Abstract
The genus Leontopodium comprises 30–41 species. The centre of diversity is the Sino-Himalayan region in south-western China, where about 15 species occur. The two species native to Europe, L. alpinum (known as the common ‘Edelweiss’) and L. nivale, are part of the cultural heritage of the people living there. Despite its importance, very little is known about the systematics of the genus. Because recent molecular studies have shown that species within this genus are closely related and difficult to distinguish with rDNA and cpDNA data, we used AFLPs to obtain a more detailed understanding of the phylogeny of the genus. Our main aims were as follows: (1) to clarify species relationships within the genus; and (2) to reveal information about the biogeography of the genus. We used AFLPs with six primer combinations to investigate 216 individuals in 38 populations of 16 different species. With AFLPs, we were able to recognize 10 different groups, all of which had strong bootstrap support. These results were also congruent with the morphology-based taxonomy of the genus. Most private and rare fragments were found in the Yunnan region (south-western China) relative to Europe and Mongolia/central China, suggesting a long-lasting in situ history of populations in the centre of diversity of the genus. Our results illustrate the utility of AFLPs to resolve phylogenetic relationships between these closely related species.
Keywords: Edelweiss, genetic variation, Qinghai–Tibetan plateau, taxonomy
INTRODUCTION
The genus Leontopodium R.Br. ex Cassini comprises 30–41 species (Fig. 1). The main distribution of the genus is in central and eastern Asia, including Russia, Japan, South Korea, Mongolia, China and along the Himalaya to the borders of Afghanistan and Pakistan. The centre of diversity is the Sino-Himalayan region in south-western China, where 15–18 different species can be found. Two species also occur in Europe: Leontopodium alpinum Cass. grows in the Pyrenees, throughout the Alps, the Carpathians and the Balkan Peninsula, whereas Leontopodium nivale (Ten.) Huet ex Hand.-Mazz. is a local and disjunct endemic of the central Apennines in Italy and the Pirin Mountains in Bulgaria (Meusel & Jaeger, 1992). For people living in the European Alps, Leontopodium alpinum, known as the common ‘Edelweiss’, is a very important part of their cultural heritage.
Figure 1.
Species of Leontopodium investigated, highlighting morphological similarities within groups found in the neighbor-net (Fig. 2) and neighbor-joining (Fig. 3) analyses. Numbers in the top left-hand corner correspond to the groups as follows: 1, A1 (L. alpinum); 2, A2 (L. nivale); 3, C1 [L. sp. (SSG-07)]; 4, C2 (L. himalayanum); 5, C3 (L. calocephalum); 6, C4 (L. souliei); 7, C5 (L. caespitosum); 8, D (L. franchetii); 9, E [L. sp. (SSG-16)]; 10, F (L. dedekensii); 11, G (L. sinense); 12, H (L. artemisiifolium); 13, I (L. andersonii); 14, J (L. cf. stracheyi).
The Alpine Edelweiss [Leontopodium alpinum Cass. or L. nivale subsp. alpinum (Cass.) Greuter] has a long tradition in folk medicine. References from the year 1582 mentioned the use of Edelweiss for the treatment of diarrhoea and dysentery (Tabernaemontanus, 1582). Several other applications for extracts and plant parts of Edelweiss have been described throughout the years, and recent phytochemical research has resulted in the detection of unknown and uncommon secondary metabolites, some with strong biological activities (Stuppner et al., 2002; Dobner et al., 2003a, b, 2004; Schwaiger et al., 2004, 2005; Speroni et al., 2006; Hornick et al., 2008; Reisinger et al., 2009). These promising results have increased the interest in other species of this genus for pharmaceutical research, and thus the need for a more predictive system of classification within the genus.
Despite its importance, relatively little is known about the systematics of the genus. The last classification was by Heinrich Handel-Mazzetti in 1927. He based his monograph on earlier works by Franchet (1892) and Beauverd (1909, 1910, 1911, 1912, 1914), accepting 41 species divided into two sections and two subgenera (Handel-Mazzetti, 1927). A recent taxonomic review of the genus Leontopodium (W. B. Dickoré, Botanische Staatssammlung Munich, Germany, unpubl. data), however, suggests a relatively lower total number of c. 35 accepted species. Most taxonomic treatments in modern Asiatic floras, such as the Flora Iranica (Rechinger, 1980), Flora of USSR (Shishkin, 1990), Flora of China (Wu, Raven & Hong, 1994), Flora of Japan (Iwatsuki et al., 1995), Flora of Bhutan (Grierson & Long, 2001), Flora of Central Asia (Grubov, 2003) and Flora of Pakistan (Qaiser & Abid, 2003), are not satisfactory, because none provides an overall synthesis. The taxonomic status of and relationship between the two European species have also not been resolved, being treated alternatively as one species with two subspecies (L. nivale subsp. nivale and L. nivale subsp. alpinum) by some workers (Greuter, 2003), and as two distinct species by others (Blöch et al., 2010).
Blöch et al. (2010) investigated the molecular phylogenetic aspects within Leontopodium. They analysed the nuclear [internal and external transcribed spacer (ITS and ETS), both from nuclear ribosomal DNA] and plastid (matK, trnL intron combined with trnL/F spacer) markers of 22 species. Three of the four markers supported monophyly for the genus. The ITS dataset was most informative with regard to species relationships. On the basis of these data, three groups of species could be recognized (group A, L. alpinum and L. nivale; group B, L. calocephalum Beauverd, L. leontopodioides Beauverd, L. ochroleucum Beauverd and L. souliei Beauverd; group C, L. dedekensii Beauverd and L. sinense Hemsl. ex Forb. & Hemsl.), but the groups did not correspond well to the morphologically based classification of Handel-Mazzetti (1927). The chloroplast markers offered only limited information on interspecific relationships within Leontopodium. Broader infrageneric relationships were not fully resolved, indicating close species affinities and suggesting recent speciation.
To provide a more detailed understanding of relationships among species within Leontopodium, we have used AFLPs (Vos et al., 1995). This DNA finger-printing technique has been proven to be useful for investigations on diverse plant groups (e.g. Lactuca, Koopman, Zevenbergen & Van den Berg, 2001; Trollius, Després et al., 2003; Senecio, Pelser, Gravendeel & van der Meijden, 2003; Solanum, Spooner, Peralta & Knapp, 2005; Hypochaeris, Tremetsberger et al., 2006), and it has now become a standard method for revealing phylogenetic relationships among closely related taxa (Després et al., 2003; Guo et al., 2005; Guo, Saukel & Ehrendorfer, 2008). Koopman (2005) mentioned that there are properties of restriction fragment data that limit the phylogenetic interpretation of AFLPs, such as possible nonindependence of fragments, problems of homology assignment of fragments, asymmetry in the probability of losing and gaining fragments, and problems in distinguishing heterozygote from homozygote bands. Nevertheless, several different authors (including Koopman, 2005) have suggested that AFLPs can be used to reveal phylogenetic information, especially for species that have diverged quite recently or radiated within a short period of time (e.g. Bussell, Waycott & Chappill, 2005; Koopman, 2005; Tremetsberger et al., 2006).
In this study, we applied AFLPs to analyse 216 individuals from 38 populations belonging to 16 different species of Leontopodium. The aims of this study were as follows: (1) to clarify phylogenetic relationships among the species; and (2) to gain insights into their biogeography.
MATERIAL AND METHODS
Two hundred and sixteen individuals in 38 populations of 16 different Leontopodium species (Fig. 1) were analysed with AFLP, using a minimum of two and a maximum of 14 individuals per population (Table 1). Most species were collected in the centre of diversity (Hengduan-Shan mountain range, Sino-Himalayan region) in the Province of Yunnan, south-western China. The species investigated are closely related and were chosen in order to avoid high proportions of nonhomologous fragments and the related loss of phylogenetic signal within AFLP analyses (as described in Koopman, 2005). The leaves were collected in silica gel and stored until DNA isolation. Vouchers were deposited in the herbaria of the University of Vienna (WU) and the Chinese Academy of Sciences in Beijing (PE).
Table 1.
Species, populations (collector(s)-collection number), sampling location/coordinates, voucher information, number of individuals analysed for AFLPs (NAFLPs), total number of fragments (Ftot), percentage of polymorphic fragments (Fpoly%), number of private fragments (Fpriv) and rarity index (DW) for the populations and species of Leontopodium investigated and collected in Europe, Mongolia/central China and the Province of Yunnan (south-western China)
| Species | Populations | Sampling location/coordinates | Herbarium voucher | N AFLPs | Ftot ± SD* | Fpoly% ± SD* | F priv | DW |
|---|---|---|---|---|---|---|---|---|
| Europe | 650 | 96.00 | 47 | 18.15 | ||||
| L. alpinum Cass. | 352 ± 55.15 | 65.64 ± 3.86 | 10 | 25.98 | ||||
| SSSP-01 | Bulgaria, Stara Planina/ 42°32′15.1″N, 25°06′21.2″E |
Safer 0043989 (WU) | 6 | 391 | 62.92 | 6 | 15.09 | |
| EH-9604 | Switzerland, Graubünden, Rheinwald/46°33′10″N, 9°19′20″E |
Hoerandl 9604 (WU) | 3 | 313 | 68.37 | 0 | 9.03 | |
| L. nivale (Ten.) Huet ex Hand.-Mazz. | 386 | 79.02 | 7 | 23.75 | ||||
| SSP-01 | Bulgaria, Pirin/41°46′06.9″N, 23°24′47.5″E |
Safer 0043989 (WU) | 7 | 386 | 79.02 | 3 | 13.41 | |
| Mongolia/central China | 609 | 96.72 | 42 | 15.91 | ||||
| L. campestre Hand.-Mazz. | 266 ± 18.39 | 56.71 ± 9.24 | 4 | 15.90 | ||||
| OB-01 | Mongolia, Ulaanbaatar, Mt. Zurkh/ 48°09′N, 107°41′E |
Batsugkh 0043990 (WU) |
6 | 279 | 50.18 | 2 | 7.77 | |
| OB-03 | Mongolia, Ulaanbaatar, Mt. Zurkh/ 48°09′N, 107°41′E |
– | 6 | 253 | 63.24 | 0 | 5.35 | |
| L. leontopodioides Beauverd | 318 ± 65.92 | 57.04 ± 17.43 | 10 | 23.72 | ||||
| Rao Y.G.-1 | China, Inner Mongolia/ 47°27′37.7″N, 120°38′15.8″E |
Rao 0043982 (WU) | 1 | – | – | – | – | |
| Rao Y.G.-2 | China, Inner Mongolia/ 40°25′30.4″N, 112°00′13.2″E |
Rao 0043981 (WU) | 1 | – | – | – | – | |
| SHB | China, Hebei/42°26′00.0″N, 117°21′00.0″E |
Guo 0043983 (WU) | 2 | 290 | 54.48 | 1 | 11.40 | |
| OB-04 | Mongolia, Ulaanbaatar, Terelj National Park/48°09′N, 107°41′E |
Batsugkh 0043991 (WU) |
7 | 296 | 62.16 | 1 | 7.67 | |
| OB-05 | Mongolia, Ulaanbaatar, Terelj National Park/48°09′N, 107°41′E |
– | 5 | 236 | 28.39 | 0 | 7.00 | |
| OB-06 | Mongolia, Ulaanbaatar, Terelj National Park/48°09′N, 107°41′E |
– | 3 | 368 | 66.85 | 0 | 11.46 | |
| OB-07 | Mongolia, Ulaanbaatar, Terelj National Park/48°09′N, 107°41′E |
– | 8 | 401 | 73.32 | 0 | 11.13 | |
| Yunnan | 746 | 98.79 | 120 | 22.44 | ||||
| L. andersonii C.B. Clarke | 332 ± 59.06 | 68.15 ± 2.22 | 13 | 22.53 | ||||
| SSG-01 | China, Yunnan/25°57′55.3″N, 100°22′37.0″E |
Safer 0044009 (WU) | 5 | 312 | 68.27 | 1 | 7.45 | |
| SSG-03 | China, Yunnan/25°42′02.9″N, 100°07′11.5″E |
Safer 0044010 (WU) | 6 | 330 | 65.45 | 1 | 8.59 | |
| SSG-06 | China, Yunnan/27°01′26.4″N, 100°13′00.1″E |
Safer 0044003 (WU) | 14 | 418 | 71.05 | 4 | 14.48 | |
| SSG-14 | China, Yunnan/27°23′43.2″N, 100°08′28.6″E |
Safer 0043998 (WU) | 8 | 255 | 69.41 | 0 | 5.34 | |
| SSG-27 | China, Yunnan/26°03′23.9″N, 102°49′26.3″E |
Safer 0043958 (WU) | 5 | 347 | 66.57 | 1 | 9.94 | |
| L. artemisiifolium Beauverd | 280 ± 51.62 | 57.65 ± 11.11 | 4 | 16.97 | ||||
| SSG-13 | China, Yunnan/27°23′43.2″N, 100°08′28.6″E |
Safer 0043997 (WU) | 7 | 243 | 49.79 | 0 | 6.52 | |
| SSG-28 | China, Yunnan/26°02′06.6″N, 102°49′54.3″E |
Safer 0043959 (WU) | 6 | 316 | 65.51 | 1 | 9.12 | |
| L. caespitosum Diels | 266 | 66.92 | 3 | 12.74 | ||||
| SSG-26 | China, Yunnan/26°04′59.9″N, 102°51′51.7″E |
Safer 0043960 (WU) | 4 | 266 | 66.92 | 2 | 7.05 | |
| L. calocephalum Beauverd | 350 | 64.86 | 7 | 18.44 | ||||
| SSG-11 | China, Yunnan/27°43′52.0″N, 99°58′12.7″E |
Safer 0043999 (WU) | 8 | 350 | 64.86 | 2 | 9.87 | |
| L. dedekensii Beauverd | 302 ± 26.12 | 65.76 ± 4.72 | 8 | 22.49 | ||||
| SSG-17 | China, Yunnan/28°16′16.8″N, 99°11′55.5″E |
Safer 0043994 (WU) | 8 | 284 | 59.86 | 0 | 6.76 | |
| SSG-19 | China, Yunnan/28°23′20.8″N, 98°53′52.6″E |
Safer 0044004 (WU) | 8 | 341 | 66.57 | 0 | 8.61 | |
| SSG-22 | China, Yunnan/26°28′19.6″N, 99°18′21.5″E |
Safer 0043954 (WU) | 7 | 291 | 65.29 | 0 | 7.32 | |
| SSG-24 | China, Yunnan/28°02′36.2″N, 99°05′19.6″E |
Safer 0043956 (WU) | 5 | 293 | 71.33 | 2 | 7.48 | |
| L. franchetii Beauverd | 281 ± 31.82 | 54.67 ± 7.94 | 8 | 18.76 | ||||
| SSG-09 | China, Yunnan/27°36′50.0″N, 99°46′08.0″E |
Safer 0044016 (WU) | 6 | 303 | 59.74 | 1 | 9.30 | |
| SSG-15 | China, Yunnan/27°55′52.9″N, 99°05′58.4″E |
Safer 0044008 (WU) | 6 | 258 | 49.61 | 0 | 7.99 | |
| L. himalayanum DC. | 265 | 67.55 | 2 | 11.81 | ||||
| SSG-18 | China, Yunnan/28°20′29.5″N, 99°04′09.0″E |
Safer 0043993 (WU) | 8 | 265 | 67.55 | 0 | 6.01 | |
| L. sinense Hemsl. ex Forb. & Hemsl. | 371 ± 18.45 | 66.79 ± 1.40 | 6 | 22.91 | ||||
| SSG-04 | China, Yunnan/25°42′05.4″N, 100°07′25.6″E |
Safer 0043975 (WU) | 4 | 351 | 66.38 | 1 | 9.34 | |
| SSG-05 | China, Yunnan/26°19′47.1″N, 100°12′11.7″E |
Safer 0044001 (WU) | 6 | 387 | 65.63 | 0 | 10.60 | |
| SSG-29 | China, Yunnan/24°56′34.3″N, 102°37′53.6″E |
Safer 0043961 (WU) | 8 | 376 | 68.35 | 1 | 10.09 | |
| L. souliei Beauverd | 290 ± 21.92 | 59.63 ± 7.94 | 1 | 13.62 | ||||
| SSG-08 | China, Yunnan/27°36′50.0″N, 99°46′08.0″E |
Safer 0044013 (WU) | 4 | 305 | 65.25 | 0 | 7.81 | |
| SSG-12 | China, Yunnan/27°40′19.9″N, 100°01′34.0″E |
Safer 0044000 (WU) | 6 | 274 | 54.01 | 0 | 7.23 | |
| L. cf. stracheyi C.B. Clarke ex Hemsl. | 213 ± 25.03 | 38.47 ± 6.31 | 10 | 17.27 | ||||
| SSG-20 | China, Yunnan/27°17′35.0″N, 99°16′04.4″E |
Safer 0043995 (WU) | 7 | 212 | 45.75 | 0 | 5.79 | |
| SSG-21 | China, Yunnan/26°27′25.5″N, 99°18′39.9″E |
Safer 0043996 (WU) | 6 | 189 | 34.92 | 0 | 5.42 | |
| SSG-25 | China, Yunnan/27°46′28.7″N, 98°33′41.3″E |
Safer 0043957 (WU) | 5 | 239 | 34.73 | 1 | 8.72 | |
| L. sp. | 324 | 49.38 | 4 | 19.80 | ||||
| SSG-07 | China, Yunnan/27°36′50.0″N, 100°11′58.1E |
Safer 0044012 (WU) | 3 | 324 | 49.38 | 1 | 10.95 | |
| L. sp. | 275 | 59.65 | 7 | 15.31 | ||||
| SSG-16 | China, Yunnan/27°54′02.8″N, 99°38′16.2″E |
Safer 0044007 (WU) | 6 | 272 | 59.56 | 0 | 7.71 | |
SD, standard deviation for the mean values calculated for species with more than one population.
Total DNA was extracted from dried leaf material following the modified hexadecyltrimethylammonium bromide (CTAB) protocol, as used in Russell et al. (2010), and quality checked using a 1% Tris-acetate-EDTA-agarose gel. Liquid nitrogen was used to freeze the material before grinding. Analysis of AFLPs (Vos et al., 1995) was conducted for 216 individuals (plus 16 replicated samples) according to the protocol described by Dixon, Schönswetter & Schneeweiss (2008). After the screening of 86 different selective primer combinations (80 with three selective nucleotides and six with four selective nucleotides), the following six primer combinations were chosen: EcoRI-ATC/MseI-CTC (FAM), EcoRI-AGG/MseI-CAT (VIC), EcoRI-ACC/MseI-CAC (NED), EcoRI-ACA/MseI-CAT (FAM), EcoRI-AAG/MseI-CTC (VIC), EcoRI-ATC/MseI-CTA (NED). The selective PCR products were purified using Sephadex G-50 Superfine following the manufacturer’s instructions (GE Healthcare BioSciences, Uppsala, Sweden). The purified products were analysed on a 3130xl Genetic Analyzer capillary sequencer (Applied BioSystems, Foster City, CA, USA) with GeneScan 500 ROX as an internal size standard (Applied BioSystems). Each sample was aligned to the internal size standard using ABI Prism GeneScan 3.7 (Applied BioSystems). The sized GeneScan files were imported to GeneMarker Version 1.85 (SoftGenetics, State College, PA, USA). After pre-analysis using default settings, the samples were re-calibrated to the size standard. A panel of scorable markers was established for each primer combination, and fragments ranging from 90 to 500 bp were scored. High-quality size calibrations (> 90% accuracy) were obtained for all samples. The relative fluorescent unit (RFU) threshold was adjusted for each marker separately, ranging from 50 RFU for the weakest to 200 RFU for the strongest fluorescence intensities. The automatic scoring was conducted using Local Southern size call, peak saturation, baseline subtraction, spike removal, pull up correction and a stutter peak filter of 5%. The results were exported as a combined presence/absence matrix.
The presence/absence matrix obtained from Gene-Marker was imported to FAMD 1.108 (Schlüter & Harris, 2006) to calculate a distance matrix based on Nei–Li distances (Nei & Li, 1979). This matrix was used to analyse genetic relationships among and within the different populations via the neighbor-net method implemented in SplitsTree 4.8 (Huson & Bryant, 2006). PAUP* version 4.0b10 (Swofford, 2003) was used to construct a phylogenetic tree based on the neighbor-joining (NJ) method and Nei–Li distances. As a result of the lack of outgroups, midpoint rooting was performed. The support for specific nodes for the NJ tree (PAUP*) was calculated using the bootstrap method (Felsenstein, 1985) with 10 000 replicates, again with the program PAUP* version 4.0b10.
Descriptive statistics included the total number of fragments (Ftot), the percentage of polymorphic fragments (Fpoly%) and the number of private fragments (Fpriv), calculated with the program FAMD 1.108 (Schlüter & Harris, 2006). In addition, the ‘frequency down-weighted marker values’ (DW; Schönswetter & Tribsch, 2005) were determined using R-script AFLPdat (Ehrich, 2006). For this measure of divergence, the value of DW is expected to be high in long-term isolated populations, whereas newly established populations are expected to exhibit low values (Schönswetter & Tribsch, 2005). All of these parameters were calculated on three levels: (1) for biogeographical ranges (Europe, Mongolia/central China and Yunnan); (2) for the different species; and (3) for the different populations. For the calculations of Fpriv and DW, subsets of 16 samples per biogeographical range and three individuals per species and population, respectively, were used (for the population SHB only the two available samples were used; the two single individuals Rao Y.G.-1 and Rao Y.G.-2 were excluded from these analyses). The reduced datasets were applied to even out the unequal sample sizes (as suggested by Schönswetter & Tribsch, 2005); the samples were chosen to represent the whole dataset as broadly as possible. For the determination of Ftot and Fpoly% on the specific level, mean values including standard deviations of the associated populations were calculated. The results are summarized in Table 1.
RESULTS
The six primer combinations used for the analysis generated 946 scorable fragments between 90 and 500 bp: EcoRI-ATC/MseI-CTC (FAM), 184 fragments; EcoRI-AGG/MseI-CAT (VIC), 149 fragments; EcoRI-ACC/MseI-CAC (NED), 140 fragments; EcoRI-ACA/MseI-CAT (FAM), 178 fragments; EcoRI-AAG/MseI-CTC (VIC), 160 fragments; EcoRI-ATC/MseI-CTA (NED), 135 fragments. Of the 946 fragments, 942 were polymorphic (99.57%) The error rate (Bonin et al., 2004) among the replicated samples amounted to 7.37%.
Descriptive statistics were generated to reveal information about the genetic diversity of the genus. The results of all analyses are summarized in Table 1. The number of total fragments (Ftot) was 746 for the Yunnan group, 650 for the European group and 609 for the Mongolian/central Chinese group. At the specific level, Ftot ranged from 213 for L. cf. stracheyi to 386 for L. nivale, and, at the populational level, from 189 for population SSG-21 (L. cf. stracheyi) to 418 for population SSG-06 (L. andersonii C.B. Clarke). The percentage of polymorphic fragments (Fpoly%) was 98.79% for the Yunnan group, 96.72% for the Mongolian/central Chinese group, 96.00% for the European group, and varied from 38.47% (L. cf. stracheyi C.B. Clarke ex Hemsl.) to 84.03% (L. leontopodioides) at the specific level and from 28.39% (OB-05) to 79.02% (SSP-01) at the populational level. The group with the largest number of private fragments (Fpriv) was the Yunnan group (120), followed by the European group (47) and the Mongolian/central Chinese group (42). At the specific level, Fpriv was quite variable, ranging between unity for L. souliei and 13 for L. andersonii; at the populational level, Fpriv was generally low, with the highest value of six for the L. alpinum population SSSP-01. The determination of the ‘frequency down-weighted marker values’ (DW) revealed the same results at the biogeographical level, where the Yunnan group had the highest value (22.44), followed by the European group (18.15) and the Mongolian/central Chinese group with the lowest value (15.91). The species with the highest value of DW was L. alpinum at 25.98 and the species with the lowest value of DW was L. himalayanum DC. at 11.81. At the populational level, the DW values ranged between 5.34 (L. andersonii SSG-14) and 15.09 (L. alpinum SSSP-01).
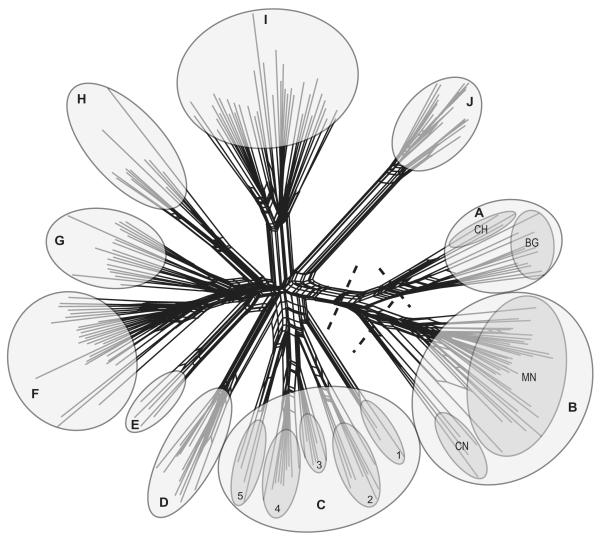
The neighbor-net network shows three large groups (Fig. 2). The first group (A) comprises the three European populations investigated (ACH, Switzerland; ABG, Bulgaria). The second group (B) represents the closely related Mongolian (BMN) and central Chinese (BCN) species. The third and largest group (including groups C–J) contains exclusively populations from the centre of diversity (Yunnan, China). On the specific level, 10 different groups can be distinguished. Seven groups are monospecific (groups D–J). Group A consists of the two European species, L. alpinum and L. nivale. Group B includes L. campestre Hand.-Mazz. and L. leontopodioides, two very common species in central China and Mongolia. Within the Mongolian group (BMN), the two species L. leontopodioides and L. campestre are not clearly separated from each other; geographical differentiation in group B (Mongolia BMN/central China BCN) is more obvious than differentiation at the specific level. Group C includes five different species [L. sp. (SSG-07; C1), L. himalayanum (C2), L. calocephalum (C3), L. souliei (C4) and L. caespitosum Diels (C5)]. The monospecific groups are D (L. franchetii Beauverd), E [L. sp. (SSG-16)], F (L. dedekensii), G (L. sinense), H (L. artemisiifolium Beauverd), I (L. andersonii) and J (L. cf. stracheyi).
Figure 2.
Neighbor-net network, highlighting groups and subgroups among species of Leontopodium: ACH, L. alpinum (Switzerland); ABG, L. alpinum and L. nivale (Bulgaria); BMN, L. leontopodioides and L. campestre (Mongolia); BCN, L. leontopodioides (central China); C1, L. sp. (SSG-07); C2, L. himalayanum; C3, L. calocephalum; C4, L. souliei; C5, L. caespitosum; D, L. franchetii; E, L. sp. (SSG-16); F, L. dedekensii; G, L. sinense; H, L. artemisiifolium; I, L. andersonii; J, L. cf. stracheyi (BG, Bulgaria; CH, Switzerland; CN, central China; MN, Mongolia). The broken lines indicate the three large groups recognized with AFLP analyses: the European group (A), the Mongolian/central Chinese group (B) and the Yunnan group (C–J).
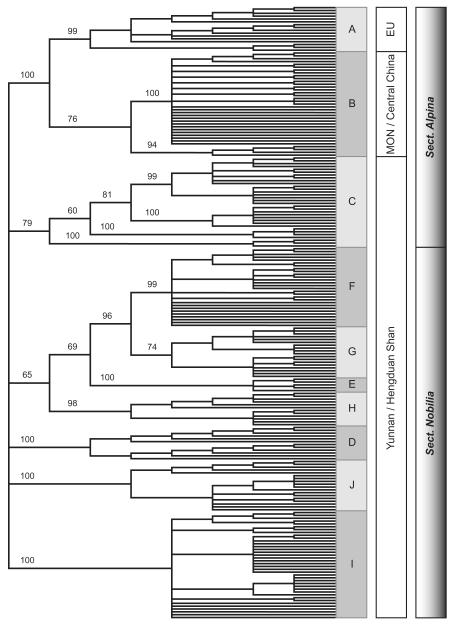
Similar results were obtained with the NJ analysis performed in PAUP*. The NJ bootstrap consensus tree (50% majority rule; Fig. 3) provides some additional information with regard to specific affinities. The European/Mongolian/central Chinese clade is clearly separated from the Yunnan species with a bootstrap support (BS) of 100%. Within this clade, the European group is supported with 99% BS. The Mongolian/central Chinese clade (BS 76%) is divided into the Mongolian populations (BS 100%) and the central Chinese populations (BS 94%). Group C consists of the five closely related species described above; this group is supported with BS 79%. The closely related L. dedekensii, L. sinense, L. artemisiifolium and L. sp. (SSG-16) (groups E, F, G and H) fall into one clade (BS 65%). Within this clade, L. dedekensii has a BS of 99%, L sinense is supported with 74%, L. sp. (SSG-16) with 100% and L. artemisiifolium with 98%. Groups D (L. franchetii), I (L. andersonii) and J (L. cf. stracheyi) have BS of 100% each.
Figure 3.
Neighbor-joining bootstrap consensus tree (50% majority rule) obtained with the program PAUP*; numbers above each branch are bootstrap percentages. The European species and the Mongolian/central Chinese species (groups A and B) are monophyletic (BS 100%). Group C consists of five species [L. sp. (SSG-07), L. himalayanum, L. calocephalum, L. souliei and L. caespitosum]; these species appear in the same clade (BS 79%). The groups E [L. sp. (SSG-16)], F (L. dedekensii), G (L. sinense) and H (L. artemisiifolium) form a weakly supported clade (BS 65%) in the neighbor-joining tree. Groups D, I and J are monospecific and supported with BS 100% each.
DISCUSSION
Species relationships within the genus
We found AFLPs to be very useful for the determination of relationships within the genus Leontopodium. Different groups can be recognized, and the boundaries between the groups are clear. The species are all closely related and, as suggested by the low sequence variability within cpDNA (Blöch et al., 2010), the genus has a short evolutionary history. Phylogenetic relationships within the whole tribe Gnaphalieae are not well resolved. Ward et al. (2009) suggested that relationships among most genera within the tribe may be difficult to discern. Within the tribe, the genus Leontopodium is part of the ‘crown radiation’, which represents the main clade of Gnaphalieae; this clade, in particular, lacks phylogenetic structure (Ward et al., 2009).
In general, the groupings found in the study of Blöch et al. (2010) can only be partially supported. Blöch et al.’s group A, consisting of the two European species L. alpinum and L. nivale (group A in our results; see Figs 2, 3), can also be recovered with AFLPs. Group C of Blöch et al. includes the two closely related species L. dedekensii and L. sinense (groups F and G, respectively, in our results; see Figs 2, 3). These species can be clearly distinguished from each other with AFLP analysis, in contrast with the results of Blöch et al. (2010), where their clade was not fully resolved. Group B of Blöch et al. (L. calocephalum, L. leontopodioides, L. ochroleucum and L. souliei) is not supported by our results. Two of the species of this group B, namely L. calocephalum and L. souliei, are part of our group C, which consists of five closely related species. In contrast, our results show that L. leontopodioides appears not to be closely related to the other species within Blöch et al.’s group B. Using AFLPs, L. leontopodioides groups in the Mongolian clade (our group B) with a BS of 100% (details see below), and is clearly separated from the other species. Unfortunately, we were not able to conclude on the position of L. ochroleucum (also included in group B of Blöch et al.).
With AFLP analysis, we were able to identify 10 different groups within Leontopodium (Figs 2, 3). Group A consists of three European populations. The group is well separated from the other groups with a BS of 99%. One population was collected in Switzerland (ACH) and two in Bulgaria (ABG). For Bulgaria, L. nivale has been described from the Pirin Mountains, and population SSP-01 was collected there. The second population from Bulgaria, however, was collected in Stara Planina in central Bulgaria, but the morphology of this population is closer to L. alpinum (and was treated as such). Therefore, we were unable to conclude on the relationship between the two European species, L. alpinum and L. nivale; an AFLP study including many European populations from wide distributional ranges would be needed to resolve this issue.
Group B includes populations of L. leontopodioides and L. campestre from Mongolia (BMN) and central China (BCN). The group has a BS of 76%. Both species belong to section Alpina/subsection Alpinoidea in Handel-Mazzetti’s monograph and are closely related. They have a wide distributional range (north, central and east Asia) and are a sister group to the European species, sharing several morphological similarities. In general, species diversity is quite low throughout this large geographical area (Meusel & Jaeger, 1992).
Groups C–J include exclusively populations collected around the Hengduan-Shan mountain range in the Province of Yunnan in south-western China. This area is recognized as the centre of diversity and, according to Meusel & Jaeger (1992), 15–18 different species are native there. The groups are well separated from the European/Mongolian/central Chinese populations with a BS of 100% (Figs 2, 3).
Group C consists of five different species [L. caespitosum, L. calocephalum, L. himalayanum, L. souliei and L. sp. (SSG-07)], which are morphologically similar to each other (small herbs with unbranched stems, linear-lanceolate leaves, a more or less densely congested inflorescence, composed of usually just a few calathidia; see also Fig. 1). Within this group, species relationships are not fully resolved, and possible hybridization between the species must be considered because of their similar distributional areas within the centre of diversity. All species are part of Handel-Mazzetti’s section Alpina and, within this section, they are divided into the two subsections Haastioidea [L. caespitosum (=L. jacotianum var. caespitosum in Handel-Mazzetti’s monograph) and L. himalayanum] and Alpinoidea (L. souliei and L. calocephalum). As the five species form a well-separated group (C) in our analysis, we recommend keeping them together in one section/subsection.
Groups D–J belong to Handel-Mazzetti’s section Nobilia, and can be clearly delimited from groups A–C (section Alpina). Group D comprises two populations of L. franchetii. The group has a BS of 100%. Leontopodium franchetii is one of the few species that can be identified easily because of its specific morphology (a woody, tall, glandular herb, with simple or occasionally branching shoots and many linear leaves). Although L. franchetii occurs sympatrically with other Leontopodium species, it is monophyletic (Fig. 3).
In group E, just one population is included [L. sp. (SSG-16)]. We initially considered this population to be L. dedekensii, but the result of the neighbor-net analysis (Fig. 2) suggested that it should be excluded and treated separately. With the NJ analysis, where population SSG-16 is sister to L. sinense and L. dedekensii, and shares a clade with L. sinense, L. dedekensii and L. artemisiifolium (Fig. 3), this finding could be confirmed, although the whole clade is not well supported (BS 65%). Because SSG-16 shares several morphological characters, especially with L. dedekensii, but also with other Leontopodium species, possible hybridization must also be considered.
Groups F and G consist of the two closely related species L. sinense (F) and L. dedekensii (G). They are difficult to distinguish using morphological characters. They have similar growth forms and occupy similar habitats, and thus are often incorrectly identified. With AFLPs, we were able to clearly distinguish the two species. In the NJ tree, their relationship as sister species is obvious, and the clade is supported with BS 96%.
Group H comprises two L. artemisiifolium populations. Leontopodium artemisiifolium is a robust fasciculate herb which is sometimes confused with L. sinense because of the similar habitat and growth form. The results of the NJ analysis (Fig. 3) indicates that this species is sister to L. sinense and L. dedekensii, and highlights a close phylogenetic relationship among the three species.
Group I comprises four populations of the widespread and morphologically distinct species L. andersonii. This species is present throughout the whole area of the centre of diversity, and occurs sympatrically with several other species. Regardless of this sympatry, gene flow between L. andersonii and other species should be low or even nonexistent because of its undoubted and highly supported clade within the NJ tree. This can also be observed in L. franchetii (group D). As a conclusion, sympatric speciation might also play a role in the genus Leontopodium, but this could not be confirmed clearly within this study.
Group J consists of three populations (SSG-20, SSG-21 and SSG-25), which we identified as L. cf. stracheyi. This species is morphologically different from other Leontopodium species, and occupies a distinct position within the genus, being supported with a BS of 100%. These populations were found in the Nujiang valley close to the border to Myanmar, where this species appears to be restricted. The Nu-Shan mountain chain divides the Nujiang valley from the other parts of the centre of diversity, which may result in genetic isolation; they have relatively low Ftot (213) and Fpoly% (38.47%); all other species have Ftot greater than 260 and Fpoly% higher than 49.00% (Table 1).
Biogeography
Our results also provide insights into the biogeography of Leontopodium. The Sino-Himalayan region in south-western China (Province of Yunnan) is the centre of diversity and perhaps also the centre of origin of the genus. Private and rare fragments accumulate through time and can be taken as a measure of population antiquity (Ortiz et al., 2009). In our study, the number of private fragments for the Yunnan group is 120, whereas, in the European group and in the Mongolian/central Chinese group, only 47 and 42 private fragments, respectively, have been accumulated (Table 1). These results suggest a longlasting in situ history of populations in the centre of diversity of the genus. In addition, we calculated the DW value (Table 1) as described in Schönswetter & Tribsch (2005), which is expected to be high in longterm isolated populations and low in newly established populations, thus helping to distinguish old vicariance from recent dispersal (Schönswetter & Tribsch, 2005). In this analysis, the Yunnan group has the highest value (Table 1), which also suggests the Sino-Himalayan region as the centre of origin for the genus.
ACKNOWLEDGEMENTS
The authors thank V. Vladimirov (Bulgarian Academy of Sciences, Bulgaria), S. Schwaiger and O. Batsugkh (University of Innsbruck, Austria), G.-Y. Rao (Peking University, China) and E. Hörandl (University of Vienna, Austria) for help with plant material collection and advice. We thank W. B. Dickoré (Botanische Staatssammlung Munich, Germany) for voucher identifications and morphological investigations. We thank the Institutes of Botany (Beijing and Kunming) of the Chinese Academy of Sciences (China), the Bulgarian Academy of Sciences (Sofia, Bulgaria) and the Austrian Academy of Sciences (Vienna, Austria) for supporting the collection trips. We thank the Pirin National Park (Bansko, Bulgaria) for collecting permission. We also thank M. H. J. Barfuss, C. Blöch, C. A. Rebernig and A. Russell (University of Vienna, Austria) for technical assistance and advice. This study was financed by the Austrian Science Fund (FWF) Grant No. P19480.
REFERENCES
- Beauverd G. Nouvelles espèces euroasiatiques du genre Leontopodium. Bulletin de la Société Botanique de Genève. 1909;1:185–196. [Google Scholar]
- Beauverd G. Contribution à l’étude des Composées. Suite IV. Bulletin de la Société Botanique de Genève. 1910;2:244–252. [Google Scholar]
- Beauverd G. Contribution à l’étude des Composées. Suite V. Bulletin de la Société Botanique de Genève. 1911;3:353–359. [Google Scholar]
- Beauverd G. Contribution à l’étude des Composées. Suite VI: Nouveaux Leontopodium et Raoulia. Bulletin de la Société Botanique de Genève. 1912;5:12–55. [Google Scholar]
- Beauverd G. Contribution à l’étude des Composées. Suite IX. Bulletin de la Société Botanique de Genève. 1914;6:142–148. [Google Scholar]
- Blöch C, Dickoré WB, Samuel R, Stuessy T. Molecular phylogeny of the Edelweiss (Leontopodium, Asteraceae – Gnaphalieae) Edinburgh Journal of Botany. 2010;67:235–264. [Google Scholar]
- Bonin A, Bellemain E, Bronken Edeisen P, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetics studies. Molecular Ecology. 2004;13:3261–3273. doi: 10.1111/j.1365-294X.2004.02346.x. [DOI] [PubMed] [Google Scholar]
- Bussell JD, Waycott M, Chappill JA. Arbitrarily amplified DNA markers as characters for phylogenetic inference. Perspectives in Plant Ecology, Evolution and Systematics. 2005;7:3–26. [Google Scholar]
- Després L, Gielly L, Redoutet W, Taberlet P. Using AFLP to resolve phylogenetic relationships in a morphologically diversified plant species complex when nuclear and chloroplast sequences fail to reveal variability. Molecular Phylogenetics and Evolution. 2003;27:185–196. doi: 10.1016/s1055-7903(02)00445-1. [DOI] [PubMed] [Google Scholar]
- Dixon CJ, Schönswetter P, Schneeweiss GM. Morphological and geographical evidence are misleading with respect to the phylogenetic position and origin of the narrow endemic polyploid Androsace cantabrica (Primulaceae) Systematic Botany. 2008;33:384–389. [Google Scholar]
- Dobner MJ, Ellmerer EP, Schwaiger S, Batsugkh O, Narantuya S, Stutz M, Stuppner H. New lignan, benzofuran, and sesquiterpene derivatives from the roots of Leontopodium alpinum and L. leontopodioides. Helvetica Chimica Acta. 2003a;86:733–738. [Google Scholar]
- Dobner MJ, Schwaiger S, Jenewein IH, Stuppner H. Antibacterial activity of Leontopodium alpinum (Edelweiss) Journal of Ethnopharmacology. 2003b;89:301–303. doi: 10.1016/j.jep.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Dobner MJ, Sosa S, Schwaiger S, Altinier G, Loggia RD, Kaneider NC, Stuppner H. Anti-inflammatory activity of Leontopodium alpinum and its constituents. Planta Medica. 2004;70:502–508. doi: 10.1055/s-2004-827148. [DOI] [PubMed] [Google Scholar]
- Ehrich D. AFLPDAT: a collection of R functions for convenient handling of AFLP data. Molecular Ecology Notes. 2006;6:603–604. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Franchet AR. Observations sur le groupe des Leontopodium. Bulletin de la Société Botanique de France. 1892;39:126–136. [Google Scholar]
- Greuter W. The Euro+Med treatment of Gnaphalieae and Inuleae (Compositae) – generic concepts and required new names. Willdenowia. 2003;33:239–244. [Google Scholar]
- Grierson AJC, Long DG. Flora of Bhutan (including a record of plants from Sikkim & Darjeeling) Royal Botanical Garden Edinburgh & Royal Government of Bhutan; Edinburgh: 2001. [Google Scholar]
- Grubov V. Conspectus generis Leontopodium (Pers.) R. Br. (Compositae) Asiae centralis [in Russian] Novosti Sistematiki Vysshikh i Nizshikh Rastenii. 2003;35:188–197. [Google Scholar]
- Guo YP, Saukel J, Mittermayr R, Ehrendorfer F. AFLP analyses demonstrate genetic divergence, hybridization, and multiple polyploidization in the evolution of Achillea (Asteraceae-Anthemideae) New Phytologist. 2005;166:273–289. doi: 10.1111/j.1469-8137.2005.01315.x. [DOI] [PubMed] [Google Scholar]
- Guo YP, Saukel J, Ehrendorfer F. AFLP trees versus scatterplots: evolution and phylogeography of the polyploid complex Achillea millefolium agg. (Asteraceae) Taxon. 2008;57:153–169. [Google Scholar]
- Handel-Mazzetti H. Systematische Monographie der Gattung Leontopodium. Beihefte zum Botanischen Centralblatt. 1927;44:1–178. [Google Scholar]
- Hornick A, Schwaiger S, Rollinger JM, Vo NP, Prast H, Stuppner H. Extracts and constituents of Leontopodium alpinum enhance cholinergic transmission: brain ACh increasing and memory improving properties. Biochemical Pharmacology. 2008;76:236–248. doi: 10.1016/j.bcp.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Iwatsuki K, Yamazaki T, Boufford DE, Ohba H. Flora of Japan. Kondasha; Tokyo: 1995. [Google Scholar]
- Koopman WJM. Phylogenetic signal in AFLP data sets. Systematic Biology. 2005;54:197–217. doi: 10.1080/10635150590924181. [DOI] [PubMed] [Google Scholar]
- Koopman WJM, Zevenbergen MJ, Van den Berg RG. Species relationships in Lactuca s.l. (Lactuceae, Asteraceae) inferred from AFLP fingerprints. American Journal of Botany. 2001;88:1881–1887. [PubMed] [Google Scholar]
- Meusel H, Jaeger EJ. Vergleichende chorologie der zentraleuropaeischen Flora. Fischer Verlag; Jena, Stuttgart, New York: 1992. [Google Scholar]
- Nei M, Li WH. Mathematical-model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz MA, Tremetsberger K, Stuessy TF, Terrab A, García-Castano JL, Talavera S. Phylogeographic patterns in Hypochaeris section Hypochaeris (Asteraceae, Lactuceae) of the western Mediterranean. Journal of Biogeography. 2009;36:1384–1397. [Google Scholar]
- Pelser PB, Gravendeel B, van der Meijden R. Phylogeny reconstruction in the gap between too little and too much divergence: the closest relatives of Senecio jacobaea (Asteraceae) according to DNA sequences and AFLPs. Molecular Phylogenetics and Evolution. 2003;29:613–628. doi: 10.1016/s1055-7903(03)00139-8. [DOI] [PubMed] [Google Scholar]
- Qaiser M, Abid R. Flora of Pakistan. Department of Botany, University of Karachi & Missouri Botanical Garden Press; Karachi, St. Louis, MO: 2003. [Google Scholar]
- Rechinger KH. Flora iranica: compositae iv – inuleae. Akademische Drucks- und Verlagsgesellschaft; Graz: 1980. [Google Scholar]
- Reisinger U, Schwaiger S, Zeller I, Messner B, Stigler R, Wiedemann D, Mayr T, Seger C, Schachner T, Dirsch VM, Vollmar AM, Bonatti JO, Stuppner H, Laufer G, Bernhard D. Leolignin, the major lignan from Edelweiss, inhibits intimal hyperplasia of venous bypass grafts. Cardiovascular Research. 2009;82:542–549. doi: 10.1093/cvr/cvp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A, Samuel R, Rupp B, Barfuss MHJ, Safran M, Besendorfer V, Chase MW. Phylogenetics and cytology of a pantropical orchid genus Polystachya (Polystachyinae, Vandeae, Orchidaceae): evidence from plastid DNA sequence data. Taxon. 2010;59:389–404. [Google Scholar]
- Schlüter PM, Harris SA. Analysis of multilocus finger-printing data sets containing missing data. Molecular Ecology Notes. 2006;6:569–572. [Google Scholar]
- Schönswetter P, Tribsch A. Vicariance and dispersal in the alpine perennial Bupleurum stellatum L. (Apiaceae) Taxon. 2005;54:725–732. [Google Scholar]
- Schwaiger S, Adams M, Seger C, Ellmerer EP, Bauer R, Stuppner H. New constituents of Leontopodium alpinum and their in vitro leukotriene biosynthesis inhibitory activity. Planta Medica. 2004;70:978–985. doi: 10.1055/s-2004-832625. [DOI] [PubMed] [Google Scholar]
- Schwaiger S, Cervellati R, Seger C, Ellmerer EP, About N, Renimel I, Godenir C, Andre P, Gafner F, Stuppner H. Leontopodic acid – a novel highly substituted glucaric acid derivative from Edelweiss (Leontopodium alpinum Cass.) and its antioxidative and DNA protecting properties. Tetrahedron. 2005;61:4621–4630. [Google Scholar]
- Shishkin BK. Flora of the U.S.S.R. Vol. XXV – Compositae. Koeltz Scientific Books; Koenigstein: 1990. [Google Scholar]
- Speroni E, Schwaiger S, Egger P, Berger AT, Cervellati R, Govoni P, Guerra MC, Stuppner H. In vivo efficacy of different extracts of Edelweiss (Leontopodium alpinum Cass.) in animal models. Journal of Ethnopharmacology. 2006;105:421–426. doi: 10.1016/j.jep.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Spooner DM, Peralta IE, Knapp S. Comparison of AFLPs with other markers for phylogenetic inference in wild tomatoes [Solanum L. section Lycopersicon (Mill.) Wettst.] Taxon. 2005;54:43–61. [Google Scholar]
- Stuppner H, Ellmerer EP, Ongania KH, Dobner M. Bisabolane derivatives from Leontopodium alpinum. Helvetica Chimica Acta. 2002;85:2982–2989. [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods) version 4.0b10 Sinauer Associates; Sunderland, MA: 2003. [Google Scholar]
- Tabernaemontanus JT. Das Ander Buch von Kreutern. In: Bauhin H, editor. D. Jacobi theodori tabernaemontani neu vollkommen kraeuter-buch. Vol. 1993. Verlag Kölbl, Grünwald bei München; 1582. pp. 779–782. Reprint Basel: König, 1731. [Google Scholar]
- Tremetsberger K, Stuessy TF, Kadlec G, Urtubey E, Baeza CM, Beck SG, Valdebenito HA, Ruas CDF, Matzenbacher NI. AFLP phylogeny of South American species of Hypochaeris (Asteraceae, Lactuceae) Systematic Botany. 2006;31:610–626. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA finger-printing. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J, Bayer RJ, Breitwieser I, Smissen R, Galbany-Casals M, Unwin M. Gnaphalieae. In: Funk VA, Susanna A, Stuessy TF, Bayer RJ, editors. Systematics, evolution and biogeography of Compositae. International Association for Plant Taxonomy; Vienna: 2009. pp. 539–588. [Google Scholar]
- Wu ZY, Raven PH, Hong DY. Flora of China. Science Press, Missouri Botanical Garden Press; Beijing, St. Louis, MO: 1994. [Google Scholar]