Abstract
Interstrand crosslinks covalently link complementary DNA strands, block replication and transcription, and can trigger cell death. In eukaryotic systems several pathways, including the Fanconi Anemia pathway, are involved in repairing interstrand crosslinks, but their precise mechanisms remain enigmatic. The lack of functional homologs in simpler model organisms has significantly hampered progress in this field. Two recent studies have finally identified a Fanconi-like interstrand crosslink repair pathway in yeast. Future studies in this simplistic model organism promise to greatly improve our basic understanding of complex interstrand crosslink repair pathways like the Fanconi pathway.
Keywords: Fanconi anemia, Interstrand crosslink repair, Mph1, Chl1, Slx4, Msh2, Msh6, Mhf1, Mhf2
Background
DNA damaging agents such as nitrogen mustard [1,2], formaldehyde [3], and cisplatin [4] generate many lesions that inhibit proper DNA replication and transcription. One such lesion, the interstrand crosslink (ICL), covalently links two complementary DNA strands and prevents their separation. Importantly, since both strands are damaged, an undamaged template strand is not available for repair. Due to these blocks and repair challenges, ICLs are considered one of the most toxic DNA lesions. It is estimated that the presence of just one unrepaired ICL is sufficient to kill yeast or bacteria [5] and approximately 40 unrepaired ICLs can kill mammalian cells [6]. As a result of this high cytotoxicity, crosslinking agents are common anticancer agents [7]. Outside of chemotherapies, ICLs can be induced by exposures in the environment [8] and byproducts of normal metabolic processes [9,10]. Thus, a clearer understanding of the mechanisms of ICL repair will inform our knowledge of both normal and cancer cells. This article and another recent review [11] describe novel findings in yeast that provide insight into the mechanisms of eukaryotic ICL repair.
A yeast fanconi-like pathway emerges
Cells have the capacity to repair ICLs through highly complex DNA repair mechanisms. ICL repair in the prokaryotic system is relatively well defined. In Escherichia coli, nucleotide excision repair (NER) creates incisions on each side of the ICL. The resulting short oligonucleotide is attached through the ICL, but is displaced from the helix, revealing a gap. The gap is filled in by homologous recombination (HR) or translesion bypass synthesis (TLS), and the displaced oligonucleotide/ICL adduct is removed by NER [12].
In lower eukaryotes, defects in most known DNA repair pathways result in ICL sensitivity suggesting that eukaryotic mechanisms are much more complex, involve multiple repair pathways, and can occur in multiple phases of the cell cycle. Several recent reviews address this complexity in detail [13-23]. In the budding yeast Saccharomyces cerevisiae, a G1-specific repair pathway involves NER and TLS similar to the E. coli system [24]. Additionally, three independent epistasis groups (PSO2, RAD52, and RAD18) are implicated in ICL repair [25], but each pathway mechanism is not fully defined. Pso2 is an exonuclease that may be important for cleaving ICL repair intermediates [26-30]. HR proteins, including Rad52 and Rad51, likely fill in gaps post-incision and/or repair double strand breaks (DSBs) that arise during ICL repair. The post replication repair (PRR) pathway may help fill in the gaps after the incision and unhooking of ICLs.
In higher eukaryotes the Fanconi anemia (FA) DNA repair pathway has emerged as a master-regulator of downstream checkpoints and pathways of ICL repair [13]. This pathway was named for patients with the heritable, recessive disorder caused by mutations in FA repair genes. These mutations confer developmental defects, cancer predisposition, and marked sensitivity to ICL-forming agents [31]. In the FA repair pathway, FANCM and FAAP24 are thought to recognize blocked forks, activate checkpoint responses, and recruit the FA core complex (FANC A, B, C, E, F, G, L, FAAP100) [32-34]. FANCM is additionally stabilized by interactions with the MHF1/MHF2 complex [35,36]. After recruitment, the FA core complex ubiquitinates FANCD2 and FANCI [32,37]. These ubiquitinated proteins likely promote HR repair and other poorly understood downstream repair events mediated by FANCD1, FANCN, FANCP/SLX4, FANCO/RAD51C and/or FANCJ [13].
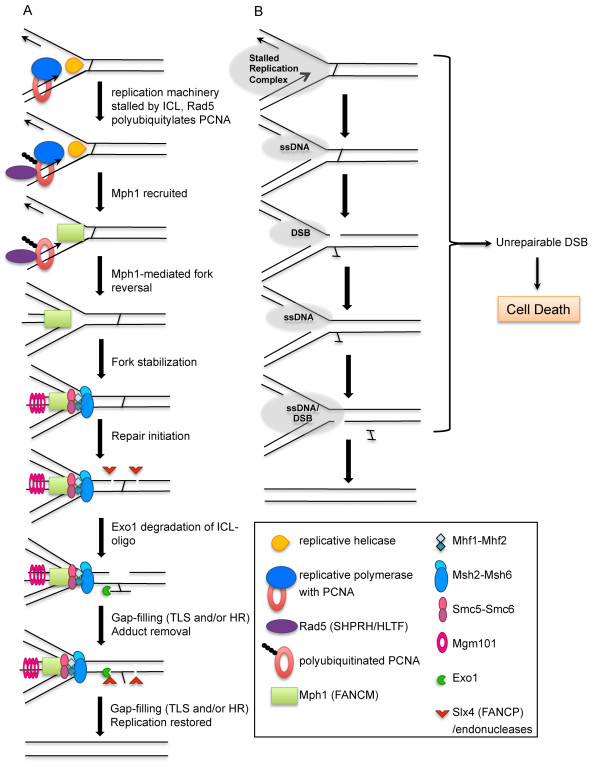
Studies in lower eukaryotic model organisms, like yeasts, have greatly improved our understanding of most DNA repair pathways. The single-celled yeast model is genetically tractable and provides a simplistic system for the study of complex DNA repair problems. Until recently, a yeast FA-like ICL repair pathway had not been functionally validated. Mph1, Mhf1/Mhf2, Chl1, and Slx4 are putative homologs to FANCM, MHF1/MHF2, FANCJ, and FANCP, respectively [34-36,38-41]. Although previous work established that the yeast proteins Mph1 [42-45], Mhf1/Mhf2 [35,36], Chl1 [46-48], and Slx4 [49] all play an important role in maintaining genomic integrity, a role in ICL repair (as indicated by mutant sensitivity to ICL agents) was not apparent. Recent publications from our group [50] and the McHugh group [51] have demonstrated that these proteins play a previously unappreciated role in ICL repair. Their function is important for ICL survival when either the Pso2 exonuclease or the PRR helicase Srs2 pathways are inactivated. These studies also revealed roles for additional proteins in the yeast FA-like pathway including Mgm101, MutSα (Msh2-Msh6), Exo1, proliferating cell nuclear antigen (Pol30/PCNA), Smc5/6 and Rad5. These studies provided key mechanistic insights that confirm, clarify, and bolster our knowledge of the FA pathway, allowing us to formulate the following model (Figure 1A):
Figure 1.
Model for replication-associated interstrand crosslink repair in yeast. (A) Replication is stalled by an ICL, Rad5 polyubiquitylates PCNA, and Mph1-mediated fork-reversal stabilizes the fork for repair (with Smc5/6 and Mhf1/2) and protects the repair intermediates from collapsing into double strand breaks (DSBs). Downstream events of repair are mediated by Slx4 and Exo1. HR and TLS are important for gap-filling steps. The figure key shows the putative human homologs in brackets. (B) The basic steps of ICL repair lead to various fragile intermediates (ssDNA, single strand DNA) that can collapse into DSBs. Cell death is triggered if the DSB cannot be repaired.
ICL-induced replication stalling recruits or activates Rad5, which polyubiquitylates PCNA. The helicase Mph1 is recruited to reverse and stabilize the fork. Although their precise ICL-repair functions are unknown, Chl1, Mhf1/Mhf2, Smc5/6, and Mgm101 likely help stabilize Mph1 and/or the ICL repair intermediates. Slx4 may coordinate incisions surrounding the ICL with its associated endonucleases. Also in this pathway, the canonical mismatch repair complex Msh2-Msh6 (MutSα) potentially senses the aberrant DNA structure at the fork and/or recruits Exo1 to digest the tethered ICL-containing oligonucleotide. Oligonucleotide degradation produces a substrate for downstream processing events such as gap-filling by TLS polymerases or HR. Once the crosslinked adduct is removed, the DNA replication fork can be restored. Importantly, this reversed-fork pathway protects the fragile intermediates of repair (Figure 1B), which can collapse into double strand breaks and trigger cell death.
The foundational studies by our group and the McHugh group have validated the yeast FA-like pathway proteins [50,51]. Despite this, many questions remain about the precise functions of each protein, particularly Chl1, Smc5/6, and Mgm101. Chl1 and Smc5/6 have been implicated in sister chromatid interactions [52-54], so it is possible that these interactions create a stable intermediate for engagement by HR. Mgm101 has been implicated in mitochondrial recombination [55], so this role may extend to the nuclear compartment as well. Future genetic studies and the examination of ICL repair intermediates in different mutant backgrounds will hopefully shed light on these open questions.
In addition to the FA-like ICL repair pathway in yeast, Pso2 and Srs2 participate in ICL repair. The Pso2 nuclease functions after initial ICL recognition and incision, which is likely mediated by NER factors [56]. Srs2 is a helicase that directs the PRR pathway by preventing substrate engagement by recombination proteins [57,58]. Since PRR is a damage tolerance it is not clear how the ICL is excised through this pathway. It is entirely possible that, rather than forming independent pathways, the Pso2- and Srs2-mediated pathways represent the early (Pso2) and late (PRR) actions of a single pathway.
Conclusion
Mechanistically, these studies confirm the existence of a yeast ICL repair pathway that is reminiscent of the mammalian FA pathway. Like the mammalian system [59], mismatch repair proteins contribute to the yeast FA-like pathway. These studies also clarify the controversial role of the PRR pathways [60-62] by demonstrating that, while the PRR proteins Srs2 and Rad18 are distinct from the FA pathway, Rad5 and PCNA are important mediators. Finally, in both yeast [50,51] and mammalian pathways [35,63], Mph1/FANCM-mediated fork regression or stabilization likely protects ICL repair intermediates from inappropriate processing or repair.
Despite the presence of a large core complex in the mammalian FA repair pathway, the yeast pathway appears to be substantially stripped down. It remains to be seen whether a core-like complex will be identified in yeast or whether evolutionary divergence was sparked by the need for the large complex in mammals. Furthermore, since the mammalian FA pathway appears to be a master regulator of repair it is surprising that the yeast pathway is secondary to Pso2- or Srs2-mediated events. Despite these differences, the simplified yeast model offers significant advantages for the FA repair field to address fundamental mechanistic questions in the future.
Abbreviations
ICL: Interstrand crosslink; TLS: Translesion synthesis; NER: Nucleotide excision repair; HR: Homologous recombination; PRR: Postreplication repair; FA: Fanconi anemia; ssDNA: Single strand DNA; DSB: Double strand break.
Competing interests
The authors declare no competing interests with the contents of this manuscript.
Authors’ contributions
The manuscript was prepared by D.L.D with editorial and substantive advice from K.M. Both authors read and approved the final manuscript.
Contributor Information
Danielle L Daee, Email: danidaee@gmail.com.
Kyungjae Myung, Email: kmyung@mail.nih.gov.
Acknowledgements
We thank members in Myung laboratory for helpful discussions and comments on the manuscript; and K.M. especially thanks E. Cho. This research was supported by the Intramural Research Programs of the National Human Genome Research Institute to KM. We apologize to researchers whose studies we could not discuss or cite due to space limitations.
References
- Geiduschek EP. “Reversible” DNA. Proc Natl Acad Sci USA. 1961;47:950–955. doi: 10.1073/pnas.47.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes P, Lawley PD. The reaction of mono- and di-functional alkylating agents with nucleic acids. Biochem J. 1961;80:496–503. doi: 10.1042/bj0800496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Hopkins PB. DNA Interstrand Cross-Linking by Formaldehyde: Nucleotide Sequence Preference and Covalent Structure of the Predominant Cross-link Formed in Synthetic Oligonucleotides. J Am Chem Soc. 1993;115:9402–9408. doi: 10.1021/ja00074a005. [DOI] [Google Scholar]
- Roberts JJ, Friedlos F. The frequency of interstrand cross-links in DNA following reaction of cis-diamminedichloroplatinum(II) with cells in culture or DNA in vitro: stability of DNA cross-links and their repair. Chem Biol Interact. 1982;39:181–189. doi: 10.1016/0009-2797(82)90120-X. [DOI] [PubMed] [Google Scholar]
- Magana-Schwencke N, Henriques JA, Chanet R, Moustacchi E. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc Natl Acad Sci USA. 1982;79:1722–1726. doi: 10.1073/pnas.79.6.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley PD, Phillips DH. DNA adducts from chemotherapeutic agents. Mutat Res. 1996;355:13–40. doi: 10.1016/0027-5107(96)00020-6. [DOI] [PubMed] [Google Scholar]
- McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483–490. doi: 10.1016/S1470-2045(01)00454-5. [DOI] [PubMed] [Google Scholar]
- Rahman A, Shahabuddin, Hadi SM. Formation of strand breaks and interstrand cross-links in DNA by methylglyoxal. J Biochem Toxicol. 1990;5:161–166. doi: 10.1002/jbt.2570050305. [DOI] [PubMed] [Google Scholar]
- Summerfield FW, Tappel AL. Detection and measurement by high-performance liquid chromatography of malondialdehyde crosslinks in DNA. Anal Biochem. 1984;143:265–271. doi: 10.1016/0003-2697(84)90662-6. [DOI] [PubMed] [Google Scholar]
- Summerfield FW, Tappel AL. Cross-linking of DNA in liver and testes of rats fed 1,3-propanediol. Chem Biol Interact. 1984;50:87–96. doi: 10.1016/0009-2797(84)90134-0. [DOI] [PubMed] [Google Scholar]
- McHugh PJ, Ward TA, Chovanec M. A prototypical Fanconi anemia pathway in lower eukaryotes? Cell Cycle. 2012;11:3739–3744. doi: 10.4161/cc.21727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage C, de Padula M, de Alencar TA, da Fonseca Goncalves SR, da Silva Vidal L, Cabral-Neto J, Leitao AC. New insights on how nucleotide excision repair could remove DNA adducts induced by chemotherapeutic agents and psoralens plus UV-A (PUVA) in Escherichia coli cells. Mutat Res. 2003;544:143–157. doi: 10.1016/j.mrrev.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Muniandy PA, Liu J, Majumdar A, Liu ST, Seidman MM. DNA interstrand crosslink repair in mammalian cells: step by step. Crit Rev Biochem Mol Biol. 2010;45:23–49. doi: 10.3109/10409230903501819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace-Aime G, Couve S, Khassenov B, Rosselli F, Saparbaev MK. The Fanconi anemia pathway promotes DNA glycosylase-dependent excision of interstrand DNA crosslinks. Environ Mol Mutagen. 2010;51:508–519. doi: 10.1002/em.20548. [DOI] [PubMed] [Google Scholar]
- McVey M. Strategies for DNA interstrand crosslink repair: insights from worms, flies, frogs, and slime molds. Environ Mol Mutagen. 2010;51:646–658. doi: 10.1002/em.20551. [DOI] [PubMed] [Google Scholar]
- Vasquez KM. Targeting and processing of site-specific DNA interstrand crosslinks. Environ Mol Mutagen. 2010;51:527–539. doi: 10.1002/em.20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Li L. Mutagenic repair of DNA interstrand crosslinks. Environ Mol Mutagen. 2010;51:493–499. doi: 10.1002/em.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD. Mammalian nucleotide excision repair proteins and interstrand crosslink repair. Environ Mol Mutagen. 2010;51:520–526. doi: 10.1002/em.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legerski RJ. Repair of DNA interstrand cross-links during S phase of the mammalian cell cycle. Environ Mol Mutagen. 2010;51:540–551. doi: 10.1002/em.20566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TV, Scharer OD. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51:552–566. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- Rahn JJ, Adair GM, Nairn RS. Multiple roles of ERCC1-XPF in mammalian interstrand crosslink repair. Environ Mol Mutagen. 2010;51:567–581. doi: 10.1002/em.20583. [DOI] [PubMed] [Google Scholar]
- Hinz JM. Role of homologous recombination in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51:582–603. doi: 10.1002/em.20577. [DOI] [PubMed] [Google Scholar]
- Hlavin EM, Smeaton MB, Miller PS. Initiation of DNA interstrand cross-link repair in mammalian cells. Environ Mol Mutagen. 2010;51:604–624. doi: 10.1002/em.20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Davies AA, Ulrich HD, McHugh PJ. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. EMBO J. 2006;25:1285–1294. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann KF, Ward AM, Matkovic ME, Folias AE, Moses RE. S. cerevisiae has three pathways for DNA interstrand crosslink repair. Mutat Res. 2001;487:73–83. doi: 10.1016/S0921-8777(01)00106-9. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Moshous D, Mornon JP, de Villartay JP. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20:7980–7990. doi: 10.1128/MCB.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/S0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- McHugh PJ, Sones WR, Hartley JA. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:3425–3433. doi: 10.1128/MCB.20.10.3425-3433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilborn F, Brendel M. Formation and stability of interstrand cross-links induced by cis- and trans-diamminedichloroplatinum (II) in the DNA of Saccharomyces cerevisiae strains differing in repair capacity. Curr Genet. 1989;16:331–338. doi: 10.1007/BF00340711. [DOI] [PubMed] [Google Scholar]
- Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE. et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Levitus M, Xue Y, Medhurst AL, Zwaan M, Ling C, Rooimans MA, Bier P, Hoatlin M, Pals G. et al. X-linked inheritance of Fanconi anemia complementation group B. Nat Genet. 2004;36:1219–1224. doi: 10.1038/ng1458. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG. et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Delannoy M, Ling C, Daee D, Osman F, Muniandy PA, Shen X, Oostra AB, Du H, Steltenpool J. et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y. et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims AE, Spiteri E, Sims RJ 3rd, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H. et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- Gerring SL, Spencer F, Hieter P. The CHL 1 (CTF 1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 1990;9:4347–4358. doi: 10.1002/j.1460-2075.1990.tb07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Suhasini AN, Brosh RM Jr. Welcome the family of FANCJ-like helicases to the block of genome stability maintenance proteins. Cell Mol Life Sci. 2009;66:1209–1222. doi: 10.1007/s00018-008-8580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, Oostra AB, Eirich K, Korthof ET, Nieuwint AW. et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, Ira G. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian KD, Schuster T, Hegemann JH, Becher D, Feldmann H, Guldener U, Gotz R, Hansen M, Hollenberg CP, Jansen G. et al. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol Gen Genet. 1999;262:683–702. doi: 10.1007/PL00013817. [DOI] [PubMed] [Google Scholar]
- Schurer KA, Rudolph C, Ulrich HD, Kramer W. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from Homologous recombination, but not from postreplicative repair. Genetics. 2004;166:1673–1686. doi: 10.1534/genetics.166.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Krejci L, Van Komen S, Anke Schurer K, Kramer W, Sung P. Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J Biol Chem. 2005;280:7854–7860. doi: 10.1074/jbc.M413898200. [DOI] [PubMed] [Google Scholar]
- Ansbach AB, Noguchi C, Klansek IW, Heidlebaugh M, Nakamura TM, Noguchi E. RFCCtf18 and the Swi1-Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe. Mol Biol Cell. 2008;19:595–607. doi: 10.1091/mbc.E07-06-0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laha S, Das SP, Hajra S, Sau S, Sinha P. The budding yeast protein Chl1p is required to preserve genome integrity upon DNA damage in S-phase. Nucleic Acids Res. 2006;34:5880–5891. doi: 10.1093/nar/gkl749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara H, Ui A, Lai MS, Enomoto T, Seki M. Chl1 and Ctf4 are required for damage-induced recombinations. Biochem Biophys Res Commun. 2007;354:222–226. doi: 10.1016/j.bbrc.2006.12.185. [DOI] [PubMed] [Google Scholar]
- Fricke WM, Brill SJ. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daee DL, Ferrari E, Longerich S, Zheng XF, Xue X, Branzei D, Sung P, Myung K. Rad5-dependent DNA repair functions of the Saccharomyces cerevisiae FANCM homolog Mph1. J Biol Chem. 2012;287:26563–26575. doi: 10.1074/jbc.M112.369918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T, Dudášová Z, Sarkar S, Bhide M, Vlasáková D, Chovanec M, McHugh PJ. Components of a Fanconi-like pathway control Pso2-independent DNA interstrand crosslink repair in yeast. PLoS Genet. 2012. in press. [DOI] [PMC free article] [PubMed]
- Chen YH, Choi K, Szakal B, Arenz J, Duan X, Ye H, Branzei D, Zhao X. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc Natl Acad Sci USA. 2009;106:21252–21257. doi: 10.1073/pnas.0908258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Kimata Y, Nomaguchi K, Watanabe K, Kohno K. Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5-SMC6 complex involved in DNA repair. J Biol Chem. 2002;277:21585–21591. doi: 10.1074/jbc.M201523200. [DOI] [PubMed] [Google Scholar]
- Skibbens RV. Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics. 2004;166:33–42. doi: 10.1534/genetics.166.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbantenkhu M, Wang X, Nardozzi JD, Wilkens S, Hoffman E, Patel A, Cosgrove MS, Chen XJ. Mgm101 is a Rad52-related protein required for mitochondrial DNA recombination. J Biol Chem. 2011;286:42360–42370. doi: 10.1074/jbc.M111.307512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehoczky P, McHugh PJ, Chovanec M. DNA interstrand cross-link repair in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2007;31:109–133. doi: 10.1111/j.1574-6976.2006.00046.x. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Friedl AA, Liefshitz B, Steinlauf R, Kupiec M. Deletion of the SRS2 gene suppresses elevated recombination and DNA damage sensitivity in rad5 and rad18 mutants of Saccharomyces cerevisiae. Mutat Res. 2001;486:137–146. doi: 10.1016/S0921-8777(01)00086-6. [DOI] [PubMed] [Google Scholar]
- Williams SA, Wilson JB, Clark AP, Mitson-Salazar A, Tomashevski A, Ananth S, Glazer PM, Semmes OJ, Bale AE, Jones NJ, Kupfer GM. Functional and physical interaction between the mismatch repair and FA-BRCA pathways. Hum Mol Genet. 2011;20:4395–4410. doi: 10.1093/hmg/ddr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Trepat M, Boyse J, Richard P, Papadopoulo D, Moustacchi E. Frequencies of HPRT- lymphocytes and glycophorin A variants erythrocytes in Fanconi anemia patients, their parents and control donors. Mutat Res. 1993;289:115–126. doi: 10.1016/0027-5107(93)90137-5. [DOI] [PubMed] [Google Scholar]
- Evdokimova VN, McLoughlin RK, Wenger SL, Grant SG. Use of the glycophorin A somatic mutation assay for rapid, unambiguous identification of Fanconi anemia homozygotes regardless of GPA genotype. Am J Med Genet A. 2005;135:59–65. doi: 10.1002/ajmg.a.30687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Blackford AN, Schwab RA, Nieminuszczy J, Deans AJ, West SC, Niedzwiedz W. The DNA translocase activity of FANCM protects stalled replication forks. Hum Mol Genet. 2012;21:2005–2016. doi: 10.1093/hmg/dds013. [DOI] [PubMed] [Google Scholar]