Abstract
Mutations causing metaphyseal chondrodysplasia type Schmid (MCDS) (e.g., Col10a1p.N617K) induce the pathology by a mechanism involving increased endoplasmic reticulum (ER) stress triggering an unfolded protein response (UPR) in hypertrophic chondrocytes (Rajpar et al. 2009). Here we correlate the expression of mutant protein with the onset of the UPR and disease pathology (hypertrophic zone [HZ] expansion) in MCDS and ColXTg cog mouse lines from E14.5 to E17.5. Embryos homozygous for the Col10a1p.N617K mutation displayed a delayed secretion of mutant collagen X accompanied by a UPR at E14.5, delayed ossification of the primary center at E15.5, and an expanded HZ at E17.5. Heterozygote embryos expressed mutant collagen X from E14.5 but exhibited no evidence of a UPR or an HZ expansion until after E17.5. Embryos positive for the ER stress-inducing ColXTg cog allele expressed Tgcog at E14.5, but the onset of the UPR was not apparent until E15.5 in homozygous and E17.5 in hemizygous embryos. Only homozygous embryos exhibited an HZ expansion at E17.5. The differential onset of the UPR and pathology, dependent on mutation type and gene dosage, indicates that hypertrophic chondrocytes have a latent capacity to deal with ER stress, which must be exceeded to trigger the UPR and HZ expansion.
Keywords: endoplasmic reticulum stress, unfolded protein response, BiP (Grp78), collagen X, chondrodysplasia, cartilage, growth plate, hypertrophic chondrocyte
Long bones are formed by the process of endochondral ossification (Karsenty 2003; Kronenberg 2003; Provot and Schipani 2005, 2007; Mackie et al. 2008). Chondrocytes first form a cartilaginous template called the anlagen, and following vascular invasion and formation of the primary center of ossification, the chondrocytes become organized into highly structured growth plates. Within the growth plate, chondrocytes proliferate, align into columns, and adopt a flattened appearance. Next, these chondrocytes exit the cell cycle and undergo hypertrophy. The hypertrophic chondrocytes can be distinguished from other chondrocytes by their increased cell size and expression of collagen X—the only specific molecular marker of hypertrophy (Schmid and Linsenmayer 1985). During terminal differentiation, the collagen X-rich matrix becomes mineralized, and the hypertrophic chondrocytes undergo apoptosis as the mineralized base of the growth plate is invaded by blood vessels. The vascular endothelial growth factor (VEGF)-driven process of vascular invasion introduces osteoclasts that degrade the mineralized extracellular matrix (ECM) and osteoblast precursors that differentiate and replace the eroded collagen II- and collagen X-rich cartilage with collagen I-rich bone.
Differentiation across the growth plate is governed by a complex cascade of growth factors and cytokines (Karsenty 2008; Karsenty et al. 2009). Longitudinal bone growth is dependent on the strict temporal regulation and control of chondrocyte differentiation, proliferation, hypertrophy, apoptosis, and vascular invasion within the growth plate. Disturbances to this coordinated process cause a large number of skeletal growth disorders.
More than 400 skeletal disorders have been classified, and several are the direct result of mutations in cartilage ECM genes critical to endochondral ossification (Newman and Wallis 2003; Warman et al. 2011). Although individually rare, collectively they are relatively common and cause a significant impact on the quality of life for patients suffering from skeletal abnormalities. For instance, mutations in collagen X cause metaphyseal chondrodysplasia type Schmid (MCDS), a relatively mild autosomal dominant skeletal dysplasia (Warman et al. 1993; Wallis et al. 1994). Collagen X is a homotrimer of three α1 (X) chains, each containing a 463–amino acid Gly-X-Y collagenous domain (COL1) flanked by a 38-residue N-terminal non-collagenous domain (NC2) and a 161-residue C-terminal non-collagenous domain (NC1). The majority of mutations found in collagen X cluster within the C-terminal NC1 domain (Bateman et al. 2005).
Many experimental approaches have been adopted to try to elucidate the disease mechanisms responsible for the development of MCDS, and recent evidence has converged to reveal involvement of endoplasmic reticulum (ER) stress and the resulting unfolded protein response (UPR) (Mclaughlin et al. 1999; Wilson et al. 2002; Wilson et al. 2005; Ho et al. 2007; Tsang et al. 2007; Rajpar et al. 2009).
Briefly, the UPR is triggered by increasing levels of ER stress, which ultimately impairs ER function (for reviews, see Malhotra and Kaufman 2007; Ron and Walter 2007; Lin et al. 2008; Boot-Handford and Briggs 2010). ER stress can be caused by many different factors, physiological and pathological, and the UPR is a protective response evolved to restore homeostasis in terms of protein folding in the ER. The UPR is mediated through three ER resident transmembrane proteins that act as ER stress sensors: the basic leucine zipper pancreatic ER kinase (PKR)-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1 (IRE1). Under stressed conditions (i.e., increasing concentrations of unfolded proteins in the ER lumen), activation of signaling cascades results in attenuation of protein translation reducing the load of nascent proteins arriving at the ER, upregulation of genes involved in ER-associated degradation (ERAD), and upregulation of chaperones, such as BiP (Grp 78). The upregulation of BiP has been used extensively as a key marker of the UPR to increases in ER stress (Lee 2005; Samali et al. 2010). The UPR involves a set of protective measures aimed at alleviating the increased ER stress. However, if the UPR fails to restore protein homeostasis, apoptosis can be triggered (Tabas and Ron 2011).
Studies on transgenic mouse models of MCDS have produced significant advances in our understanding of the disease pathogenesis. In a transgenic mouse line expressing an MCDS-causing 13-base pair (bp) deletion in Col10a1 (13del), the mutant collagen X protein was intracellularly retained and an ER stress response was elicited within hypertrophic chondrocytes, which subsequently led to altered chondrocyte differentiation (Tsang et al. 2007). Similar results were also observed in a knock-in Col10a1 p.N617K MCDS mouse model (Rajpar et al. 2009) and in another transgenic mouse line bearing the mouse equivalent of a human MCDS p.P620fsX621 mutation (Ho et al. 2007). To functionally characterize the putative role of ER stress in the disease mechanism of MCDS, a transgenic mouse line was generated in which ER stress was targeted to hypertrophic chondrocytes (Rajpar et al. 2009). In this ColXTg cog mouse, expression of an ER stress-inducing protein (cog mutant form of thyroglobulin—Tgcog) was driven by the Col10a1 promoter. Interestingly, the targeted stimulation of ER stress in hypertrophic chondrocytes was sufficient to replicate and closely “phenocopy” the MCDS phenotype independently of any Col10a1 mutations, thereby illustrating the pivotal role played by increases in ER stress in the disease mechanism of MCDS.
In the Col10a1 p.N617K and ColXTg cog mouse models, a disease phenotype (expanded HZ of growth plate) was observed in newborn mice homozygous for the mutant allele, but the onset of the disease phenotype earlier in development has not been explored (Rajpar et al. 2009). The objective of this study was to characterize, correlate, and ultimately compare the onset of mutant gene expression with the onset of the UPR and pathology in Col10a1 p.N617K and ColXTg cog mouse embryos. We demonstrate that the onset of the UPR and tissue pathology did not always correlate with the spatial and temporal expression of the ER stress-inducing mutant protein but was dependent on the nature of the mutant protein and the gene dosage. The results provide new insights into the capacity of mouse hypertrophic chondrocytes to tolerate threshold levels of ER stress that need to be exceeded to induce the UPR and the resulting pathology.
Materials and Methods
Ethics Statement
All mice used in this study were handled and sacrificed in accordance with Home Office regulations.
Generation and Genotyping of Mutant Mice
The generation and genotyping of the Col10a1 p.N617K and ColXTg cog mouse lines have been described previously (Rajpar et al. 2009). Briefly, the Col10a1 p.N617K mouse line was generated via gene targeting technology, and mice carrying the Col10a1 mutation were maintained on an SV129/C57Bl6 background. Genotyping was performed using primers GAT TTA TGG TGA GTT AGA GTC (forward) and GTG AGC ACT TCC TGT CAA GC (reverse) flanking the LoxP site. The ColXTg cog mouse line was generated by pronuclear injection, and mice carrying the ColXTg cog transgene were maintained on an FVB/N/C57Bl6 background. Genotyping was performed using primers GGA CTG TTG TGT GAG TGG (forward) and TTC CAT CTT CAG AGC ACT GG (reverse) between the collagen X promoter and the Tgcog.
Timed matings of heterozygous mice were used to generate embryos at 14.5, 15.5, and 17.5 days of development. Pregnant female mice were sacrificed by carbon dioxide overdose, and embryos were immersed in ice-cold PBS followed by ice-cold fixative.
Histology and Immunohistochemistry
Embryonic samples were fixed overnight in RNase-free ice-cold 4% paraformaldehyde (PFA) or 95% ethanol/5% acetic acid. The samples were dehydrated in a graded series of ethanol solutions, cleared twice in xylene, embedded in paraffin wax, and sectioned (5 µm). For hematoxylin and eosin (H&E) staining, the slides were dewaxed in xylene, rehydrated, and H&E stained using Varistain 24.4 automated stainer (Thermo Shandon, Waltham, MA). Slides were then dehydrated in increasing concentrations of ethanol and then cleared in xylene; and mounted using a xylene-based mounting solution.
Immunohistochemistry was performed on 95% ethanol/5% acetic acid-fixed sections. Antigen unmasking for collagen X immunohistochemistry was carried out in 0.5 mg trypsin (Sigma, St. Louis, MO)/ml of PBS for 4 min at room temperature followed by washes in PBS. Antigen unmasking for Tgcog and BiP immunohistochemistry was carried out in citrate buffer (pH 6.0) heated to >85C for 10 min followed by washes in PBS. Quenching of endogenous peroxidase activity was carried out in 3% (v/v) hydrogen peroxide in PBS for 5 min. Tissue sections were blocked for 1 hr at room temperature and incubated with primary antibody at 4C overnight.
Primary antibodies used were collagen X (polyclonal rabbit anti–collagen X against recombinant mouse NC1 domain) diluted 1/500 in PBS containing 2% goat serum (Rajpar et al. 2009), thyroglobulin, which was detected via its myc-tag using an anti-myc mouse monoclonal (cat. no. 05-724; Millipore, Billerica, MA) diluted 1/300 using a mouse-on-mouse (M.O.M.) immunodetection kit (BMK2202; Vector Laboratories, Burlingame, CA), and BiP, which was detected using an anti-Grp 78 goat polyclonal (SC-1051; Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1/300 in PBS containing 2% horse serum. Secondary antibodies used were biotinylated goat anti-rabbit IgG (E0432; DakoCytomation Ltd., Carpinteria, CA) diluted 1/1000 in 2% goat serum, biotinylated anti-mouse IgG as provided with the M.O.M. immunodetection kit, and biotinylated horse anti-goat IgG (BA 9500; Vector Laboratories) diluted 1/1000 in 2% horse serum.
Sections were then incubated with ABC reagent (PK-6100; Vector Laboratories) for 30 min and developed using the Vector VIP kit (SK-4600; Vector Laboratories). Slides were dehydrated in increasing concentrations of ethanol, then cleared in xylene and mounted using a xylene-based mounting solution. Negative control sections for immunohistochemistry were performed with the appropriate serum minus the primary antibody. Tissue sections that were known to express the protein of interest were included as positive controls.
In Situ Hybridization
DIG-labeled colorimetric in situ was performed as described (Rajpar et al. 2009). Embryonic tissue was fixed in ice-cold 4% PFA and sectioned as described above. The Col10a1 probe was a 900-bp fragment of the 3′ end of the coding sequence; the BiP probe was a 350-bp fragment from I.M.A.G.E clone 6334883. cDNA probes were cloned into pT7T3, linearized, and transcribed using the appropriate restriction enzyme and RNA polymerase, respectively. Negative control sections were performed without the RNA probe. Tissue sections that were known to express the gene of interest were included as positive controls.
Tartrate-Resistant Acid Phosphatase (TRAP) Staining
Osteoclasts were stained using the Acid Phosphatase kit from Sigma-Aldrich (387A). The protocol was performed as per the manufacturer’s instructions on PFA-fixed sections. For each animal, the data from three separate sections, spaced at least 50 µm apart, were averaged. Measurements were analyzed by one-way ANOVA using online software (http://faculty.vassar.edu/lowry/anova1u.html) for statistical significance.
Imaging
Images were captured using the Carl Zeiss Axiovision microscope fitted with an Axiocam color CCD camera and associated Axiovision software (Carl Zeiss, Jena, Germany), then processed and analyzed using Adobe Photoshop (Adobe, San Jose, CA).
HZ width measurements were performed using the Photoshop Ruler Tool on images of known magnification, as described previously (Rajpar et al. 2009). For each animal, the data from three separate sections, spaced at least 50 µm apart, were averaged. The start of the HZ was defined as the point at which the disk-shaped cells of the proliferative zone started to round up and become larger. Measurements were analyzed by one-way ANOVA using online software (http://faculty.vassar.edu/lowry/anova1u.html) for statistical significance.
Results
E14.5 Col10a1 p.N617K Embryos Exhibit Disrupted Expression and Secretion of Collagen X and an Induced UPR
There were no morphological abnormalities in the cartilage anlagen of wild-type (wt/wt), heterozygous (wt/m), and homozygous (m/m) Col10a1 p.N617K embryos at E14.5 (Fig. 1A). Col10a1 mRNA expression was first apparent in hypertrophic chondrocytes at E14.5 in all genotypes (Fig. 1B). In wt/wt and wt/m embryos, hypertrophic chondrocytes in the central region of the HZ were downregulating Col10a1 mRNA levels (Fig. 1B), a sign of terminal hypertrophic differentiation in preparation for vascular invasion and formation of the primary center of ossification. In contrast, m/m embryos exhibited a more evenly distributed but “speckled” pattern of expression of Col10a1 mRNA with no obvious downregulation toward the center of the anlagen (Fig. 1B).
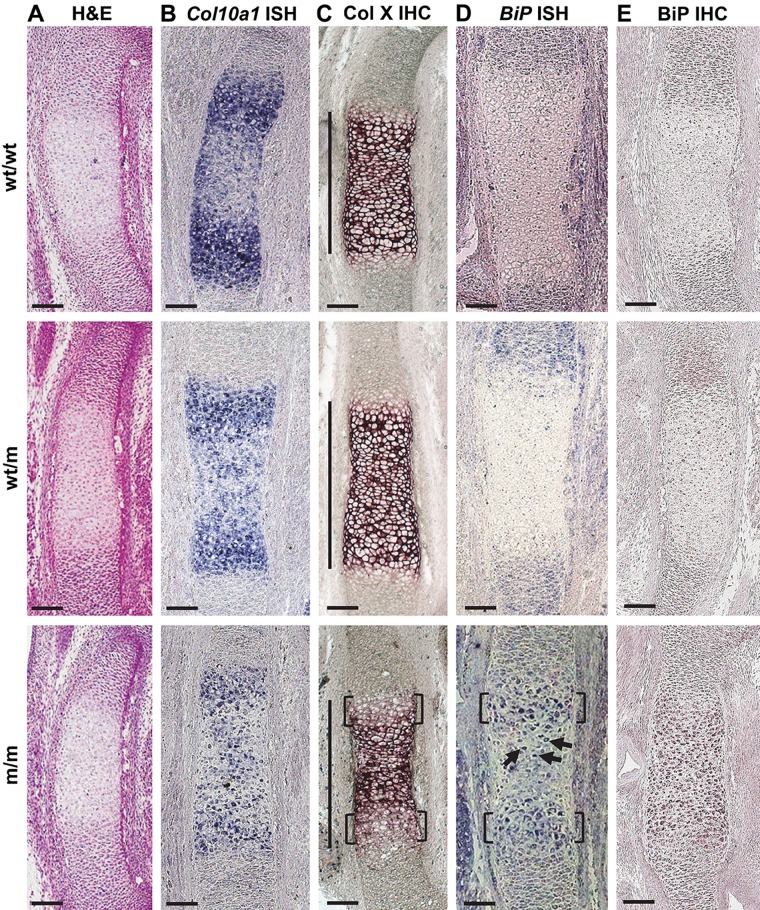
Figure 1.
Histological and in situ characterization of E14.5 Col10a1 p.N617K embryonic growth plates. (A) Hematoxylin and eosin (H&E) staining on sections from the tibial growth plate of wild type (wt/wt), heterozygous (wt/m), and homozygous (m/m) embryos for the Col10a1 p.N617K mutation. (B) In situ hybridization (ISH) for Col10a1 mRNA on sections from the tibial growth plates of wt/wt, wt/m, and m/m embryos. The presence of the transcript is indicated by the dark blue staining. (C) Immunohistochemistry (IHC) for collagen X on sections from the tibial growth plate of wt/wt, wt/m, and m/m embryos. The dark purple staining indicates collagen X localization within the extracellular matrix in wt/wt and wt/m embryos. The brackets indicate delayed collagen X secretion in m/m embryos. The hypertrophic zone (HZ) is indicated by the vertical lines. (D) ISH for BiP mRNA on sections from the tibial growth plates of wt/wt, wt/m, and m/m embryos. The presence of the transcript is indicated by the dark blue staining. BiP mRNA transcript was below the level of detection in wt/wt and wt/m embryos. The brackets indicate BiP mRNA expression in the upper HZ of m/m embryos. The arrows indicate low levels of BiP mRNA in the central region of the HZ of m/m embryos. (E) IHC for BiP on sections from the tibial growth plate of wt/wt, wt/m, and m/m embryos. The dark purple staining represents the accumulation of BiP in the HZ of m/m embryos. Scale bar = 100 µm.
Immunolocalization of collagen X in E14.5 wt/wt and wt/m embryos revealed the expected extracellular staining in the HZ (Fig. 1C). In contrast, E14.5 m/m embryos displayed delayed secretion of collagen X by the most recently formed hypertrophic chondrocytes at the borders of the HZ adjacent to the proliferative zones (Fig. 1C, brackets). There was no detectable intracellular accumulation of mutant collagen X in the m/m hypertrophic chondrocytes, suggesting that the rate of mutant protein synthesis was matched by its rate of secretion and/or intracellular degradation at this stage of development (Fig. 1C).
BiP mRNA expression was below the level of detection in the hypertrophic chondrocytes of E14.5 wt/wt and wt/m embryos despite synthesis of some mutant collagen X by the latter (Fig. 1D). However, increases in BiP expression, indicative of UPR activation, were detected within the most recently differentiated hypertrophic chondrocytes of E14.5 m/m embryos that bordered the proliferative zones (Fig. 1D, brackets). In addition, some hypertrophic chondrocytes in the central region of the HZ were also expressing increased levels of BiP mRNA in the m/m embryos (Fig. 1D, arrows). BiP immunohistochemistry revealed minimal staining in E14.5 wt/wt and wt/m embryos, whereas an accumulation of BiP protein was apparent throughout the HZ of E14.5 m/m embryos (Fig. 1E). Increased BiP expression and accumulation in hypertrophic chondrocytes demonstrated that these cells in the E14.5 m/m embryos were exhibiting a UPR, whereas those expressing half the level of ER stress–inducing mutant collagen X (wt/m) were not.
E15.5 Col10a1 p.N617K Embryos Exhibit Delayed Terminal Differentiation, Delayed Formation of the Primary Center of Ossification, and an Induced UPR
H&E staining of growth plate sections from E15.5 wt/wt and wt/m embryos revealed that vascular invasion and bone formation were apparent in the diaphyses of the developing tibia (Fig. 2A, asterisks). Col10a1 mRNA expression (Fig. 2B) and immunostaining (Fig. 2C) were largely confined to the HZ bordering the bony diaphyses of the wt/wt and wt/m embryos.
Figure 2.
Histological and in situ characterization of E15.5 Col10a1 p.N617K embryonic growth plates. (A) Hematoxylin and eosin (H&E) staining on sections from the tibial growth plate of wild type (wt/wt), heterozygous (wt/m), and homozygous (m/m) embryos for the Col10a1 p.N617K mutation. (B) In situ hybridization (ISH) for Col10a1 mRNA on sections from the tibial growth plates of wt/wt, wt/m, and m/m embryos. The presence of the transcript is indicated by the dark blue staining. (C) Immunohistochemistry (IHC) for collagen X on sections from the tibial growth plate of wt/wt, wt/m, and m/m embryos. The dark purple staining indicates collagen X localization within the extracellular matrix in wt/wt and wt/m embryos. The brackets indicate delayed collagen X secretion in m/m embryos. The hypertrophic zone (HZ) is indicated by the vertical lines. (D) ISH for BiP mRNA on sections from the tibial growth plates of wt/wt, wt/m, and m/m embryos. The presence of the transcript is indicated by the dark blue staining. BiP mRNA transcript was below the level of detection in wt/wt and wt/m embryos. The brackets indicate elevated BiP mRNA expression in the upper HZ of m/m embryos. (E) IHC for BiP on sections from the tibial growth plate of wt/wt, wt/m, and m/m embryos. The dark purple staining represents the accumulation of BiP in the HZ of m/m embryos. *Bone. Scale bar = 100 µm.
In contrast, E15.5 m/m embryos exhibited hypertrophic chondrocytes throughout the diaphyses (Fig. 2A) with no bone formation apparent. m/m embryos displayed Col10a1 mRNA expression not only in the newly differentiated hypertrophic regions at each end of the diaphysis but also in a sporadic pattern throughout the central region of the diaphysis (Fig. 2B). In addition, collagen X immunostaining was apparent throughout the central ECM of the diaphysis. Furthermore, the E15.5 m/m embryos exhibited a similar delayed secretion of mutant collagen X by the newly differentiated hypertrophic chondrocytes at each end of the diaphysis (Fig. 2C, brackets), as shown above in E14.5 m/m embryos. The ossification of the primary center in all wt/m embryos examined exhibited an intermediate phenotype with some evidence of delayed vascular invasion apparent based on the retention of a thin core of collagen X matrix extending through the diaphyses (Fig. 2C).
As described above for E14.5 embryos, BiP mRNA expression was below the level of detection in the hypertrophic chondrocytes of E15.5 wt/wt and wt/m growth plates. In contrast, m/m embryos displayed elevated BiP mRNA expression in the upper HZ (Fig. 2D, brackets), where Col10a1 mRNA was first induced (Fig. 2B) and collagen X secretion delayed (Fig. 2C). Furthermore, BiP immunohistochemistry revealed accumulation of BiP protein throughout the HZ of E15.5 m/m embryos (Fig. 2E), similar to the staining described above for E14.5 m/m embryos (Fig. 1E). Minimal staining was observed in the E15.5 wt/wt or in the wt/m embryos (Fig. 2E).
E17.5 Col10a1 p.N617K Embryos Displayed Characteristic HZ Expansion, Altered Chondrocyte Differentiation, Intracellular Retention of Mutant Collagen X, and an Induced UPR
The HZ of E17.5 m/m tibial growth plates was significantly expanded compared with wt/wt embryos (Fig. 3A, F). E17.5 wt/wt and wt/m embryos had HZ widths of 217 µm and 230 µm, respectively, whereas the m/m embryos were significantly expanded in comparison (377 µm; p<0.01; Fig. 3F).
Figure 3.
Histological and in situ characterization of E17.5 Col10a1 p.N617K embryonic growth plates. (A) Hematoxylin and eosin (H&E) staining on sections from the tibial growth plate of wild type (wt/wt), heterozygous (wt/m), and homozygous (m/m) embryos for the Col10a1 p.N617K mutation. (B) In situ hybridization (ISH) for Col10a1 mRNA on sections from the tibial growth plates of wt/wt, wt/m, and m/m embryos. The presence of the transcript is indicated by the dark blue staining. (C) Immunohistochemistry (IHC) for collagen X on sections from the tibial growth plate of wt/wt, wt/m, and m/m embryos. The dark purple staining indicates collagen X localization within the extracellular matrix in wt/wt and wt/m embryos. In the m/m embryos, the secretion of mutant collagen X is severely disrupted with signs of intracellular accumulation of mutant collagen X (indicated by the arrows). The hypertrophic zone (HZ) is indicated by the vertical lines. (D) ISH for BiP mRNA on sections from the tibial growth plates of wt/wt, wt/m, and m/m embryos. The presence of the transcript is indicated by the dark blue staining. Low levels of BiP mRNA transcript could be seen in wt/wt and wt/m embryos (indicated by the arrows). In comparison, considerably high levels of BiP mRNA expression were detected in the HZ of m/m embryos. (E) IHC for BiP on sections from the tibial growth plate of wt/wt, wt/m, and m/m embryos. The dark purple staining represents the accumulation of BiP in the upper HZ of wt/m embryos and throughout the HZ of m/m embryos. Scale bar = 100 µm. (F) HZ widths from the tibial growth plate of wt/wt, wt/m, and m/m embryos. *p<0.01 versus control (wt/wt).
Col10a1 mRNA expression was induced in the newly differentiated hypertrophic chondrocytes in all embryos (Fig. 3B). However, in both wt/m and m/m embryos, collagen X expression was more sporadic in the lower part of the HZ (Fig. 3B).
As with previous developmental time points, immunolocalization of collagen X revealed the expected extracellular staining within the HZ of E17.5 wt/wt and wt/m embryos (Fig. 3C). In comparison, immunohistochemistry for collagen X revealed aberrant staining in the growth plate of E17.5 m/m embryos (Fig. 3C). As with E14.5 and E15.5 m/m embryos (Figs. 1C and 2C, respectively), E17.5 m/m embryos displayed delayed secretion of mutant collagen X by the most recently formed hypertrophic chondrocytes within the upper region of the HZ (Fig. 3C). However, in contrast to results seen at earlier time points, there was also evidence of intracellular accumulation of mutant collagen X within these cells in the upper region of the HZ of the m/m embryos (Fig. 3C, arrows).
Low levels of BiP mRNA transcript were detected in some hypertrophic chondrocytes of E17.5 wt/wt and wt/m embryos (Fig. 3D). In comparison, m/m embryos exhibited considerably elevated BiP mRNA expression in the upper region of the HZ (Fig. 3D). Subsequently, BiP mRNA expression was downregulated in the lower portion of the m/m HZ. BiP immunohistochemistry revealed no apparent BiP accumulation in the HZ of E17.5 wt/wt embryos (Fig. 3E). However, an accumulation of BiP protein was observed in the upper portion of the HZ of E17.5 wt/m embryos and throughout the HZ of E17.5 m/m embryos (Fig. 3E).
Delayed Recruitment of Osteoclasts to the Vascular Invasion Front in Col10a1 p.N617K Embryos
TRAP staining for osteoclasts revealed no major delays in the recruitment of osteoclasts in E14.5 wt/wt, wt/m, or m/m embryos (Fig. 4A). In contrast, recruitment of osteoclasts to the HZ of E15.5 m/m embryos was delayed compared with wt/wt embryos in which osteoclasts had clearly penetrated the wild-type HZ (Fig. 4A). E15.5 wt/m embryos displayed an intermediate phenotype. By E17.5, TRAP staining revealed significantly fewer osteoclasts located at the vascular invasion front (VIF) in m/m embryos compared with wt/wt (p<0.01; Fig. 4A, B).
Figure 4.
Tartrate-resistant acid phosphatase (TRAP) staining and analysis of E14.5, E15.5, and E17.5 Col10a1 p.N617K embryonic growth plates. (A) TRAP staining on sections from the tibial growth plate of wild type (wt/wt), heterozygous (wt/m), and homozygous (m/m) embryos for the Col10a1 p.N617K mutation at E14.5, E15.5, and E17.5. The red/brown color represents positively stained osteoclast cells (indicated by the arrows). (B) The number of positive cells per mm of the vascular invasion front (VIF) in E17.5 wt/wt, wt/m, and m/m embryos. MCDS, metaphyseal chondrodysplasia type Schmid. *p<0.01 versus control (wt/wt). Scale bar = 100 µm.
Expression of ER Stress-Inducing Tgcog Protein was Detected in E14.5 ColXTgcog Embryos with No Induction of the UPR
H&E staining revealed no morphological abnormalities in the embryonic growth plates between embryos hemizygous (+/c), homozygous (c/c), or lacking (+/+) the ColXTg cog transgene at E14.5 (Fig. 5A). Intracellular retention of the ER stress-inducing Tgcog protein was detected in E14.5 +/c and c/c embryos, and expression was limited to hypertrophic chondrocytes (Fig. 5B, arrows). Tgcog expression was not detected in +/+ controls (Fig. 5B). Tgcog expression did not alter the localization of collagen X in +/c and c/c compared with +/+ embryos (Fig. 5C). In addition, expression of the ER stress-inducing Tgcog protein did not result in increased expression of BiP mRNA in either +/c or c/c embryos at E14.5 (Fig. 5D). In addition, immunohistochemical analyses revealed no evidence for the accumulation of BiP protein in hypertrophic chondrocytes of E14.5 +/+, +/c, and c/c embryos (Fig. 5E).
Figure 5.
Histological and in situ characterization of E14.5 ColXTg cog embryonic growth plates. (A) Hematoxylin and eosin (H&E) staining on sections from the tibial growth plate of wild type (+/+), hemizygous (+/c), and homozygous (c/c) embryos for the ColXTg cog transgene. (B) Immunohistochemistry (IHC) for Tgcog using a myc antibody on sections from the embryonic tibial growth plate of +/+, +/c, and c/c embryos. The dark purple staining indicates the intracellular accumulation of Tgcog protein in the hypertrophic zone (HZ) of +/c and c/c embryos (indicated by the arrows). (C) IHC for collagen X on sections from the tibial growth plates of +/+, +/c, and c/c. The dark purple staining indicates collagen X localization within the extracellular matrix. The HZ is indicated by the vertical lines. (D) In situ hybridization (ISH) for BiP mRNA on sections from the tibial growth plate of +/+, +/c, and c/c embryos. Levels of BiP mRNA expression were below the level of detection at E14.5 in all embryos. (E) IHC for BiP on sections from the tibial growth plate of +/+, +/c, and c/c embryos. Levels of accumulated BiP protein were below the level of detection at E14.5 in all embryos. Scale bar = 100 µm.
E15.5 ColXTgcog Embryos Exhibit Onset of UPR Activation
No morphological abnormalities were apparent in the growth plates of +/+, +/c, and c/c embryos at E15.5 (Fig. 6A). Tgcog expression was apparent in E15.5 +/c and c/c embryos, and again, expression was limited to hypertrophic chondrocytes, with no expression in the +/+ controls (Fig. 6B, arrows). The onset of vascular invasion was apparent as erosion of the mineralized collagen X-rich cartilage was observed at the edges of the developing anlagen in all embryos (Fig. 6C, asterisks). The induction of Tgcog expression did not alter the localization of endogenous collagen X in +/c or c/c embryos (Fig. 6C).
Figure 6.
Histological and in situ characterization of E15.5 ColXTg cog embryonic growth plates. (A) Hematoxylin and eosin (H&E) staining on sections from the tibial growth plate of wild type (+/+), hemizygous (+/c), and homozygous (c/c) embryos for the ColXTg cog transgene. (B) Immunohistochemistry (IHC) for Tgcog using a myc antibody on sections from the tibial growth plate of +/+, +/c, and c/c embryos. The dark purple staining indicates the intracellular accumulation of Tgcog protein in the hypertrophic zone (HZ) of +/c and c/c embryos (indicated by the arrows). (C) IHC for collagen X on sections from the tibial growth plates of +/+, +/c, and c/c embryos. The dark purple staining indicates collagen X localization within the extracellular matrix in all embryos. The HZ is indicated by the vertical lines. (D) In situ hybridization (ISH) for BiP mRNA on sections from the tibial growth plate of +/+, +/c, and c/c embryos. Levels of BiP mRNA transcript were below the level of detection in +/+ and +/c embryos. In comparison, BiP mRNA expression was upregulated in the newly differentiating hypertrophic chondrocytes of c/c embryos (indicated by the brackets). (E) IHC for BiP on sections from the tibial growth plate of +/+, +/c, and c/c embryos. The dark purple staining represents the accumulation of BiP in the HZ of c/c embryos. *Bone. Scale bar = 100 µm.
BiP mRNA expression was below the level of detection in the hypertrophic chondrocytes of +/+ and +/c embryos (Fig. 6D). However, there was a clear induction of BiP mRNA in newly differentiating hypertrophic chondrocytes at the borders of the proliferative zones of E15.5 c/c embryos (Fig. 6D, brackets). There was no BiP accumulation in the HZ of E15.5 +/+ embryos, but BiP accumulation was apparent in hypertrophic chondrocytes of c/c and, to a lesser extent, +/c embryos (Fig. 6E). Increased BiP expression and accumulation in hypertrophic chondrocytes demonstrate that these cells in the E15.5 c/c (and +/c) embryos were exhibiting signs of ER stress-induced UPR that was not evident in Tgcog expressing E14.5 embryos (Fig. 5).
E17.5 ColXTgcog Embryos Exhibit a Characteristic HZ Expansion and an Induced UPR
The HZ of tibial growth plates was significantly expanded in E17.5 c/c embryos compared with +/+ embryos (Fig. 7A, F). E17.5 +/+ and +/c embryos had an HZ width of 221 µm and 235 µm, respectively, whereas the c/c embryos were significantly expanded in comparison to +/+ (265 µm; p<0.05; Fig. 7F). Tgcog expression was detected in E17.5 +/c and c/c embryos, and expression was limited to hypertrophic chondrocytes (Fig. 7B) and, as with the earlier time points, the induction of Tgcog expression did not alter the localization of collagen X (Fig. 7C).
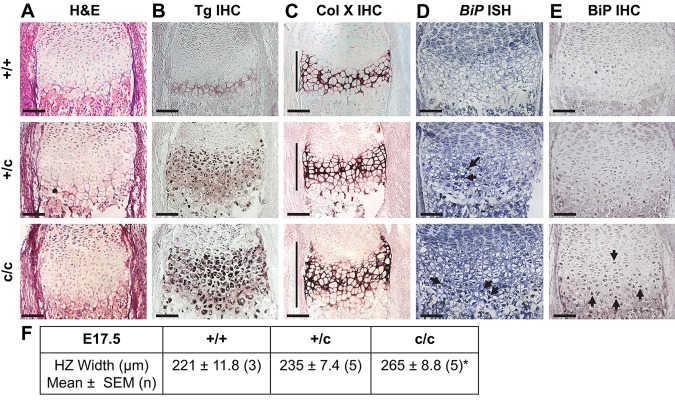
Figure 7.
Histological and in situ characterization of E17.5 ColXTg cog embryonic growth plates. (A) Hematoxylin and eosin (H&E) staining on sections from the tibial growth plate of wild type (+/+), hemizygous (+/c), and homozygous (c/c) embryos for the ColXTg cog transgene. (B) Immunohistochemistry (IHC) for Tgcog using a myc antibody on sections from the tibial growth plate of +/+, +/c, and c/c embryos. The dark purple staining indicates the intracellular accumulation of Tgcog protein in the hypertrophic zone (HZ) of +/c and c/c embryos. (C) IHC for collagen X on sections from the tibial growth plate of +/+, +/c, and c/c embryos. The dark purple staining indicates collagen X localization within the extracellular matrix in all embryos. The HZ is indicated by the vertical lines. (D) In situ hybridization (ISH) for BiP mRNA on sections from the tibial growth plate of +/+, +/c, and c/c embryos. Levels of BiP mRNA transcript were below the level of detection in +/+ embryos. In comparison, BiP mRNA expression was upregulated in the HZ of +/c and c/c embryos (indicated by the arrows). (E) IHC for BiP on sections from the tibial growth plate of +/+, +/c, and c/c embryos. The dark purple staining represents the accumulation of BiP in the HZ of +/c and c/c embryos. Scale bar = 100 µm. (F) HZ widths from the tibial growth plate of +/+, +/c, and c/c embryos. *p<0.05 versus control (+/+). The arrows indicate the accumulation of BiP in hypertrophic chondrocytes of c/c embryos.
BiP mRNA levels were below the level of detection in +/+ embryos. However, elevated BiP mRNA expression was detected in the lower portion of the HZ of +/c and c/c embryos (Fig. 7D, arrows). BiP protein accumulation was not apparent in the HZ of E17.5 +/+ embryos, whereas accumulation of BiP was apparent in c/c and, to a lesser extent, in the +/c embryos (Fig. 7E).
Delayed Recruitment of Osteoclasts to the VIF in ColXTgcog Embryos
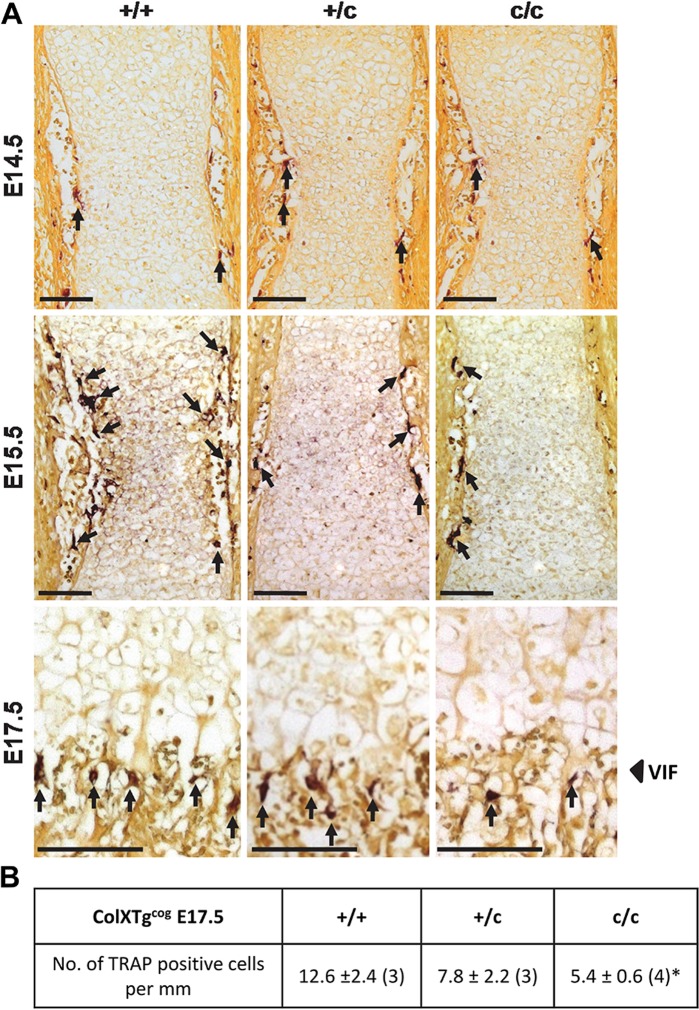
TRAP staining revealed no major changes in osteoclast recruitment to the developing tibia in E14.5 +/+, +/c, or c/c embryos, whereas at E15.5, c/c embryos exhibited a slight decrease in recruitment compared with +/+ controls (Fig. 8A). By E17.5, osteoclast recruitment to the VIF in c/c embryos was significantly delayed compared with +/+ embryos (p<0.05; Fig. 8A, B).
Figure 8.
Tartrate-resistant acid phosphatase (TRAP) staining and analysis of E14.5, E15.5, and E17.5 ColXTg cog embryonic growth plates. (A) TRAP staining on sections from the tibial growth plate of wild type (+/+), hemizygous (+/c), and homozygous (c/c) embryos for ColXTg cog at E14.5, E15.5, and E17.5. The red/brown color represents positively stained osteoclast cells (indicated by the arrows). (B) The number of positive cells per mm of the vascular invasion front (VIF) in E17.5 +/+, +/c, and c/c embryos. *p<0.05 versus control (+/+). Scale bar = 100 µm.
Discussion
The postnatal growth plate phenotypes of the Col10a1 p.N617K and ColXTg cog mouse lines were characterized previously (Rajpar et al. 2009). Rajpar et al. (2009) and other studies (Ho et al. 2007; Tsang et al. 2007) revealed that the hypertrophic chondrocytes expressing mutant forms of collagen X experienced a profound UPR, a disrupted differentiation pattern including decreased expression of VEGF, and a decreased recruitment of osteoclasts to the VIF, causing an expansion of the hypertrophic zone. Changes in proliferation or apoptotic rates in affected growth plates played no part in the pathological expansion of the hypertrophic zone in any of the lines examined (Tsang et al. 2007; Rajpar et al. 2009; Cameron et al. 2011). Furthermore, the key pathological features of MCDS were replicated in vivo by directly triggering a UPR in hypertrophic chondrocytes through the targeted expression of Tgcog, demonstrating the central role of increased ER stress in the disease mechanism (Rajpar et al. 2009). The current investigation was undertaken to characterize, correlate, and compare the onset of mutant gene expression, UPR, and pathology in the Col10a1 p.N617K and ColXTg cog mouse lines during embryonic development.
Collagen X expression was not detected at E13.5 (data not shown) but was apparent by E14.5 (Fig. 1B,C), in agreement with previously published data (Tsang et al. 2007). In the Col10a1 p.N617K m/m embryos, the expression of mutant collagen X at E14.5 had immediate effects in terms of altered spatial and temporal expression of mutant Col10a1 mRNA and the induction of BiP mRNA (Fig. 1). The “speckled” appearance of mutant Col10a1 expression accompanied by a marked induction of BiP mRNA was apparent in E15.5 and E17.5 m/m embryos (Figs. 2 and 3 respectively), as previously described in 3-week-old m/m mice, and is indicative of a robust UPR and a disrupted pattern of differentiation (Tsang et al. 2007; Rajpar et al. 2009). We therefore examined when the first signs of disruption of ossification became apparent. The central diaphyses had ossified by E15.5 in wt/wt embryos, whereas in the m/m equivalents, the diaphyses remained hypertrophic cartilage based on collagen X mRNA expression and collagen X immunolocalization, with no signs of vascular invasion and primary center ossification (Fig. 2). Indeed, this developmental delay in the E15.5 m/m embryos correlated with a lack of osteoclast invasion (Fig. 4A), the invasion being required to form the primary center of ossification. It is of particular note that throughout this period of development, the wt/m embryos, which express half the level of the ER stress–inducing mutant collagen X compared with m/m littermates, showed no signs of either a UPR or disrupted differentiation in terms of collagen X and BiP mRNA expression, ossification of the diaphysis, and osteoclast recruitment (Figs. 1 –4). Interestingly, the 13del transgenic model of MCDS has a more severe phenotype than the models described above but does not have a delay in formation of the primary center because the transgene’s relatively short Col10a1 promoter only becomes active around E15, and so induction of the UPR is delayed until after primary center formation (Tsang et al. 2007).
In the transgenic ColXTg cog mouse, collagen X and the Tgcog expression were both evident at E14.5 in the HZ of the developing growth plate (Fig. 5B,C), demonstrating that the ColXTg cog transgene, with its relatively long Col10a1 promoter, recapitulated the expression of the endogenous Col10a1 gene. However, the level of ER stress–inducing Tgcog expression at E14.5 was not sufficient to provoke a UPR in terms of the upregulation of BiP expression (Fig. 5D) in contrast to the m/m Col10a1 p.N617K embryos described above. However, by E15.5, more than 24 hr after Tg cog expression was instigated, BiP expression was upregulated in newly differentiated hypertrophic chondrocytes but only in the mice that were homozygous for the transgene (Fig. 6D). By E17.5, the HZ had expanded significantly in c/c growth plates and the +/c hypertrophic chondrocytes were beginning to upregulate BiP expression, although their growth plates were not significantly expanded (Fig. 7). The delay in onset of the UPR in c/c embryos meant that the primary ossification process, starting around E15, was little affected. Collectively, these data emphasize that it is the induction of the UPR rather than the timing of the expression of the ER stress-inducing protein that is critical in determining the pathology in MCDS. In some cases, expression of the ER stress-inducing protein and triggering of the UPR are coincidental, but in other cases, expression of the protein can precede induction of a UPR by many hours to days.
The delayed onset of BiP mRNA upregulation, with respect to the expression of the ER stress-inducing protein in wt/m Col10a1 mutant embryos and in both c/c and +/c Tg cog embryos, supports the contention that hypertrophic chondrocytes have a finite capacity to deal with ER stress that must be exceeded before the UPR is triggered. There are several possible explanations for why the ColXTg cog c/c embryos were delayed in comparison with the m/m Col10a1 p.N617K embryos in terms of UPR activation. First, as described previously (Rajpar et al. 2009), the Tg cog mRNA concentration in c/c mice was approximately one-third of the mRNA concentration of endogenous collagen X transcripts within hypertrophic chondrocytes, suggesting that the rate of Tgcog protein synthesis may be significantly lower than that of collagen X. Second, the level of ER stress caused by the misfolding of the two mutant proteins may differ significantly. This is clearly the case when comparing the consequences of expressing the Col10a1 p.N617K mutation reported above with that of the Col10a1 13del mutation (Tsang et al. 2007). As described above, the heterozygote wt/m mice did not exhibit BiP expression above that of the wt/wt controls at E17.5, although BiP expression is clearly elevated after birth in these animals (Rajpar et al. 2009). In contrast, as soon as the 13del allele was expressed in hemizygous E15.5 embryos, BiP expression became markedly upregulated (Tsang et al. 2007). Therefore, a tissue’s response to the expression of a mutant protein depends critically on two factors: first, the amount of mutant protein expressed and, second, the ER stress-inducing nature of the specific mutation, as different mutations within the same gene can have the potential to elicit different levels of stress as in the case of N617K and 13del Col10a1 mutations described above. Different mutations in the same protein will influence the nature of the misfolding, which will affect the cells’ ability to either refold it successfully or process it for degradation.
Furthermore, several analyses in addition to the current study have revealed different levels of UPR activation in Col10a1 p.N617K and ColXTg cog mice (Rajpar et al. 2009; Cameron et al. 2011). For instance, activation of ATF6 and IRE1 pathways was reported in both mutants by biochemical and microarray analyses (Rajpar et al. 2009; Cameron et al. 2011). However, increased phosphorylation of PERK-activated eIF2α was apparent only in Col10a1 p.N617K mice, which, together with the microarray data, supports our conclusion that Tgcog elicits a milder ER stress and, consequently, a milder phenotype than Col10a1 p.N617K (Cameron et al. 2011; Rajpar et al. 2009). MCDS is a dominant disease in humans and usually has a relatively mild phenotype that is not apparent at birth but becomes evident as a child starts to walk. This correlates well with our findings in the Col10a1 p.N617K and ColXTg cog lines in which a mild growth plate phenotype and bone growth defects only become apparent in heterozygotes or hemizygotes, respectively, as maximum growth rates are achieved at around 3 weeks of age (Rajpar et al. 2009). These data demonstrate for the first time that hypertrophic chondrocytes have a latent capacity to deal with increases in ER stress prior to eliciting the UPR and BiP upregulation. In the heterozygote MCDS mouse and, by implication, in human, the latent capacity of the hypertrophic chondrocyte to deal with ER stress appears only to be exceeded after birth as longitudinal bone growth accelerates. These data add further to our understanding of the importance of the ER and regulation of ER stress in chondrocytes. Recent studies describing the effects of knocking out ER stress-related factors, such as BBF2H7 (Saito et al. 2009) and site-1 protease (Patra et al. 2007), have reported the most severe effects on chondrogenesis. Chondrocytes clearly require a well-developed ER capable of processing and secreting the large quantities and variety of proteins required to assemble the abundant extracellular matrix of cartilage tissue. The data presented here illustrate that chondrocytes have a little “spare” capacity to cope with fluctuations in ER stress and that this spare capacity needs to be used up to trigger a UPR. As a consequence, variations in phenotypic severity in MCDS may arise in part because of the different levels of ER stress provoked by distinct mutations in collagen X coupled to genetic variations affecting the capacity of individuals to effectively deal with ER stress. Of particular hope in terms of disease therapy, strategies designed to reduce the level of ER stress through administration of chemical/pharmacological chaperones or by increasing the rate at which the unfolded protein is degraded (for reviews, see Papp and Csermely 2006; Engin and Hotamisligil 2010) are likely to be of potential benefit in either preventing or limiting the pathological consequences in MCDS and other ER stress-related connective tissue disorders (Boot-Handford and Briggs 2010).
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by the Wellcome Trust (084353/Z/07/Z), EU (LSHM-CT-2007-03747), BBSRC, and Certus Technology Associates Limited (Exeter, UK).
References
- Bateman JF, Wilson R, Freddi S, Lamande SR, Savarirayan R. 2005. Mutations of COL10A1 in Schmid metaphyseal chondrodysplasia. Hum Mutat. 25(6):525–534 [DOI] [PubMed] [Google Scholar]
- Boot-Handford RP, Briggs MD. 2010. The unfolded protein response and its relevance to connective tissue diseases. Cell Tissue Res. 339:197–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron TL, Bell KM, Tatarczuch L, Mackie EJ, Rajpar MH, McDermott BT, Boot-Handford RP, Bateman JF. 2011. Transcriptional profiling of chondrodysplasia growth plate cartilage reveals adaptive ER-stress networks that allow survival but disrupt hypertrophy. PLoS ONE. 6(9):e24600. 10.1371/journal.pone.0024600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin F, Hotamisligil GS. 2010. Restoring endoplasmic reticulum function by chemical chaperones: an emerging therapeutic approach for metabolic diseases. Diabetes Obes Metab. 12:108–115 [DOI] [PubMed] [Google Scholar]
- Ho MSP, Tsang KY, Lo RL, Susic M, Makitie O, Chan TW, Ng VC, Sillence DO, Boot-Handford RP, Gibson G, et al. 2007. COL10A1 nonsense and frame-shift mutations have a gain-of-function effect on the growth plate in human and mouse metaphyseal chondrodysplasia type Schmid. Hum Mol Genet. 16(10):1201–1215 [DOI] [PubMed] [Google Scholar]
- Karsenty G. 2003. The complexities of skeletal biology. Nature. 423(6937):316–318 [DOI] [PubMed] [Google Scholar]
- Karsenty G. 2008. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 9:183–196 [DOI] [PubMed] [Google Scholar]
- Karsenty G, Kronenberg HM, Settembre C. 2009. Genetic control of bone formation. Ann Rev Cell Dev Biol. 25:629–648 [DOI] [PubMed] [Google Scholar]
- Kronenberg HM. 2003. Developmental regulation of the growth plate. Nature. 423(6937):332–336 [DOI] [PubMed] [Google Scholar]
- Lee AS. 2005. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 35:373–381 [DOI] [PubMed] [Google Scholar]
- Lin JH, Walter P, Yen TB. 2008. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol-Mech. 3(1): 399–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie EJ, Ahmed YA, Tatarczuch L, Chen KS, Mirams M. 2008. Endochondral ossification: how cartilage is converted into bone in the developing skeleton. Int J Biochem Cell Biol. 40(1):46–62 [DOI] [PubMed] [Google Scholar]
- Malhotra JD, Kaufman RJ. 2007. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 18(6):716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclaughlin SH, Conn SN, Bulleid NJ. 1999. Folding and assembly of type X collagen mutants that cause metaphyseal chondrodysplasia-type Schmid: evidence for co-assembly of the mutant and wild-type chains and binding to molecular chaperones. J Biol Chem. 274:7570–7575 [DOI] [PubMed] [Google Scholar]
- Newman B, Wallis GA. 2003. Skeletal dysplasias caused by a disruption of skeletal patterning and endochondral ossification. Clin Genet. 63(4):241–251 [DOI] [PubMed] [Google Scholar]
- Papp E, Csermely P. 2006. Chemical chaperones: mechanisms of action and potential use. Handb Exp Pharmacol. 172:405–416 [DOI] [PubMed] [Google Scholar]
- Patra D, Xing X, Davies S, Bryan J, Franz C, Hunziker EB, Sandell LJ. 2007. Site-1 protease is essential for endochondral bone formation in mice. J Cell Biol. 179(4):687–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provot S, Schipani E. 2005. Molecular mechanisms of endochondral bone development. Biochem Biophys Res Commun. 328(3):658–665 [DOI] [PubMed] [Google Scholar]
- Provot S, Schipani E. 2007. Fetal growth plate: a developmental model of cellular adaptation to hypoxia. Ann N Y Acad Sci. 1117:26–39 [DOI] [PubMed] [Google Scholar]
- Rajpar MH, McDermott B, Kung L, Eardley R, Knowles L, Heeran M, Thornton DJ, Wilson R, Bateman JF, Poulsom R, et al. 2009. Targeted induction of endoplasmic reticulum stress induces cartilage pathology. PLoS Genet. 5(10):e1000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 8(7):519–529 [DOI] [PubMed] [Google Scholar]
- Saito A, Hino S, Murakami T, Kanemoto S, Kondo S, Saitoh M, Nishimura R, Yoneda T, Furuichi T, Ikegawa S, et al. 2009. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat Cell Biol. 11:1197–1204 [DOI] [PubMed] [Google Scholar]
- Samali A, FitzGerald U, Deegan S, Gupta S. 2010. Methods for monitoring endoplasmic reticulum stress and the unfolded protein response. Int J Cell Biol. 2010:830307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid TM, Linsenmayer TF. 1985. Immunohistochemical localization of short chain cartilage collagen (type X) in avian tissues. J Cell Biol. 100(2):598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, Ron D. 2011. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 13:184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang KY, Chan D, Cheslett D, Chan WCW, So CL, Melhado IG, Chan TWY, Kwan KM, Hunziker EB, Yamada Y, et al. 2007. Surviving endoplasmic reticulum stress is coupled to altered chondrocyte differentiation and function. PLoS Biol. 5(3):e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis GA, Rash B, Sweetman WA, Thomas JT, Super M, Evans G, Grant ME, Boot-Handford RP. 1994. Amino acid substitutions of conserved residues in the carboxyl-terminal domain of the a1 chain of type X collagen occur in two unrelated families with metaphyseal chondrodysplasia type Schmid. Am J Hum Genet. 54:169–178 [PMC free article] [PubMed] [Google Scholar]
- Warman ML, Abbott M, Apte SS, Hefferon T, McIntosh I, Cohn DH, Hecht JT, Olsen BR, Francomano CA. 1993. A type X collagen mutation causes Schmid metaphyseal chondrodysplasia. Nat Genet. 5:79–82 [DOI] [PubMed] [Google Scholar]
- Warman ML, Cormier-Daire V, Hall C, Krakow D, Lachman R, LeMerrer M, Mortier G, Mundlos S, Nishimura G, Rimoin DL, et al. 2011. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet. 155A:943–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Freddi S, Bateman JF. 2002. Collagen X chains harboring Schmid metaphyseal chondrodysplasia NC1 domain mutations are selectively retained and degraded in stably transfected cells. J Biol Chem. 277:12516–12524 [DOI] [PubMed] [Google Scholar]
- Wilson R, Freddi S, Chan D, Cheah KS, Bateman JF. 2005. Misfolding of collagen X chains harboring Schmid metaphyseal chondrodysplasia mutations results in aberrant disulfide bond formation, intracellular retention, and activation of the unfolded protein response. J Biol Chem. 280:15544–15552 [DOI] [PubMed] [Google Scholar]