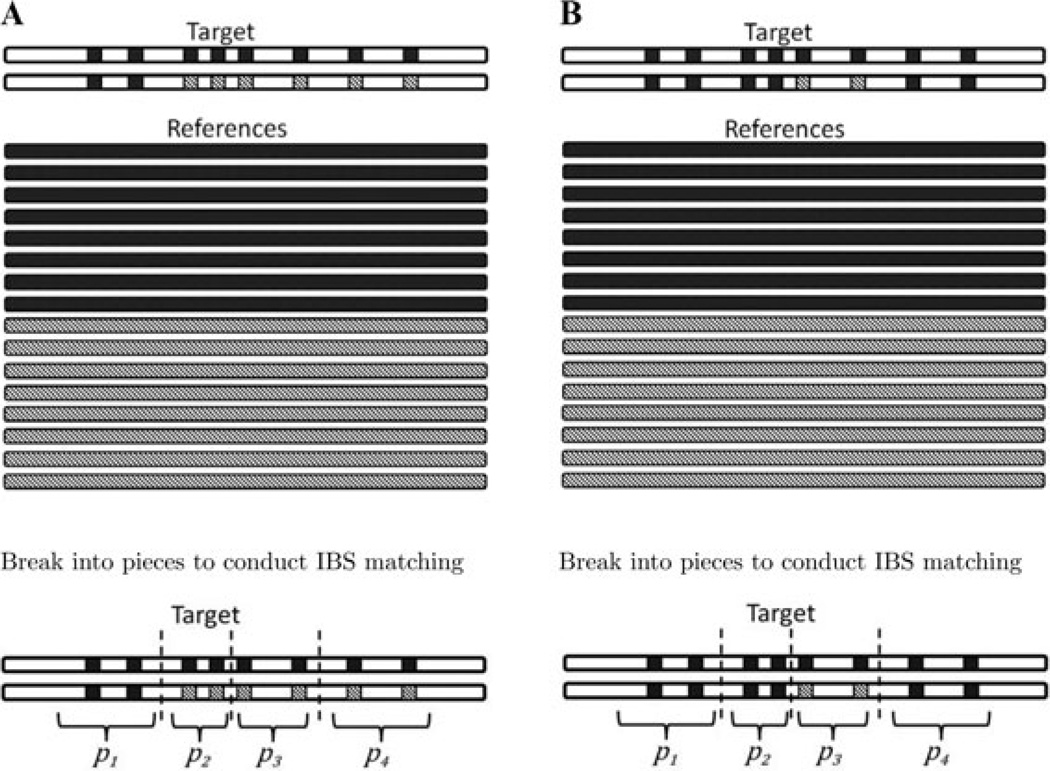
Figure 1 explains the three IBS strategies under two simple scenarios. In both scenarios, there are eight markers measured in both target and reference with color indicating the allelic status where the same color at the same locus implies the same allele. In both Figure 1A and B, the first chromosome of the target individual shares all eight alleles with the dark-colored reference haplotypes and zero alleles with the light-shaded reference haplotypes. In Figure 1A, the second chromosome of the target individual shares two alleles with the dark-colored reference haplotypes and the remaining six alleles with the light-shaded reference haplotypes; whereas in Figure 1B, the second chromosome shares six alleles with the dark-colored reference haplotypes and the remaining two alleles with the light-shaded reference haplotypes.
Fig. 1.
A cartoon illustration of two scenarios where the three IBS-based selection methods perform differently. The two lines on the top panel represent the two chromosomes of a target individual and the lines on the bottom panel represent the pool of H = 16 reference haplotypes. Color determines the allelic status such that the same color at the same locus implies the same allele. The bottom parts show how our piecewise selection method breaks the imputation region into four pieces with . Here we assume no constraint on the minimum piece size (i.e., ν = 0).
Suppose . Figure 1A illustrates a scenario where the whole-haplotype Single Queue strategy is not optimal because only dark-colored haplotypes will be selected into the effective reference panel. By combining two sets selected from two separate queues, the whole-haplotype Double Queue strategy is advantageous in the scenario. On the other hand, neither the whole-haplotype Single Queue nor the whole-haplotype Double Queue strategy can handle the scenario in Figure 1B well because both strategies would only select the dark-colored reference haplotypes. Ideally, the selected reference haplotypes should, when possible, contain information to represent every part of both chromosomes carried by the target individual. In the scenario presented in Figure 1B, because the target individual carries segment of the light-shaded haplotype, it is desirable to have some representation of the light-shaded haplotypes in the effective reference panel. Our piecewise IBS method achieves this by breaking the whole region into pieces and selecting some reference haplotypes according to genetic matching in each piece (illustrated in the bottom part of Figure 1A and B). By conducting local IBS matching and choosing a few reference haplotypes within each piece, it is able to have some representation of the light-shaded reference haplotypes. As a result, all parts of the target chromosomes are well represented by the selected reference haplotypes. In general, we believe that selecting a small number of reference haplotypes for each piece locally performs better than selecting globally at the whole-haplotype level. Note that the piecewise IBS method has the same computational complexity as the two whole-haplotype IBS methods.
ANCESTRY-WEIGHTED APPROACH
Besides IBS-based methods, we also evaluate an ancestry-weighted selection method, which is motivated by the idea of weighted cosmopolitan panel discussed in the Introduction section. This method concerns the scenario where the reference panel consists of haplotypes from several populations, for instance CEU and YRI, such that the H reference haplotypes are naturally decomposed into several groups. Let Q denote the number of populations included and Hq denote the number of haplotypes from reference population q, q = 1, 2, …, Q. We first consider the issue of weight determination for each contributing reference population, that is, the fraction of reference haplotypes to be selected from that population. Intuitively, the weights should depend on the proportions of ancestry from these reference populations for the target admixed individual(s). The weights can be, on one extreme, the same for all individuals in the target population (e.g., when the admixture makeup is similar across all individuals), or different for subpopulations within the target population, or on the other extreme, specific for each target individual. For presentation clarity, we suppress the individual index i and denote w = (w1, w2, …, wQ) as the vector of weights, under the constraint that w1 + w2 + ⋯ wQ = 1. In this work, we consider the same set of weights for all target individuals. The weights are to represent the average contributions over the imputation region and for all target individuals. We choose to use such average weights over weights specific to each single individual because the average weights can be more stably estimated.
There are several natural ways to estimate the weights. One could prespecify the weights according to estimates of ancestry proportion. For example, it is reasonable to use a ~2:8 CEU:YRI weighting scheme for African Americans who are estimated to have about 20% Caucasian and 80% African ancestries [Lind et al., 2007; Parra et al., 1998; Reiner et al., 2007; Stefflova et al., 2011]. Alternatively, one can estimate the ancestry proportions for the target individuals under investigation. We have implemented an imputation-based approach within MaCH-Admix to infer ancestry proportions, according to the contributions of reference haplotypes from each population to the constructed mosaics of the target individuals so that the weights can be estimated by MaCH-Admix internally. We use the software package structure [Pritchard et al., 2000], specifically its Admix+LocPrior model, on LD-pruned set of single nucleotide polymorphisms (SNPs) to confirm our internal ancestry inference.
Having determined the weights, we are interested in constructing a set of t effective reference haplotypes within each Markov iteration from the pool of H reference haplotypes according to the ancestry proportions. We achieve this by sampling without replacement t × wq haplotypes from the Hq haplotypes in reference population q. For each target individual, we sample a different reference panel under the same set of weights.
MACH-ADMIX
We have implemented the aforementioned methods (three IBS-based and one ancestry-weighted) in our software package MaCH-Admix. MaCH-Admix breaks the one-step imputation in MaCH into three steps: phasing, model parameter (including error rate and recombination rate parameters) estimation, and haplotype-based imputation. The splitting into phasing and haplotype-based imputation is similar to IMPUTE2. Our software can accommodate both regional and whole-chromosome imputation and allows both data-dependent and data-independent model parameter estimation. The flexibility regarding model parameter estimation allows one to perform imputation with standard reference panels such as those from the HapMap or the 1000 Genomes Projects with precalibrated parameters in a data-independent fashion, similar to IMPUTE2, which uses recombination rates estimated from the HapMap data and a constant mutation rate. Alternatively, if one works with study-specific reference panels, or suspects the model parameters differ from those precalibrated (e.g., when target individuals are of unknown ethnicity or from an isolated population), one has the option to simultaneously estimate these model parameters while performing imputation.
DATASETS
We assessed the reference selection methods in the following six target sets:
3,587 WHI Hispanic Americans (WHI-HA),
8,421 WHI African Americans (WHI-AA),
200 randomly sampled WHI-HA individuals,
200 randomly sampled WHI-AA individuals,
49 HapMap III African Americans (ASW),
50 HapMap III Mexican individuals (MEX).
The WHI SHARe consortium offers one of the largest genetic studies in admixed populations. WHI [Anderson et al., 2003; The WHI Study Group, 1998] recruited a total of 161, 808 women with 17% from minority groups (mostly African Americans and Hispanics) from 1993 to 1998 at 40 clinical centers across the United States. The WHI SHARe consortium genotyped all the WHI-AA and WHI-HA individuals using the Affymetrix 6.0 platform. Detailed demographic and recruitment information of these genotyped samples are previously described [Qayyum et al., 2012]. Besides standard quality control (details described previously in [Liu et al., 2012]), we removed SNPs with minor allele frequency (MAF) below 0.5%. To evaluate the imputation performance on target sets of smaller size, we randomly sampled 200 individuals from WHI-HA and WHI-AA separately.
For the two HapMapIII datasets, our target individuals are ASW (individuals of African ancestry in Southwest USA) and MEX (individuals of Mexican ancestry in Los Angeles, California) respectively from the phase III of the International HapMap Project [The International HapMap Consortium, 2010]. These individuals (83 ASW and 77 MEX) were all genotyped using two platforms: the Illumina Human1M and the Affymetrix 6.0. We restricted our analysis to founders only: 49 ASW and 50 MEX.
The main focus of our work is imputation with large reference panel. Thus, we first evaluated the imputation performance of all six target sets with reference from the 1000 Genomes Project (release 20101123, H = 2, 188 haplotypes). For the WHI datasets, the number of markers overlapping between the target and reference, bounded by the number of markers typed in target samples, is smaller than that in the HapMap individuals. Therefore, we performed imputation 10 times, each time masking a different 5% of the Affymetrix 6.0 markers. This masking strategy allowed us to evaluate imputation quality at 50% of Affymetrix 6.0 SNPs. For HapMap III ASW and MEX individuals, we randomly masked 50% of the overlapping markers and evaluated the performance at these markers. We used two different masking schemes for the HapMap and WHI samples because we have ~1.5 million typed markers in the HapMap samples and thus can still achieve reasonable imputation accuracy by masking 50% of the markers in a single trial. In the WHI samples, masking 50% of the ~0.8 million markers in a single trial would substantially reduce imputation accuracy and using one trial with a small percentage of markers masked would lead to insufficient number of markers for evaluation. Therefore, we used multiple trials with 5% masking for the WHI datasets.
To provide a comprehensive evaluation, we also conducted imputation on all six target sets using HapMapII or HapMapIII haplotypes as reference. We used HapMap II CEU+YRI (H = 240) for WHI-AA individuals and HapMapII CEU+YRI+JPT+CHB (H = 420) for WHI-HA individuals. The evaluation is based on masking 50% of the overlapping markers. For HapMap III ASW target set, we considered three different reference panels: HapMapII CEU+YRI (H = 240), HapMapIII CEU+YRI (H = 464), and HapMapIII CEU+YRI+LWK+MKK (H = 930), where LWK (Luhya in Webuye, Kenya) and MKK (Maasai in Kinyawa, Kenya) are two African populations from Kenya. For HapMap III MEX target set, we considered HapMapII CEU+YRI+JPT+CHB (H = 420), and HapMapIII CEU+YRI+JPT+CHB (H = 804). For the HapMap target sets with HapMap references, we used genotypes at SNPs on the Illumina HumanHap650 BeadChip for imputation input and reserved other genotypes for evaluation. We have posted the HapMap data and our command lines used in this work on MaCH-Admix website (see Web Resources).
We picked five 5 Mb regions across the genome to represent a wide spectrum of LD levels. We first calculated median half life of r2, defined as the physical distance at which the median r2 between pairs of SNPs is 0.5, for every 5 Mb region using a sliding window of 1 Mb, in CEU, YRI, and JPT+CHB, respectively. We used HapMapII phased haplotypes for the calculation. The five regions we picked are: chromosome3: 80–85 Mb, chromosome1: 75–80 Mb, chromosome4: 57–62 Mb, chromosome14: 50–55 Mb, and chromosome8: 18–23 Mb in a decreasing order of LD level. The median half life of r2 is around 90th, 70th, 50th, 30th, and 10th percentile within each of the three HapMap populations, for the five regions, respectively (Supplementary Table S1). Supplementary Figure S1 shows the LD levels for the five residing chromosomes. For each region, we treat the middle 4 Mb as the core region and the 500 kb on each end as flanking regions. Only SNPs imputed in the core region were evaluated to gauge imputation accuracy.
METHODS COMPARED
We evaluated the following reference selection approaches implemented in MaCH-Admix:
random selection (MaCH-Admix Random or original MaCH),
IBS Piecewise selection (MaCH-Admix IBS-PW),
IBS Single-Queue selection (MaCH-Admix IBS-SQ),
IBS Double-Queue selection (MaCH-Admix IBS-DQ),
Ancestry-weighted selection (MaCH-Admix AW) (for HapMapIII datasets).
We also included IMPUTE2 [Howie et al., 2009] and BEAGLE [Browning and Browning, 2009] for comparison. We used IMPUTE 2.1.2 and BEAGLE 3.3.1 with default settings (-k_hap 500 -iter 30 for IMPUTE2; niterations = 10 nsamples = 4 for BEAGLE). As aforementioned, MaCH-Admix can conduct imputation with precalibrated parameters (similar to IMPUTE2); alternatively, MaCH-Admix can perform imputation together with data-dependent parameter estimation in an integrated mode. The integrated mode generates slightly better results at the cost of increased computing time. Here, we report results from the precalibrated mode.
MEASURE OF IMPUTATION QUALITY
We and others have proposed multiple statistics to measure imputation quality [Browning and Browning, 2009; Li et al., 2009; Lin et al., 2010; Marchini and Howie, 2010], measuring either the concordance rate, correlation, or agreement between the imputed genotypes or estimated allele dosages (the fractional counts of an arbitrary allele at each SNP for each individual, ranging continuously from 0 to 2) and their experimental counterpart. We opt to report the dosage r2 values, which are the squared Pearson correlation between the estimated allele dosages and the true experimental genotypes (recoded as 0, 1, and 2 corresponding to the number of minor alleles), because it is a better measure for uncommon variants by taking allele frequency into account and directly related to the effective sample size for downstream association analysis [Pritchard and Przeworski, 2001]. For the remainder of the work, with no special note, average dosage r2 values will be plotted as a function of approximation level (measured by the effective reference panel size, i.e., t described in Methods section, corresponding to MaCH-Admix’s –states option and IMPUTE2’ s -k option). Hereafter, we use approximation level, effective reference size, t, and #states/-k interchangeably. We note that for standard haplotypes-to-genotype imputation (i.e., using reference haplotypes to imputed target individuals with genotypes), computational costs increase quadratically with the approximation level. MaCH-Admix and IMPUTE2 both also have an approximation parameter at the haplotype-based imputation step, MaCH-Admix’s –imputeStates and IMPUTE2’s -k_hap, which increases the computation time linearly and is by default set at a large value (500). We kept both at the default value because increasing beyond the default has rather negligible effects on imputation quality and that total computing time attributable to the haplotype-based imputation step is typically much smaller compared to –states and -k.
RESULTS
WHI-AA AND WHI-HAWITH THE 1000G REFERENCE
Figures 2 and 3 show results for full WHI-HA and WHIAA sets using 2,188 haplotypes from 20101123 release of the 1000 Genomes Project as the reference (selected three out of the five 5 Mb regions: the first, third, and fifth regions according to level of LD). The remaining results under the default or middle settings are presented in Tables I and II (all five regions for WHI-HA and WHI-AA, respectively). Note that BEAGLE’s performance remains constant because it does not have a parameter analogous to MaCH-Admix’s –states or IMPUTE2’s -k.
Fig. 2.
Imputation of 3,587 WHI-HA with the 1000G reference panel. Imputation quality (measured by dosage r2) is plotted as a function of the effective reference panel size (i.e., #states), for WHI-HA individuals in three selected 5 Mb regions (ordered by LD from high to low). (A) Imputation quality of WHI-HA with the 1000G reference panel. (B) Uncommon SNP imputation quality of WHI-HA with the 1000G reference panel. We set the maximum plotting range on y-axis to be 5%. IMPUTE2 in (c) is lower than the lower bound of the plotting range.
Fig. 3.
Imputation of 8,421 WHI-AA with the 1000G reference panel. Imputation quality (measured by dosage r2) is plotted as a function of the effective reference panel size (i.e., #states), for WHI-AA individuals in three selected 5 Mb regions (ordered by LD from high to low). (A) Imputation quality of WHI-AA with the 1000G reference panel. (B) Uncommon SNP imputation quality of WHI-AA with the 1000G reference panel. Note that WHI-AA has significantly less number of SNPs in this category than WHI-HA does. Also, we set the maximum plotting range on y-axis to be 5%. MaCH-Admix Random in (b),(c) and BEAGLE in (a),(b),(c) are lower than the lower bound of the plotting range.
TABLE I.
Imputation results of WHI-HA individuals over five 5 Mb regions with the 1000G reference
| All 3,587 individuals | Random 200 subset | |||||
|---|---|---|---|---|---|---|
| Overall dosage r2 (std dev) |
Uncommon SNPs dosage r2 (std dev) |
Running time |
Overall dosage r2 (std dev) |
Uncommon SNPs dosage r2 (std dev) |
Running time |
|
| Chromosome3: 80–85 Mb | ||||||
| MaCH-Admix Random | 0.935 (0.107) | 0.796 (0.189) | 35,968 | 0.921 (0.121) | 0.794 (0.231) | 841 |
| MaCH-Admix IBS-PW | 0.942 (0.101) | 0.817 (0.189) | 40,422 | 0.923 (0.111) | 0.814 (0.210) | 1,041 |
| MaCH-Admix IBS-SQ | 0.939 (0.104) | 0.799 (0.190) | 38,208 | 0.923 (0.119) | 0.796 (0.231) | 988 |
| MaCH-Admix IBS-DQ | 0.941 (0.102) | 0.806 (0.191) | 38,439 | 0.924 (0.119) | 0.799 (0.232) | 995 |
| IMPUTE2 | 0.939 (0.104) | 0.797 (0.191) | 40,722 | 0.925 (0.119) | 0.799 (0.233) | 2,076 |
| BEAGLE | 0.931 (0.107) | 0.799 (0.190) | 162,888 | 0.912 (0.128) | 0.779 (0.231) | 6,614 |
| Chromosome1: 75–80 Mb | ||||||
| MaCH-Admix Random | 0.918 (0.130) | 0.821 (0.190) | 50,108 | 0.924 (0.129) | 0.855 (0.211) | 1,214 |
| MaCH-Admix IBS-PW | 0.927 (0.123) | 0.841 (0.186) | 57,671 | 0.927 (0.121) | 0.873 (0.197) | 1,490 |
| MaCH-Admix IBS-SQ | 0.923 (0.122) | 0.823 (0.187) | 53,908 | 0.926 (0.125) | 0.861 (0.209) | 1,443 |
| MaCH-Admix IBS-DQ | 0.926 (0.121) | 0.830 (0.185) | 57,321 | 0.928 (0.123) | 0.866 (0.207) | 1,452 |
| IMPUTE2 | 0.921 (0.121) | 0.809 (0.183) | 51,362 | 0.921 (0.127) | 0.845 (0.204) | 2,545 |
| BEAGLE | 0.917 (0.124) | 0.815 (0.184) | 229,514 | 0.917 (0.129) | 0.851 (0.209) | 9,194 |
| Chromosome4: 57–62 Mb | ||||||
| MaCH-Admix Random | 0.904 (0.148) | 0.761 (0.208) | 53,960 | 0.918 (0.137) | 0.813 (0.213) | 1,239 |
| MaCH-Admix IBS-PW | 0.913 (0.139) | 0.783 (0.202) | 61,827 | 0.922 (0.134) | 0.824 (0.212) | 1,527 |
| MaCH-Admix IBS-SQ | 0.907 (0.141) | 0.757 (0.195) | 60,806 | 0.918 (0.135) | 0.807 (0.209) | 1,460 |
| MaCH-Admix IBS-DQ | 0.911 (0.138) | 0.770 (0.195) | 59,088 | 0.921 (0.133) | 0.817 (0.210) | 1,455 |
| IMPUTE2 | 0.906 (0.142) | 0.751 (0.198) | 62,272 | 0.908 (0.147) | 0.773 (0.225) | 2,991 |
| BEAGLE | 0.900 (0.150) | 0.751 (0.218) | 360,545 | 0.907 (0.155) | 0.787 (0.244) | 14,888 |
| Chromosome14: 50–55 Mb | ||||||
| MaCH-Admix Random | 0.921 (0.132) | 0.800 (0.202) | 57,082 | 0.936 (0.122) | 0.847 (0.202) | 1,600 |
| MaCH-Admix IBS-PW | 0.932 (0.120) | 0.826 (0.184) | 60,800 | 0.940 (0.119) | 0.859 (0.198) | 1,876 |
| MaCH-Admix IBS-SQ | 0.927 (0.118) | 0.807 (0.175) | 61,112 | 0.938 (0.119) | 0.849 (0.199) | 1,877 |
| MaCH-Admix IBS-DQ | 0.930 (0.115) | 0.819 (0.176) | 61,175 | 0.939 (0.118) | 0.854 (0.197) | 1,876 |
| IMPUTE2 | 0.924 (0.120) | 0.793 (0.180) | 52,818 | 0.931 (0.125) | 0.828 (0.216) | 2,579 |
| BEAGLE | 0.926 (0.121) | 0.806 (0.189) | 332,586 | 0.929 (0.130) | 0.824 (0.218) | 14,182 |
| Chromosome8: 18–23 Mb | ||||||
| MaCH-Admix Random | 0.896 (0.155) | 0.793 (0.212) | 75,511 | 0.901 (0.150) | 0.821 (0.225) | 1,899 |
| MaCH-Admix IBS-PW | 0.911 (0.143) | 0.824 (0.198) | 84,885 | 0.906 (0.147) | 0.833 (0.221) | 2,302 |
| MaCH-Admix IBS-SQ | 0.903 (0.145) | 0.797 (0.200) | 83,051 | 0.903 (0.149) | 0.820 (0.227) | 2,270 |
| MaCH-Admix IBS-DQ | 0.906 (0.143) | 0.805 (0.200) | 80,794 | 0.904 (0.147) | 0.822 (0.224) | 2,285 |
| IMPUTE2 | 0.900 (0.145) | 0.773 (0.206) | 75,001 | 0.893 (0.159) | 0.781 (0.247) | 3,647 |
| BEAGLE | 0.905 (0.142) | 0.807 (0.201) | 498,822 | 0.894 (0.154) | 0.800 (0.232) | 17,146 |
Note: All results were generated using default or suggested parameter values: MaCH-Admix: –rounds 30, –states 120, –imputeStates 500; IMPUTE2: -iter 30, -k 120, -k_hap 500; BEAGLE: niterations = 10 nsamples = 4. Running time is measured in seconds. Best performance in each comparison is highlighted by bold font.
TABLE II.
Imputation results of WHI-AA individuals over five 5 Mb regions with the 1000G reference
| All 8,421 individuals | Random 200 subset | |||||
|---|---|---|---|---|---|---|
| Overall dosage r2 (std dev) |
Uncommon SNPs dosage r2 (std dev) |
Running time |
Overall dosage r2 (std dev) |
Uncommon SNPs dosage r2 (std dev) |
Running time |
|
| Chromosome3: 80–85 Mb | ||||||
| MaCH-Admix Random | 0.912 (0.100) | 0.782 (0.150) | 161,637 | 0.932 (0.091) | 0.824 (0.194) | 897 |
| MaCH-Admix IBS-PW | 0.947 (0.073) | 0.850 (0.158) | 174,083 | 0.945 (0.083) | 0.849 (0.194) | 1,026 |
| MaCH-Admix IBS-SQ | 0.944 (0.075) | 0.844 (0.161) | 176,147 | 0.942 (0.086) | 0.835 (0.198) | 1,035 |
| MaCH-Admix IBS-DQ | 0.946 (0.074) | 0.849 (0.160) | 169,442 | 0.944 (0.083) | 0.851 (0.198) | 1,021 |
| IMPUTE2 | 0.943 (0.075) | 0.847 (0.151) | 111,307 | 0.943 (0.085) | 0.836 (0.187) | 2,017 |
| BEAGLE | 0.921 (0.088) | 0.795 (0.170) | 23,082a | 0.915 (0.107) | 0.784 (0.217) | 6,435 |
| Chromosome1:75–80 Mb | ||||||
| MaCH-Admix Random | 0.873 (0.143) | 0.703 (0.219) | 214,385 | 0.886 (0.141) | 0.726 (0.241) | 1,240 |
| MaCH-Admix IBS-PW | 0.921 (0.106) | 0.802 (0.176) | 226,019 | 0.906 (0.128) | 0.770 (0.232) | 1,530 |
| MaCH-Admix IBS-SQ | 0.915 (0.109) | 0.794 (0.174) | 232,880 | 0.900 (0.130) | 0.756 (0.224) | 1,504 |
| MaCH-Admix IBS-DQ | 0.918 (0.106) | 0.803 (0.168) | 232,858 | 0.903 (0.131) | 0.762 (0.235) | 1,476 |
| IMPUTE2 | 0.917 (0.103) | 0.810 (0.157) | 138,080 | 0.898 (0.135) | 0.760 (0.240) | 2,412 |
| BEAGLE | 0.892 (0.119) | 0.759 (0.173) | 25,618a | 0.875 (0.145) | 0.713 (0.242) | 8,621 |
| Chromosome4: 57–62 Mb | ||||||
| MaCH-Admix Random | 0.883 (0.126) | 0.688 (0.187) | 241,045 | 0.905 (0.111) | 0.749 (0.169) | 1,290 |
| MaCH-Admix IBS-PW | 0.927 (0.092) | 0.795 (0.159) | 260,231 | 0.922 (0.100) | 0.792 (0.175) | 1,508 |
| MaCH-Admix IBS-SQ | 0.920 (0.094) | 0.782 (0.148) | 254,002 | 0.915 (0.105) | 0.777 (0.180) | 1,545 |
| MaCH-Admix IBS-DQ | 0.924 (0.090) | 0.796 (0.138) | 248,524 | 0.920 (0.100) | 0.793 (0.175) | 1,478 |
| IMPUTE2 | 0.918 (0.091) | 0.787 (0.129) | 166,642 | 0.912 (0.104) | 0.778 (0.168) | 2,939 |
| BEAGLE | 0.898 (0.109) | 0.735 (0.167) | 43,573a | 0.892 (0.131) | 0.738 (0.222) | 14,528 |
| Chromosome14: 50–55 Mb | ||||||
| MaCH-Admix Random | 0.875 (0.140) | 0.726 (0.216) | 240,789 | 0.908 (0.120) | 0.807 (0.198) | 1,663 |
| MaCH-Admix IBS-PW | 0.921 (0.105) | 0.823 (0.171) | 254,530 | 0.927 (0.104) | 0.852 (0.167) | 1,900 |
| MaCH-Admix IBS-SQ | 0.914 (0.108) | 0.809 (0.172) | 253,231 | 0.919 (0.112) | 0.835 (0.191) | 1,918 |
| MaCH-Admix IBS-DQ | 0.918 (0.105) | 0.818 (0.168) | 254,555 | 0.924 (0.107) | 0.850 (0.175) | 1,900 |
| IMPUTE2 | 0.912 (0.106) | 0.815 (0.157) | 143,772 | 0.913 (0.116) | 0.820 (0.186) | 2,575 |
| BEAGLE | 0.893 (0.118) | 0.775 (0.176) | 27,666a | 0.899 (0.127) | 0.786 (0.216) | 14,139 |
| Chromosome8: 18–23 Mb | ||||||
| MaCH-Admix Random | 0.830 (0.177) | 0.682 (0.235) | 343,104 | 0.857 (0.163) | 0.735 (0.235) | 1,977 |
| MaCH-Admix IBS-PW | 0.889 (0.142) | 0.798 (0.207) | 357,858 | 0.884 (0.148) | 0.800 (0.218) | 2,377 |
| MaCH-Admix IBS-SQ | 0.882 (0.145) | 0.789 (0.207) | 347,473 | 0.877 (0.152) | 0.786 (0.224) | 2,393 |
| MaCH-Admix IBS-DQ | 0.885 (0.144) | 0.795 (0.205) | 356,928 | 0.881 (0.149) | 0.797 (0.220) | 2,318 |
| IMPUTE2 | 0.884 (0.140) | 0.795 (0.194) | 211,879 | 0.876 (0.153) | 0.795 (0.218) | 3,618 |
| BEAGLE | 0.858 (0.151) | 0.743 (0.206) | 43,068a | 0.856 (0.158) | 0.767 (0.229) | 16,931 |
In our experiments, BEAGLE cannot finish imputation with the complete 1000G references within 7 days, which is the hard limit on our cluster server. We thus restrict the markers in the reference panel to be the set of Affymetrix 6.0 markers plus 2.5% of the remaining 1000G markers. The size of the restricted set in each region is about 10 ~ 15% of the size of original 1000G marker set.
All results were generated using default or suggested parameter values: MaCH-Admix: –rounds 30, –states 120, –imputeStates 500; IMPUTE2: -iter 30, -k 120, -k_hap 500; BEAGLE: niterations = 10 nsamples = 4. Running time is measured in seconds. Best performance in each comparison is highlighted by bold font.
Generally, we observe higher imputation accuracy in regions with higher level of LD for all approaches evaluated. In addition, in regions with higher LD, imputation accuracy reaches a plateau with smaller effective reference sizes. This is because the LD pattern can be captured fairly well by a smaller number of reference haplotypes in regions with higher level of LD. In regions with lower level of LD, accuracy plateau is reached with larger effective reference sizes. But generally an effective reference size of 80–120 is good for MaCH-Admix to perform well at all LD levels.
We found that the piecewise IBS selection approach (IBS-PW) is clearly the best among the three IBS-based methods implemented in MaCH-Admix. Its performance is stable even with a small #states value. For the other two IBS-based reference selection approaches implemented in MaCH-Admix, we observed IBS-DQ performs better than IBS-SQ. The performance order of the three MaCH-Admix IBS-based methods is expected based on our reasoning in the Material and Methods section. In addition, all three IBS-based methods show clear advantage over random selection, particularly when the effective reference size is small. IMPUTE2 has similar performance to that of IBS-DQ when the effective reference size is small. Interestingly, IMPUTE2’ s accuracy curve tends to stay relatively flat while those for MaCH-Admix’s IBS-based methods increase with the effective reference size.
Across all five regions evaluated, with effective reference size at 120, IBS-PW has consistent performance gain over other evaluated methods. Importantly, IBS-PW and IBS-DQ, particularly IBS-PW, manifest more pronounced advantage for uncommon variants (MAF < 5%) in WHI-HA. For these uncommon variants, average dosage r2 is 0.818, 0.782, and 0.794 (0.808, 0.805, and 0.756) for WHI-HA (WHI-AA) using our IBS-PW, IMPUTE2, and BEAGLE, respectively. The advantage of IBS-PW in uncommon SNPs is however smaller in WHI-AA largely because of the much smaller number of uncommon variants in WHI-AA (Supplementary Figure S2). However, the difference is highly significant (P-value ≤5.02 × 10−5) in both WHI samples. Our observation is consistent in both the full set and the subset of 200 individuals (Tables I and II).
HAPMAP ASW AND MEX WITH THE 1000G REFERENCE
In this setting, we use a large reference panel to impute two small target sets. Supplementary Figure S3 shows the imputation quality of three regions for both ASW and MEX. The complete results are presented in Supplementary Table S2. Similar to previous experiments, we found that IBS-PW is very effective in finding the most relevant reference from a large panel (1000G) and clearly outperforms the other methods. IMPUTE2 again shows a flatter curve in most regions. Random selection and BEAGLE tend to perform worse than the IBS-based methods. This again proves that IBS-based selections are very effective in working with large reference panels.
IMPUTATION PERFORMANCE WITH HAPMAP REFERENCES
First, consistent with what has been reported that imputation quality improves with reference panel size, imputation quality is indeed lower with HapMap references than with the 1000G reference. For example, average dosage r2 is 90.0–91.3% with the 1000G reference (Table I) for WHI-HA individuals in the chromosome4: 57–62 Mb region but drops to 84.4–86.2% with HapMapII references (Supplementary Table S3). Second, difference among various methods is much smaller with these smaller HapMap reference sets (H = 240 ~ 930),which is consistent with our intuition that, given fixed computational costs, reference selection makes more pronounced difference with large reference panel because only a small portion of reference can be selected.
WHI-HA and WHI-AA With HapMap References
The complete results are presented in Supplementary Tables S3 and S4. In WHI-HA (Supplementary Table S3, H = 420), IBS-PW outperforms IBS-SQ and IBS-DQ slightly and the advantage disappears in WHI-AA (Supplementary Table S4, H = 240).MaCH-Admix and IMPUTE2 yield similar imputation accuracy, and both outperform BEAGLE slightly.
HapMap ASW and MEX With HapMap References
For ASW, we experimented with three reference panels: HapMapII CEU+YRI, HapMapIII CEU+YRI, and HapMapIII CEU+YRI+LWK+MKK; for MEX two reference panels: HapMapII CEU+YRI+JPT+CHB and HapMapIII CEU+YRI+JPT+CHB. Results for ASW with HapMapIII CEU+YRI+LWK+MKK as the reference are shown in Supplementary Figure S4 (the same three selected regions). The remaining results are presented in Supplementary Tables S5A–C. Again, MaCH-Admix and IMPUTE2 yield similar imputation accuracy, both outperform BEAGLE slightly. IBS-PW is still an obvious winner in most regions and settings. But the relative difference among different methods diminishes when H is small.
We also included ancestry-weighted selection in evaluation in this setting because weights can be estimated stably given the relatively simple population structure in reference. Interestingly, we did not observe noticeable advantage of the ancestry-weighted selection method despite the obvious population structure within the reference panel and the target being admixed individuals. It however outperforms random selection slightly in most ASW experiments.
RUNNING TIME
Methods implemented in MaCH-Admix have comparable running time to that of IMPUTE2. BEAGLE has similar running time in experiments with HapMap references. It however needs significantly more computing time than MaCH-Admix and IMPUTE2 when imputing with the 1000G reference, which we believe has to do with how consecutive untyped variants are modeled. Note that, due to the large number of experiments, we conducted all experiments on a big Linux cluster with more than 1000 CPUs. This leads to moderate fluctuations in running time over short regions due to I/O competition. But we obtain largely consistent conclusions across different experimental settings.
DISCUSSION
In summary, the emergence of large reference panels calls for more efficient methods to utilize the rich resource. we have implemented two classes of reference-selection methods, namely IBS-based and ancestry-weighted approaches, to construct effective reference panels within our previously described HMM and implemented them in software package MaCH-Admix for genetic imputation in admixed populations. We have performed systematic evaluations on large (WHI-AA and WHI-HA full sample with 8,421 and 3,587 individuals), medium (subset of 200 individuals from each of the two WHI admixed cohorts), and small (HapMap ASW and MEX with 49 and 50 founders, respectively) target samples; using large (the latest 1000G with H = 2,188) and small (HapMap with H = 240–930) reference panels; and in five regions with different levels of LD. Compared with popular existing methods, MaCH-Admix demonstrates its advantage mostly because its piecewise algorithm takes potential changes in haplotype pattern sharing across regions into direct account (vs. IMPUTE2, which adopts a whole-haplotype IBS matching approach) and because it does not reduce local haplotype complexity (vs. BEAGLE, which does so to gain computational efficiency). Based on our evaluations, we recommend our proposed piecewise IBS-based method, which demonstrates the best trade-off between quality and computing time.
As the reference panel continues to grow rapidly (e.g., the 1000 Genomes Project will generate ~5,000 haplotypes within 2 years), approaches that can rapidly explore the entire reference pool will become increasingly appreciated. IBS-based approaches show such potential. As manifested by results from both WHI individuals and the HapMapIII individuals, IBS-based approaches can generate accurately imputed genotypes by preferentially selecting a small but different subset of ~100 (corresponding to ~5% for the current 1000G case where H = 2, 188) haplotypes from the entire reference pool in each iteration. As computational costs increase quadratically with the effective number of haplotypes used in each iteration, such ~95% reduction in the effective number of reference haplotypes corresponds to >99.5% reduction in computational investment.
Previous studies [Hao et al., 2009; Li et al., 2009; Seldin et al., 2011; Shriner et al., 2010; Zhang et al., 2011] have recommended the use of a combined reference panel, which pools haplotypes from all available reference populations (e.g., from the HapMap or the 1000 Genomes Projects), especially for populations that do not have a single best match reference population for increased imputation accuracy. Two forces working in opposite directions are introduced by including reference haplotypes from populations different from those in target samples in such a cosmopolitan panel: shared haplotype stretches (likely even shorter) that would increase imputation quality while noise added by including population-specific local haplotypes would harm imputation quality. Therefore, the recommendation of using a cosmopolitan panel to enhance imputation quality also applies to MaCH-Admix, conceptually more applicable because MaCH-Admix reduces the noise force by choosing local haplotypes that are most relevant into effective reference.
One key question concerns the optimal region size for imputation. From the perspective of including more LD information, particularly the long-range LD information that would be particularly critical for the imputation of uncommon variants, imputation over longer regions is desired. However, approaches that select reference haplotypes according to genetic matching between reference haplotypes and genotypes of target individuals across the entire region like whole-haplotype IBS-based methods will likely suffer from the change in genetic matching over a long region. For example, for both scenarios presented in Figure 1, there are two distinct subregions according to the matching pattern. Lumping them naively together, particularly using a single queue, may well lead to inferior performance as discussed earlier. We attempt to solve the problem by breaking the entire region into smaller pieces and within each piece selecting some reference haplotypes according to local genetic matching. This conceptually shares similarity with local ancestry adjustment in analysis of admixed populations [Wang et al., 2011a]. Pasaniuc et al. [2011] also found local ancestry increases imputation accuracy. The proposed piecewise IBS-based selection method is robust to imputation region size. We have evaluated the performance on whole chromosomes using ASW/MEX with HapMap references and found that both piecewise IBS and ancestry-weighted selection perform much better than whole-haplotype IBS based methods (data not shown). Between piecewise IBS and ancestry-weighted selections, the piecewise IBS method has advantage in most whole chromosome experiments and is very close to ancestry-weighted selection in the rest.
Ancestry-weighted approaches have been previously utilized to construct reference panels in admixed populations for tag SNP selection or imputation [Egyud et al., 2009; Pasaniuc et al., 2010; Pemberton et al., 2008]. However, such reference panels created a priori induce two problems for imputation. First, haplotypes from contributing reference populations are literally duplicated, thus substantially increasing computational burden. Second, the same fixed preconstructed reference haplotypes are to be used for all Markov iterations, preventing imputation algorithms from taking into account the uncertainty in creating the reference panel. Our ancestry-weighted approach selects reference haplotypes probabilistically according to the estimated ancestry proportions and creates a different reference panel in each Markov iteration. This strategy ensures that all reference haplotypes to be selected when we run the Markov iterations long enough, thus avoiding both problems mentioned above. An attractive feature that we have added to MaCH-Admix is a functionality to estimate ancestry proportions so that it can internally generate weights for ancestry-weighted approach without the need to install and call external programs. Although there exist many methods to infer ancestry including, for example, structure [Pritchard et al., 2000], HAPMIX [Price et al., 2009], and GEDI-ADMX [Pasaniuc et al., 2009], we believe that researchers will find this build-in feature convenient. We found our estimates reasonably close to estimates from structure and working well for imputation purpose.
In this study, we have examined the performance of our proposed and other imputation methods in both Hispanics and African Americans. Between the two, Hispanics are known to have more complex LD structure because of three ancestral populations involved as opposed to two for African Americans. The more complex LD in Hispanics indeed makes it essential to more explicitly account for the larger variability in local ancestry (e.g., using our proposed piecewise approach). The more complex LD and population substructure in Hispanics have prevented a lot of investigators from even attempting imputation. However, we observe similar if not slightly better imputation quality in the five regions examined, with an average dosage r2 of 92.5% (81.8%) vs. 92.1% (81.4%) for all (uncommon) SNPs in WHI-HA and WHI-AA respectively using our piecewise IBS approach. That imputation performance for Hispanics is comparable with that for African Americans is expected due to on average less African ancestry (where LD is the lowest and thus most challenging for imputation) in Hispanics compared to African Americans. Therefore, we highly encourage investigators working with Hispanics perform imputation as well.
Although in this work we propose the reference selection methods for imputation of admixed individuals, the methods can be directly applied to imputation in general for nonadmixed populations by finding the best genetic match for each target individual. For the same reason, IBS-based methods tend to work better than ancestry-weighted approaches when between-individual variation among the target individuals is large (data not shown). This is not surprising because IBS-based approaches select a different effective reference panel tailored for each target individual, rather than one uniform reference sampling setting for all target individuals as in the ancestry-weighted approach.
We have also attempted to examine common and uncommon genetic variants separately, using MAF 5% as cutoff. We observe more pronounced differences among the attempted methods with uncommon variants, suggesting that choice of reference selection methods matters more for uncommon variants. Due to the nature of the SNPs evaluated (either typed Affymetrix 6.0 markers for the WHI individuals, or HapMap markers) and the target sample size (49–50 for HapMapIII ASW and MEX), there are few really rare (MAF<1%) variants. Although several attempts have been made [Howie et al., 2011; Liu et al., 2012; The International HapMap Consortium, 2010;Wang et al., 2011b], imputation quality for uncommon variants is far from being fully assessed and needs to be further evaluated when data from large scale sequencing efforts become available.
Last but clearly not the least point concerns computational efficiency. MaCH-Admix is very flexible in terms of the effective number of haplotypes used in each iteration and the number of iterations. Imputation accuracy depends on both parameters. Because computational cost increases quadratically with –states and linearly with –rounds, for practical purpose, we recommend using –states 100–120 and –rounds ≥20. We also have an option analogous to IMPUTE2’s -k_hap, which increases computational costs linearly and even defaulting at a large value (500) contributes to only a small proportion of computing time. Between the two categories of approaches proposed, the ancestry-weighted approach requires only one-time upfront costs for the estimation of ancestry proportions. The IBS-based methods, on the other hand, require overhead costs at each iteration for calculating genetic similarities between individuals in the target population and the reference haplotypes. For both, the costs increase with the reference panel size. Finally, computational costs would increase only linearly with –states if we start with haplotypes of the target individuals, that is, for haplotype-to-haplotype (both reference and target are in haplotypes) imputation as performed by software minimac. We plan to extend our proposed methods to minimac in the future.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Gonçalo R. Abecasis, Noah Rosenberg, and Mike Boehnke for helpful discussions. Y.L. is supported by R01HG006292 and R01HG006703. W.W. is supported by NIH 1-RC2-MH089951-01, NIH U01 CA134240, and NSF IIS-0812464. M.L. is supported by R01HG004517 and R01HG005854. We also thank the WHI SHARe consortium for generating the data for the WHI participants.
Footnotes
Supporting Information is available in the online issue at wileyonlinelibrary.com.
WEB RESOURCES
Census fact for admixed populations, http://quickfacts.census.gov/qfd/states/00000.html
The 1000 Genomes Project, http://www.1000genomes.org/
MaCH-Admix, http://www.unc.edu/~yunmli/MaCH-Admix/
MaCH, http://www.sph.umich.edu/csg/yli/mach/
IMPUTE, http://mathgen.stats.ox.ac.uk/impute/impute.html
BEAGLE, http://faculty.washington.edu/browning/beagle/beagle.html
structure: http://pritch.bsd.uchicago.edu/software.html
REFERENCES
- Anderson GL, Manson J, Wallace R, Lund B, Hall D, Davis S, Shumaker S, Wang CY, Stein E, Prentice RL. Implementation of the Women’s Health Initiative study design. Annals Epidemiol. 2003;13(9):S5–S17. doi: 10.1016/s1047-2797(03)00043-7. [DOI] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84(2):210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson A, Wheeler E, Glazer N, Bouatia-Naji N, Gloyn AL, Lindgren CM, Mägi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparsø T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proença C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O’Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Böttcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jørgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martínez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orrù M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvänen AC, Tanaka T, Thorand B, Tichet J, Tønjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Ríos M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF, Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egyud MRL, Gajdos ZKZ, Butler JL, Tischfield S, Le Marchand L, Kolonel LN, Haiman CA, Henderson BE, Hirschhorn JN. Use of weighted reference panels based on empirical estimates of ancestry for capturing untyped variation. Hum Genet. 2009;125(3):295–303. doi: 10.1007/s00439-009-0627-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridley BL, Jenkins G, Deyo-Svendsen ME, Hebbring S, Freimuth R. Utilizing genotype imputation for the augmentation of sequence data. PLoS One. 2010;5(6):e11018. doi: 10.1371/journal.pone.0011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao K, Chudin E, McElwee J, Schadt EE. Accuracy of genome-wide imputation of untyped markers and impacts on statistical power for association studies. BMC Genet. 2009;10(1):27. doi: 10.1186/1471-2156-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3: Genes, Genomes, Genetics. 2011;1(6):457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):15. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Li Y, Singleton AB, Hardy JA, Abecasis G, Rosenberg NA, Scheet P. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet. 2009;84(2):235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10(1):387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. Mach: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P, Hartz SM, Zhang Z, Saccone SF,Wang J, Tischfield JA, Edenberg HJ, Kramer JR, M Goate A, Bierut LJ, Rice JP COGA Collaborators COGEND Collaborators, GENEVA. Anew statistic to evaluate imputation reliability. PLoS One. 2010;5(3):10. doi: 10.1371/journal.pone.0009697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind JM, Hutcheson-Dilks HB, Williams SM, Moore JH, Essex M, Ruiz-Pesini E, Wallace DC, Tishkoff SA, O’Brien SJ, Smith MW. Elevated male European and female African contributions to the genomes of African American individuals. Hum Genet. 2007;120(5):713–722. doi: 10.1007/s00439-006-0261-7. [DOI] [PubMed] [Google Scholar]
- Liu EY, Buyske S, Aragaki AK, Peters U, Boerwinkle E, Carlson C, Carty C, Crawford DC, Haessler J, Hindorff LA, Marchand LL, Manolio TA, Matise T, Wang W, Kooperberg C, North KE, Li Y. Genotype imputation of metabochipsnps using a study-specific reference panel of ~4,000 haplotypes in African Americans from the Women’s Health Initiative. Genet Epidemiol. 2012;36(2):107–117. doi: 10.1002/gepi.21603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, Forrester T, Allison DB, Deka R, Ferrell RE, Shriver MD. Estimating African American admixture proportions by use of population-sspecific alleles. Am J Hum Genet. 1998;63(6):1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B, Kennedy J, Mandoiu I. Imputation-based local ancestry inference in admixed populations. Lect Notes Comput Sci. 2009;5542:221–233. [Google Scholar]
- Pasaniuc B, Avinery R, Gur T, Skibola CF, Bracci PM, Halperin E. A generic coalescent-based framework for the selection of a reference panel for imputation. Genet Epidemiol. 2010;34(8):773–782. doi: 10.1002/gepi.20505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B, Zaitlen N, Lettre G, Chen GK, Tandon A, Kao WHL, Ruczinski I, Fornage M, Siscovick DS, Zhu X, Larkin E, Lange LA, Cupples LA, Yang Q, Akylbekova EL, Musani SK, Divers J, Mychaleckyj J, Li M, Papanicolaou GJ, Millikan RC, Ambrosone CB, John EM, Bernstein L, Zheng W, Hu JJ, Ziegler RG, Nyante SJ, Bandera EV, Ingles SA, Press MF, Chanock SJ, Deming SL, Rodriguez-Gil JL, Palmer CD, Buxbaum S, Ekunwe L, Hirschhorn JN, Henderson BE, Myers S, Haiman CA, Reich D, Patterson N, Wilson JG, Price AL. Enhanced statistical tests for gwas in admixed populations: assessment using African Americans from care and a breast cancer consortium. PLoS Genet. 2011;7(4):15. doi: 10.1371/journal.pgen.1001371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton TJ, Jakobsson M, Conrad DF, Coop G, Wall JD, Pritchard JK, Patel PI, Rosenberg NA. Using population mixtures to optimize the utility of genomic databases: linkage disequilibrium and association study design in India. Annals Hum Genet. 2008;72(Pt 4):535–546. doi: 10.1111/j.1469-1809.2008.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, Barnes KC, Rafaels N, Ruczinski I, Beaty TH, Mathias R, Reich D, Myers S. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5(6) doi: 10.1371/journal.pgen.1000519. e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qayyum R, Snively BM, Ziv E, Nalls MA, Liu Y, Tang W, Yanek LR, Lange L, Evans MK, Ganesh S, Austin MA, Lettre G, Becker DM, Zonderman AB, Singleton AB, Harris TB, Mohler ER, Logsdon BA, Kooperberg C, Folsom AR, Wilson JG, Becker LC, Reiner AP. A meta-analysis and genome-wide association study of platelet count and mean platelet volume in African Americans. PLoS Genet. 2012;8(3) doi: 10.1371/journal.pgen.1002491. e1002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Patterson N. Will admixture mapping work to find disease genes? Philos Trans R Soc Lond Ser B, Biol Sci. 2005;360(1460):1605–1607. doi: 10.1098/rstb.2005.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner AP, Carlson CS, Ziv E, Iribarren C, Jaquish CE, Nickerson DA. Genetic ancestry, population sub-structure, and cardiovascular disease-related traits among African-American participants in the CARDIA Study. Hum Genet. 2007;121(5):565–575. doi: 10.1007/s00439-007-0350-2. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11(5):356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna S, Pitzalis M, Zoledziewska M, Zara I, Sidore C, Murru R, Whalen MB, Busonero F, Maschio A, Costa G, Melis MC, Deidda F, Poddie F, Morelli L, Farina G, Li Y, Dei M, Lai S, Mulas A, Cuccuru G, Porcu E, Liang L, Zavattari P, Moi L, Deriu E, Urru MF, Bajorek M, Satta MA, Cocco E, Ferrigno P, Sotgiu S, Pugliatti M, Traccis S, Angius A, Melis M, Rosati G, Abecasis GR, Uda M, Marrosu MG, Schlessinger D, Cucca F. Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat Genet. 2010;42(6):495–497. doi: 10.1038/ng.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin MF, Pasaniuc B, Price AL. New approaches to disease mapping in admixed populations. Nat Rev Genet. 2011;12(8):523–528. doi: 10.1038/nrg3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner D, Adeyemo A, Chen G, Rotimi CN. Practical considerations for imputation of untyped markers in admixed populations. Genet Epidemiol. 2010;34(3):258–265. doi: 10.1002/gepi.20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NL, Chen M-H, Dehghan A, Strachan DP, Basu S, Soranzo N, Hayward C, Rudan I, Sabater-Lleal M, Bis JC, de Maat MP, Rumley A, Kong X, Yang Q, Williams FM, Vitart V, Campbell H, Mälarstig A, Wiggins KL, Van Duijn CM, McArdle WL, Pankow JS, Johnson AD, Silveira A, McKnight B, Uitterlinden AG, Aleksic N, Meigs JB, Peters A, Koenig W, Cushman M, Kathiresan S, Rotter JI, Bovill EG, Hofman A, Boerwinkle E, Tofler GH, Peden JF, Psaty BM, Leebeek F, Folsom AR, Larson MG, Spector TD, Wright AF, Wilson JF, Hamsten A, Lumley T, Witteman JC, Tang W, O’Donnell CJ Wellcome Trust Case Control Consortium. Novel associations of multiple genetic loci with plasma levels of factor vii, factor viii, and von Wille-brand factor: the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation. 2010;121(12):1382–1392. doi: 10.1161/CIRCULATIONAHA.109.869156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefflova K, Dulik MC, Barnholtz-Sloan JS, Pai AA, Walker AH, Rebbeck TR. Dissecting the within-Africa ancestry of populations of African descent in the Americas. PLoS One. 2011;6(1):e14495. doi: 10.1371/journal.pone.0014495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Coram M, Wang P, Zhu X, Risch N. Reconstructing genetic ancestry blocks in admixed individuals. Am J Hum Genet. 2006;79(1):1–12. doi: 10.1086/504302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The WHI Study Group. Design of the Women’s Health Initiative clinical trial and observational study. Control Clin Trials. 1998;19(1):61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhu X, Qin H, Cooper R, Ewens W, Li C, Li M. Adjustment for local ancestry in genetic association analysis of admixed populations. Bioinformatics. 2011a;27(5):670–677. doi: 10.1093/bioinformatics/btq709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jacobs K, Yeager M, Hutchinson A, Sampson J, Chatterjee N, Albanes D, Berndt S, Chung C, Diver W, Gapstur S, Teras L, Haiman C, Henderson B, Stram D, Deng X, Hsing A, Virtamo J, Eberle M, Stone J, Purdue M, Taylor P, Tucker M, Chanock S. Improved imputation of common and uncommon SNPs with a new reference set. Nat Genet. 2011b;44(1):6–7. doi: 10.1038/ng.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40(2):161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler CA, Nelson GW, Smith MW. Admixture mapping comes of age. Annu Rev Genomics Hum Genet. 2010;11:65–89. doi: 10.1146/annurev-genom-082509-141523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WTCCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zhi D, Zhang K, Gao G, Limdi NN, Liu N. Practical consideration of genotype imputation: sample size, window size, reference choice, and untyped rate. Stat Interface. 2011;4(3):339–352. doi: 10.4310/sii.2011.v4.n3.a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Cooper RS, Elston RC. Linkage analysis of a complex disease through use of admixed populations. Am J Hum Genet. 2004;74(6):1136–1153. doi: 10.1086/421329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.