Abstract
Acrolein (Acr) is a ubiquitous environmental pollutant as well as an endogenous compound. Acrolein-derived 1,N2-propanodeoxyguanosines (Acr-dG) are exocyclic DNA adducts formed following exposure to cigarette smoke or from lipid peroxidation. Acr-dG is mutagenic and potentially carcinogenic and may represent a useful biomarker for the early detection of cancers related to smoking or other oxidative conditions, such as chronic inflammation. In this study, we have developed a high-throughput, automated method using a HistoRx PM-2000 imaging system combined with MetaMorph software for quantifying Acr-dG adducts in human oral cells by immunohistochemical detection using a monoclonal antibody recently developed by our laboratory. This method was validated in a cell culture system using BEAS-2B human bronchial epithelial cells treated with known concentrations of Acr. The results were further verified by quantitative analysis of Acr-dG in DNA of BEAS-2B cells using a liquid chromatography/tandem mass spectrometry/multiple-reaction monitoring method. The automated method is a quicker, more accurate method than manual evaluation of counting cells expressing Acr-dG and quantifying fluorescence intensity. It may be applied to other antibodies that are used for immunohistochemical detection in tissues as well as cell lines, primary cultures, and other cell types.
Keywords: immunohistochemisty, acrolein, DNA adducts, biomarkers
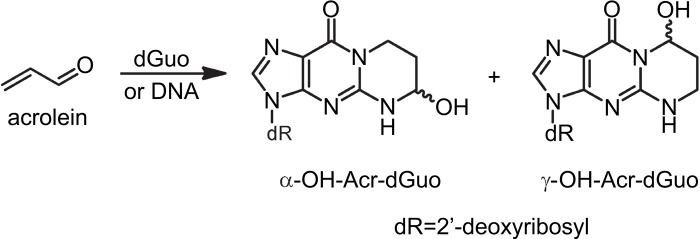
Cigarette smoking is a major source of human exposure to acrolein (Acr), a highly toxic and reactive aldehyde that is also formed endogenously through lipid peroxidation (Chung et al. 1996; Pan and Chung 2002; Stevens and Maier 2008; Thompson and Burcham 2008). Acr reacts with dGuo in DNA to form two pairs of regioisomeric 1,N2-propanodeoxyguanosine adducts: α-OH-Acr-dGuo and γ-OH-Acr-dGuo (Fig. 1) (Chung et al. 1984; Zhang et al. 2007, 2011). α-OH-Acr-dG in double-stranded DNA was found to be more mutagenic than γ-OH-Acr-dG in human cells and induced predominantly G→T transversions (Minko et al. 2009; Yang et al. 2002). The contribution of Acr-dG DNA adducts in smoking-related lung and oral cancers has recently been the subject of much research due to the finding that these adducts formed preferentially at the same dG locations in the human p53 gene that were shown to be the sites of mutational hotspots in cigarette smoke-induced lung tumors (Feng et al. 2006).
Figure 1.
Chemical structure of acrolein and acrolein-dG.
Oral squamous cell carcinoma (OSCC) is second only to lung cancer as the most prevalent smoking-related cancer worldwide. Despite recent advances in the early detection and diagnosis of the disease, the 5-year survival rate remains poor at approximately 50% (Neville and Day 2002). Therefore, there is an urgent need to develop biomarkers for the early detection and prevention of oral cancer. Previously, our laboratory found, by using a 32P-postlabeling high- performance liquid chromatography (HPLC) method, that levels of Acr-derived 1,N2-propanodeoxyguanosine (Acr-dG) were significantly higher in oral tissue from smokers than from non-smokers (Nath et al. 1998). This indicates that Acr-dG may be a useful biomarker for the early detection of oral cancer in normal-appearing oral mucosa or premalignant lesions from smokers and could be used to monitor the efficacy of chemoprevention strategies (Choudhury et al. 2004). Currently, our laboratory is investigating the chemopreventive effects of tea by measuring the DNA adducts caused by smoking in oral cells, including Acr-dG, in a randomized, double-blind, crossover clinical trial of smokers. Ingestion of tea has previously been shown in pilot trial studies to decrease DNA damage associated with cigarette smoking in human oral cells and improve oral premalignant lesion clinical response rate (Schwartz et al. 2005; Tsao et al. 2009). There is a need to develop a high-throughput, quantitative immunohistochemcial method to directly detect Acr-dG in human oral cells obtained from population-based studies. Traditionally, levels of Acr-dG and other adducts in DNA are detected and quantified by liquid chromatography/tandem mass spectrometry/multiple-reaction monitoring (LC-MS/MS-MRM) or 32P-postlabeling/HPLC methods (Emami et al. 2008; Nath et al. 1998; Zhang et al. 2011). However, these methods require large amounts of starting DNA, which is not always available in translational studies using human cells/tissues. In addition, these methods can measure the total adduct level in cells or tissue but cannot detect lesions in individual cells and require multiple steps in sample preparation and detection; thus, they are not amenable for studies involving large sample sizes. Recently, our laboratory developed the first monoclonal antibody against Acr-dG adducts that can be used in the immunohistochemical analysis of human tissues (J. Pan et al., unpublished data). Using this antibody, we have developed an immunofluorescent method for detecting Acr-dG adducts in human oral cells in a high-throughput, quantitative system.
Many research groups quantify immunohistochemical staining intensity by visual judgment of the number of stained cells and the staining intensity. This method is time-consuming, labor intensive, and unreliable among observers (Prasad and Prabhu 2012). During the course of the present study, we had made numerous attempts at applying methods relying on visual judgment, such as ImageJ (National Institutes of Health; Bethesda, MD), to quantitatively determine the positively stained oral cells, but all the efforts not only were inefficient but also failed to obtain consistent results. Thus, we sought to develop an automated image analysis methodology for quantifying DNA damage biomarkers, such as Acr-dG, in human oral cells. To this end, we combined automated imaging using a HistoRx PM-2000 system with image analysis using MetaMorph software (Molecular Devices; Sunnyvale, CA) to quantify the percentage of nuclei in a sample expressing the adduct (histomorphic analysis) as well as the intensity of the staining (proportional to the number of adducts in each cell). MetaMorph software has been successfully used to analyze such diverse processes as skeletal muscle fiber morphometry and pathological prion protein deposition (Garton et al. 2010; Maximova et al. 2006). In the study described here, we validated the computer-assisted quantification of Acr-dG in human oral cells by using a method to quantify Acr-dG–positive nuclei and intensity in BEAS-2B human bronchial cells treated with known concentrations of Acr. We also verified the findings by quantifying the levels of Acr-dG in BEAS-2B cells measured by LC-MS/MS-MRM. We found that quantification of Acr-dG–positive nuclei and staining intensity in human oral cells obtained using HistoRx PM-2000 imaging, combined with MetaMorph analysis, was in accordance with changes in Acr treatment concentrations in BEAS-2B cells and LC-MS/MS measurements of Acr-dG in DNA isolated from these cells.
Materials and Methods
Cell Culture
Human bronchial BEAS-2B cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) FBS and 1% penicillin/streptomycin. Cells were treated with 20, 35, 50, 100, 200, and 300 µM Acr (Alfa Aesar; Ward Hill, MA) for 24 hr. Control cells were treated with vehicle (PBS) alone.
Monoclonal Antibody for Acr-dG
The detailed information on the development of the monoclonal antibody against ACr-dG will be published elsewhere. Briefly, the antibody against Acr-dG was raised in BALB/C mice. Mice were immunized with Acr-derived guanosine adduct-conjugated BSA in saline emulsified with an equal amount of Freund’s complete adjuvant, given in a split dose, intraperitoneally (IP) and subcutaneously (SC). A second immunization was given 2 weeks after the first one in incomplete Freund’s adjuvant. Mice were boosted with the conjugate in saline and given IP on days 1, 2, 3, and 4 on the fourth week after the second injection. On day 5, mice were sacrificed and the spleen was removed for fusion. Test bleeds were taken to check the antibody reactivity toward immunogens using ELISA.
The mice were used for cell fusion and hybridoma production. A total of 14 hybridoma cell lines derived from seven parental clones were produced. Splenocyte and myeloma were fused, plated into 96-well culture plates, and screened by ELISA to detect the positive clones. The selected clones were then subcloned by limiting dilution until they were monoclonal and stable hybridomas. Two subclones from each parental clone were expanded into culture flasks, and four to six vials of cells for each subclonal cell line were cryopreserved.
Collection and Processing of Human Oral Cells
Samples were obtained in accordance with Georgetown University Institutional Review Board (IRB) guidelines. Oral cells were collected from the cheeks and tongue by the cytobrush technique and suspended in 1× PBS. Cells were washed two times in 1× PBS through centrifugation at 5000 rpm for 5 min after each wash and resuspended in 1 ml of clean 1× PBS. Then, 500 µ1 of this suspension was added to 500 µ1 of 2% paraformaldehyde and incubated overnight at 40C. The next morning, cells were washed two times with 1× PBS, centrifuging at 5000 rpm for 5 min after each wash. After the final wash, the supernatant was removed and melted histogel (Thermo Scientific; Waltham, MA) was added at a volume of 1:1 to the cells. Cells were carefully resuspended in the histogel by pipetting, taking care to avoid air bubbles. The cells in histogel were incubated at 40C for 30 min, and 70% ethanol was added to the top of the tube. The cells in histogel were submitted to the Georgetown Histology and Tissue Shared Resource for embedding into paraffin blocks according to standard procedures.
Immunofluorescence of Cultured Cells
BEAS-2B cells were grown on 22 × 22-mm glass coverslips in six-well plates to approximately 50% confluence and then treated with Acr (as above, for 24 hr). Cells were washed with cold 1× PBS and then fixed in 4% paraformaldehyde for 10 min, followed by permeabilization in 0.5% Triton X-100/2 M HCl in PBS for 5 min. Cells were then blocked in 10% FBS/5% BSA/0.5% Triton X-100 in PBS for 1 hr at room temperature, followed by incubation with Acr-dG antibody (1:2000) in 1% FBS/0.5% BSA/0.5% Triton X-100 in PBS for 1 hr at room temperature (J. Pan et al., unpublished data). Cells were washed in PBS and then incubated with secondary antibody (Alexa Fluor 488 goat anti-mouse; 1:200 in 1% FBS/0.5% BSA/0.5% Triton X-100 in PBS; Molecular Probes, Eugene, OR) for 1 hr at room temperature in the dark. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; 1:10,000). Cells on coverslips were mounted onto glass slides using Prolong Gold Antifade Reagent containing DAPI (Molecular Probes) and dried in the dark at room temperature for 24 hr before visualization.
Immunofluorescence of Oral Cells
Briefly, formalin-fixed paraffin-embedded (FFPE) oral cells were de-paraffinized in two sequential washings of xylene for 5 min each; dehydrated in graded ethanols of 100%, 100%, 95%, 70%, and deionized water for 2 min each; and incubated with 1% hydrogen peroxide for 20 min at room temperature. Cells were subjected to antigen retrieval in 10 mM sodium citrate (pH 6) and blocked with 10% normal goat serum (Vector Laboratories; Burlingame, CA) in PBS-Brij solution for 30 min at room temperature. Cells were then incubated overnight at 4C with Acr-dG antibody (1:4000). Cells were washed and incubated with biotinylated goat anti-mouse (1:100; Vector Laboratories) secondary antibody for 45 min at room temperature, followed by incubation with streptavidin Cy3 (1:100; Invitrogen, Carlsbad, CA) tertiary antibody for 45 min at room temperature. Following washing with PBS-Brij, coverslips were applied to slides using Prolong Gold Antifade Reagent containing DAPI (Molecular Probes) and dried in the dark for 24 hr at room temperature before visualization.
LC-MS/MS Analysis of Acr-dG in BEAS-2B Cells
BEAS-2B cells were grown in 150-mm dishes to approximately 80% confluency and then treated with Acr (as above, for 24 hr). Flasks were treated in duplicate at each concentration. Both floating and attached cells were collected by scraping. DNA was extracted using the DNA Midi Kit (Qiagen; Gaithersburg, MD). Approximately 75 µg DNA was used for measuring Acr-dG using an LC-MS/MS-MRM method with some modifications (Zhang et al. 2007). Briefly, DNA samples, mixed with [13C10,15N5]-Acr-dG as internal standards, were hydrolyzed enzymatically with DNase I, alkaline phosphatase, and phosphodiesterase I. The hydrolysate was then purified by solid-phase extraction (SPE), and the fraction containing Acr-dG adducts was collected, dried, and injected on the column for the LC-MS/MS-MRM experiment. Detection and quantification of Acr-dG adducts were carried out with an ACQUITY UPLC liquid chromatography system (Waters Corporation; Milford, MA) equipped with 50 × 2.1-mm, 1.7-µm particle size C18 column (Waters Acquity UPLC BEH C18) and coupled with Applied Biosystems/MDS SCIEX 4000 Q TRAP (Life Technologies Corporation; Carlsbad, CA) triple quadrupole mass spectrometer. The separation of Acr-dG adducts was performed isocratically eluting 3% acetonitrile (ACN), 1 mM ammonium formate buffer over 3.5 min using a 0.5-ml/min flow rate at 40C, followed by 100% ACN wash. The electrospray ionization (ESI) source was operated in positive mode. MRM experiment was performed using ion transitions of 324.2→208.1 m/z (Acr-dG) and 339.2→218.1 m/z ([13C10,15N5]-Acr-dG) for quantitation and those of 324.2→190.1 m/z (Acr-dG) and 339.2→200.1 m/z ([13C10,15N5]-Acr-dG) for structural confirmation. All other parameters were optimized to achieve maximum signal intensity. Calibration curves were constructed for all three Acr-dG regioisomers before each analysis using standard solutions of Acr-dG and [13C10,15N5]-Acr-dG. A constant concentration of [13C10,15N5]-Acr-dG (1 fmol/µl) was used with different concentrations of Acr-dG (1.68 amol/µl–220 fmol/µl) and analyzed using 37-µl injections by LC-MS/MS-MRM.
Image Acquisition for Validation Study
The HistoRx PM-2000 system from HistoRx, Inc. (New Haven, CT) interfaced on a personal computer was used to acquire high-resolution (pixel dimension 512 × 512), monochrome images. The PM-2000 system consists of an Olympus BX51 fluorescent microscope (Melville, NY), Optronics Quantifire camera (Goleta, CA), X-Cite equipped with a mercury/metal halide lamp (Ontario, Canada), and an H101A ProScan motorized stage from Prior Scientific (Rockland, MA). AQUAsition (version 2.3.3.2) software from HistoRx, Inc. was used to load sample slides, set control parameters, and acquire images. The slide type was set to Whole Tissue Section (WTS). Acquisition type was set to single, and magnification was set to ×20. DAPI, FITC, and Cy3 filters were selected. The usage was set to nuclear, target, and cytoplasm for DAPI, Alexa Fluor 488, and Cy3 channels, respectively. Cy3 channel images were not needed for our analysis but nonetheless selected because the AQUAsition software requires users to acquire images at three wavelengths. Autofocus was set to the FITC channel. Autoexposure was used for DAPI images, and 2000-msec and 100-msec exposures were used for Alexa Fluor 488 and Cy3 images, respectively. The slide was scanned prior to image acquisition to preselect areas of interest or “spot” over the sample. Depending on the pellet size of the sample, generally 10 to 35 images were acquired. Once the acquisition was complete, the image file was loaded in AQUAnalysis (version 2.3.3.2) software from HistoRx, Inc. In the AQUAnalysis software, images with defects such as blurred images or images with artifacts were discarded. The rest of the images were exported in tiff file format.
Immunofluorescence Quantification for Method Validation Study
The exported image files were loaded into MetaMorph software (version 7.7.3.0; Molecular Devices). Journals were created to automatically convert the images exported in 8-bit to 16-bit (required for the MetaMorph journals), to identify DAPI nuclei, to identify nuclei with fluorescence (in our case, Alexa Fluor 488) that were above background fluorescence, to eliminate any large or small artifacts from the image, and to export data automatically to Excel. The created journals gave data on % positive nuclei and average intensity of positive nuclei for each image.
Quantification of Acr-dG in Human Oral Cells
For image acquisition, the same parameters were used from the validation study except the usage was set to nuclear, cytoplasm, and target for DAPI, Alexa Fluor 488, and Cy3 channels, respectively. Autofocus was set to the Cy3 channel. Autoexposure was used for DAPI images, and 50-msec and 1000-msec exposures were used for Alexa Fluor 488 and Cy3 images, respectively. The image file was exported from the AQUAnalysis software in the same fashion as in the validation study. The exported image files were loaded into MetaMorph software. The same MetaMorph journals were used as in the validation study to quantify positive nuclei, but in this case, DAPI and Cy3 images were analyzed. Independent t-tests were conducted for each treatment period to analyze the treatment effects.
Results
Quantification of Fluorescence Using MetaMorph Software
Through use of the MetaMorph software, a user can program journals that can execute a set of customized functions in order. In this study, two specific journals were created to execute high-throughput quantification of immunofluorescence for the images obtained from the HistoRx PM-2000 system. The first journal converts a set of DAPI and Alexa Fluor 488 images to binary images and performs the MetaMorph application “Cell Scoring” to the images. The cell-scoring application identifies the nuclei in the DAPI channel and calculates the percentage of positively stained nuclei in the secondary channel (e.g., Alexa Fluor 488 or Cy3) based on the default parameter. The journal also measures the fluorescence intensity in the nuclear region in the Alexa Fluor 488 or Cy3 image. In addition, the journal is programmed to exclude very small and very large objects from the image. This process eliminates non-cellular debris that may interfere with the analysis. Several sets of DAPI and Alexa Fluor 488 or Cy3 images were analyzed with the naked eye to find the optimal parameters that correctly identify the location of cellular nuclei and define the intensity of the positively stained nuclei in the Alexa Fluor 488 or Cy3 channel in each sample. In this study, any visible fluorescence in the nuclear region above the background was defined as positively stained. The optimized parameters were then updated to the cell-scoring application in the journal. The second journal simply repeats the execution of the first journal to all images in a directory of exported files.
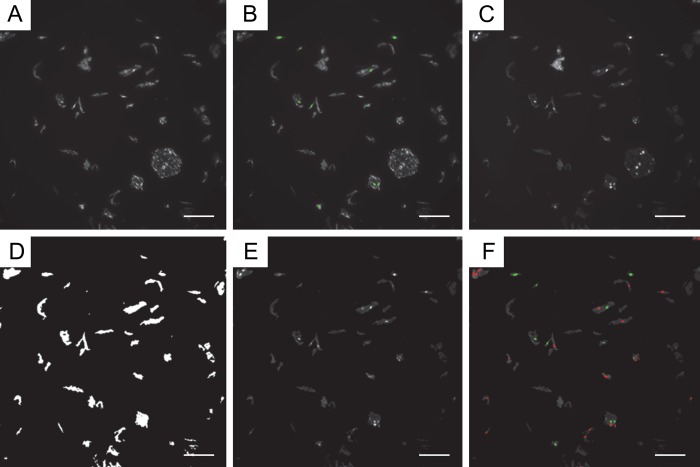
Representative images of the steps taken to quantify fluorescence in our study are shown in Fig. 2. In addition, the journal is also programmed to report the image name, the total number of cells, the number of positive cells, the number of negative cells, the percentage of positive cells, the average intensity of all the nuclei identified in the DAPI channel, the average intensity of all the nuclei identified in the Alexa Fluor 488 (or Cy3) channel, the average intensity of the positively stained nuclei in the Alexa Fluor 488 (or Cy3) channel, and the average intensity of the negatively stained nuclei in the Alexa Fluor 488 (or Cy3) channel to a Microsoft Excel (Microsoft Corp.; Redmond, WA) software spreadsheet (Table 1).
Figure 2.
Representative example of quantification of Acr-dG immunofluorescence using MetaMorph. For immunofluorescence analysis of Acr-dG, formalin-fixed paraffin-embedded (FFPE) oral cells were subjected to a standard procedure using a monoclonal antibody against Acr-dG. Cy3 was used as the secondary antibody, and nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Samples were imaged using a HistoRx PM-2000 imaging device, and images were analyzed to identify and enumerate positively stained nuclei using MetaMorph software. (A) Original image in Cy3 channel staining for Acr-dG. (B) Cy3 image; green nuclei are those identified “positive for Cy3” by MetaMorph. (C) Original image in DAPI channel. (D) DAPI image-integrated morphometry mask to eliminate large and small objects. (E) Resulting filtered DAPI image. (F) Final DAPI image; green nuclei are those identified “positive for Cy3” by MetaMorph, and red nuclei are those identified “negative for Cy3” by MetaMorph. The scale bar size shown is 100 µm.
Table 1.
Cell Scoring Application Results Reported to Excel Spreadsheet
| Image Name | Total Cells | Positive Cells | Negative Cells | % Positive Cells | All Nuclei W1 Average Intensity | All Nuclei W2 Average Intensity | Positive Nuclei W2 Average Intensity | Negative Nuclei W2 Average Intensity |
|---|---|---|---|---|---|---|---|---|
| DAPI_00002 | 38 | 24 | 14 | 63 | 90 | 59 | 64 | 47 |
| DAPI_00003 | 38 | 38 | 0 | 100 | 86 | 85 | 85 | 0 |
| DAPI_00004 | 57 | 53 | 4 | 93 | 79 | 66 | 68 | 43 |
| DAPI_00005 | 51 | 51 | 0 | 100 | 84 | 101 | 101 | 0 |
Method Validation Study with BEAS-2B Human Bronchial Cells
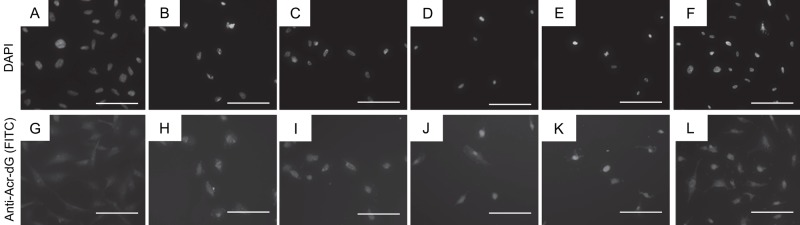
Immunohistochemical analysis of Acr-dG in BEAS-2B human bronchial cells showed a correlation between nuclear staining and increasing concentration of Acr treatment. Untreated samples (0 µM Acr) yielded no visible nuclear accumulation of Acr-dG immunofluorescence (Fig. 3A, B). This may indicate that the method is not highly sensitive because Acr can be formed endogenously and therefore, even at 0 µM Acr treatment, there should be a background level of Acr-dG. BEAS-2B cells treated with 20 µM (Fig. 3C, D) and 35 µM (Fig. 3E, F) Acr showed weak and moderate nuclear staining for Acr-dG. BEAS-2B cells treated with 50 µM (Fig. 3G, H), 100 µM (Fig. 3I, J), and 200 µM (Fig. 3K, L) Acr showed progressively increasing intensity of nuclear staining for Acr-dG, along with an increasing percentage of positively stained nuclei.
Figure 3.
Representative examples of Acr-dG immunofluorescence in BEAS-2B cells treated with varying concentrations of Acr. BEAS-2B cells were treated with Acr at varying concentrations for 24 hr as described in the Materials and Methods. Immunofluorescence was performed using an anti-Acr-dG antibody. (A) 4′,6-Diamidino-2-phenylindole (DAPI) channel image of BEAS-2B cells, no Acr treatment. (B) Alexa Fluor 488 channel image of BEAS-2B cells, no Acr treatment. (C) DAPI channel image of BEAS-2B cells, 20 µM Acr treatment. (D) FITC channel image of BEAS-2B cells, 20 µM Acr treatment. (E) DAPI channel image of BEAS-2B cells, 35 µM Acr treatment. (F) FITC channel image of BEAS-2B cells, 35 µM Acr treatment. (G) DAPI channel image of BEAS-2B cells, 50 µM Acr treatment. (H) FITC channel image of BEAS-2B cells, 50 µM Acr treatment. (I) DAPI channel image of BEAS-2B cells, 100 µM Acr treatment. (J) FITC channel image of BEAS-2B cells, 100 µM Acr treatment. (K) DAPI channel image of BEAS-2B cells, 200 µM Acr treatment. (L) FITC channel image of BEAS-2B cells, 200 µM Acr treatment. The scale bar size is 100 µm.
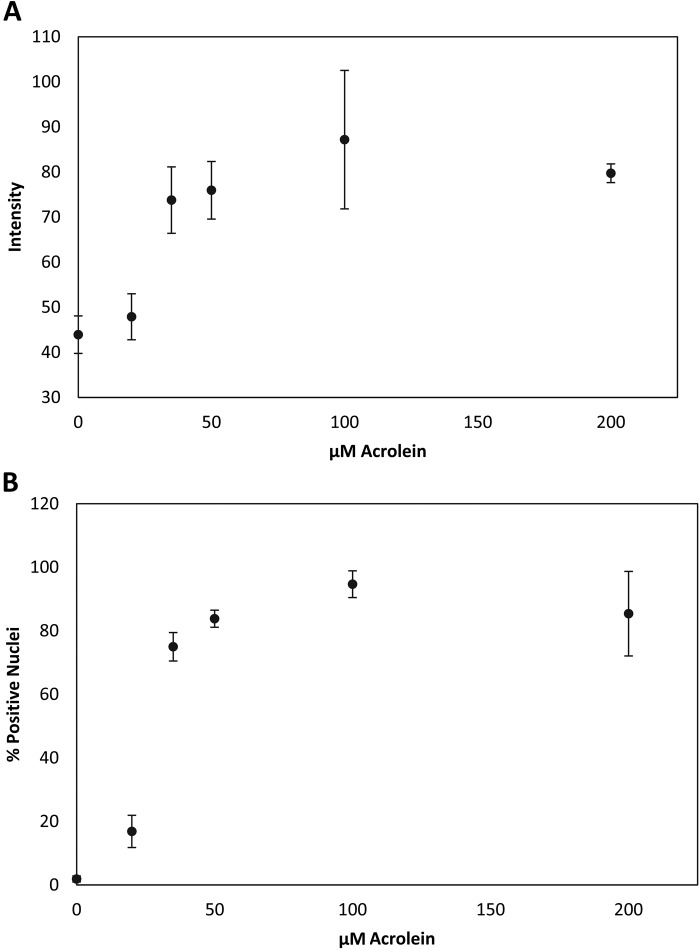
Quantitative analysis of fluorescence intensity and percent positive nuclei using MetaMorph software showed that the fluorescence intensity increased minimally between 0 and 20 µM Acr, but sharply increased between 20 and 35 µM Acr. However, the intensity reached a plateau between 50 and 200 µM Acr (Fig. 4A). The percentage of positively stained Acr-dG nuclei also increased in accordance with increasing Acr concentration (Fig. 4B). At 0 µM Acr, there was virtually no positive staining detected. At 20 µM Acr, the percentage of positively stained nuclei increased to 17%. Similar to the intensity result, the percentage of positively stained nuclei increased sharply at 35 µM Acr to 75% and reached a plateau at 50, 100, and 200 µM Acr to approximately 90% (Fig. 4B).
Figure 4.
MetaMorph quantification of Acr-dG immunofluorescence. BEAS-2B cells were treated with varying concentrations of Acr for 24 hr as described in Materials and Methods. (A) Acr-dG immunofluorescence intensity. Dots, average of triplicate experiments; bars, standard error of the mean. (B) Percentage of positively stained nuclei for Acr-dG. Dots, average of triplicate experiments; bars, standard error of the mean.
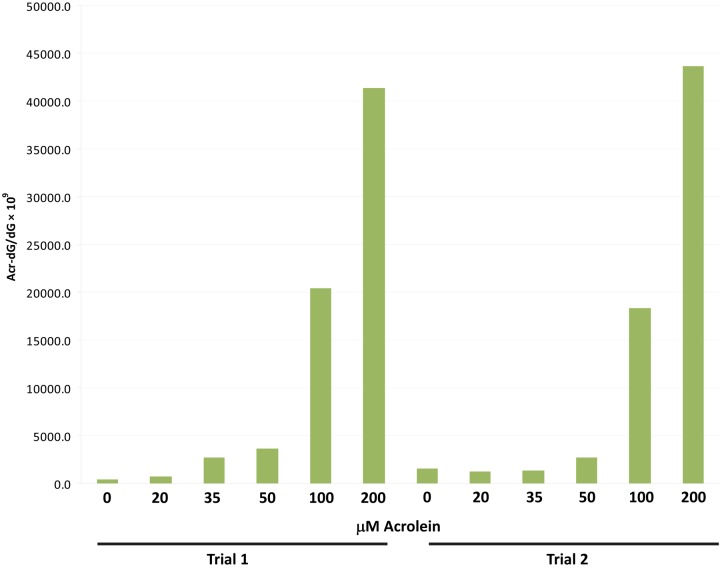
To further validate the immunofluorescence quantification using MetaMorph software, we performed LC-MS/MS-MRM analysis of Acr-dG DNA adducts in BEAS-2B cells treated with varying concentrations of Acr for 24 hr (Fig. 5). As expected, there was a steady increase in Acr-dG levels in DNA with increasing Acr treatment concentration. In contrast to the MetaMorph results, LC-MS/MS-MRM analysis showed that the formation of Acr-dG did not plateau between 50 and 200 µM Acr. The plateau of intensity and positively stained nuclei at the higher concentrations probably reflects the saturation of antibody binding to the Acr-dG formed at these concentrations.
Figure 5.
Liquid chromatography/tandem mass spectrometry/multiple-reaction monitoring (LC-MS/MS-MRM) analysis of Acr-dG levels in BEAS-2B cells. BEAS-2B cells were treated with varying concentrations of Acr for 24 hr. DNA was extracted from cells and LC-MS/MS-MRM analysis of Acr-dG levels in DNA was performed as described in the Materials and Methods. The experiment was performed in duplicate for each concentration of Acr.
Immunofluorescence Quantification of Acr-dG in Human Oral Cells
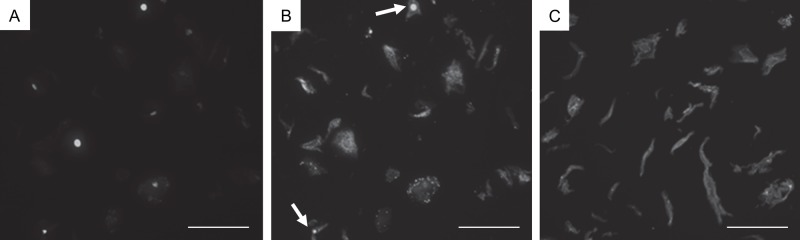
The immunofluorescence quantification method detailed in the Materials and Methods was applied to FFPE human oral cells of smokers to quantify Acr-dG adduct levels. Figure 6A shows a representative example of the oral cell morphometry. Nuclei were stained with DAPI. Figure 6B shows several positive nuclei for anti-Acr-dG antibody by immunofluorescence staining. The oral cells generally presented a high level of background fluorescence in the Cy3 channel and a lesser amount in the DAPI channel (Fig. 6A, B). This may be due to the intrinsic autofluorescence of the oral cells in the 550-nm spectral region (Onizawa et al. 2003). The negative control (no primary antibody) oral cell sample showed a high level of background fluorescence as well, although no nuclear staining was detected (Fig. 6C).
Figure 6.
Acr-dG immunofluorescence in human oral cells. Human oral cells from a smoker were subjected to immunofluorescence staining using an antibody against Acr-dG as described in the Materials and Methods. (A) 4′,6-Diamidino-2-phenylindole (DAPI) channel image indicating the location of the nuclei. (B) Acr-dG immunofluorescence (Cy3). Arrows indicate nuclei that are positive for Acr-dG adducts. (C) Negative control for Acr-dG immunofluorescence (Cy3). The procedure remained the same except for no incubation with Acr-dG antibody. Note that although there is background fluorescence, there is no nuclear staining. The scale bar size is 100 µm.
Discussion
In this study, we developed a high-throughput, computer-assisted method for quantifying nuclear immunofluorescence staining. We applied our method to measuring levels of Acr-dG adducts in human bronchial epithelial cells in culture and in human oral cells collected by the cytobrush technique, but this method conceivably can be applied to multiple antibodies and cell/tissue types. Our method was further verified by measuring Acr-dG levels in BEAS-2B cells by LC-MS/MS-MRM. This method may prove to be efficient and sensitive for quantifying data from large numbers of samples from translational and/or clinical studies that may include several hundred subjects. One of the main advantages of this method, compared with manual observation, is that by changing several detection parameters among samples in MetaMorph, one can quickly analyze many samples and retain relatively high accuracy for reproducibility.
We performed the validation studies for the quantification method by treating BEAS-2B cells with known concentrations of Acr. Although our method can quantify intensity and positive nuclei according to Acr concentration, the immunohistochemistry (IHC) results were not completely linear. This non-linearity may be due to several factors. First, the small increase in Acr-dG intensity and percent positive nuclei between 0 and 20 µM Acr concentrations can be attributed to the fact that Acr is a highly reactive electrophilic compound (Fig. 4A, B). At low concentrations, Acr may react with proteins before reaching the nucleus (Zhu et al. 2011). In addition, it is known that Acr-dG in DNA can be efficiently repaired by the nucleotide excision repair (NER) pathway (unpublished results). Therefore, it is likely that repair activity suppressed the accumulation of Acr-dG in the nuclear DNA at low concentrations. Higher concentrations of Acr (35 µM and above), however, may overwhelm the repair capacity, which could cause the sharp spike in Acr-dG intensity and percent positive nuclei. Acr-dG immunofluorescence intensity and percent positive nuclei plateaued between 50 and 200 µM Acr, whereas the adduct levels measured by LC-MS still increased. This may be primarily attributed to the antibodies that were saturated in the immunohistochemical staining assay and also possibly to the fact that high concentrations of Acr promote apoptosis. The formation of Acr-dG is known to be highly correlated to apoptosis induction (Pan et al. 2009). Unlike LC-MS/MS analysis, MetaMorph software only analyzes live cells with intact nuclei; cells that undergo apoptosis are not counted. It should be pointed out that the lower range of Acr concentrations is physiologically more relevant than the higher concentrations.
It is important to note that due to the inevitable staining variability among samples, the nuclear immunofluorescence intensity values often yield inconsistent data resulting in large standard errors (Fig. 4A). In addition, intensity values are affected by the varying level of background fluorescence. However, quantifying the percentage of positively stained nuclei using MetaMorph software depends on the size of the nuclei and the intensity of a pixel in the nucleus above background pixel intensity. This process eliminates the background intensity interference and yields much more consistent data with smaller standard errors (Fig. 4B). Furthermore, quantifying positively stained nuclei provides information on adduct levels in individual cells.
The application of the method to human oral cells collected by the cytobrush technique shows that the percentage of positively stained nuclei can be measured using MetaMorph software to quantify Acr-dG levels. Although this is a high-throughput method, the potential variables in measuring clinical oral cell samples may come from the intrinsic autofluorescence and sometimes the poor quality of the collected cells that may affect measurements. In applying this method in clinical trials, increasing the sample size analyzed may help to reduce the variability and achieve more accurate results.
From the validation study, we demonstrated that the quantification method developed with HistoRx and MetaMorph software can reliably analyze Acr-dG immunofluorescence intensity and percentage of positive nuclei. We also demonstrated that measuring the percentage of positively stained nuclei via MetaMorph is a more robust method of quantifying Acr-dG adduct levels than measuring the immunofluorescence intensity of the nuclei. We are confident that this method can be applied to quantify immunofluorescence of multiple antibodies, and multiple cell/tissue types, as with simple changes in the journal of the MetaMorph software, the method can be used to analyze immunofluorescence at other wavelengths. The most important advantage of this method is its high-throughput nature, as once appropriate parameters are set for nuclei location and what constitutes positive nuclei are defined, the program can analyze all subsequent images in an automated fashion.
Acknowledgments
We thank the Histopathology and Tissue Shared Resource and the Microscopy and Imaged Shared Resource at the Lombard Comprehensive Cancer Center and the General Clinical Research Center (GCRC) of the Georgetown University Medical Center.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Cancer Institute (NCI) grant (1R01CA113449–01).
References
- Choudhury S, Pan J, Amin S, Chung FL, Roy R. 2004. Repair kinetics of trans-4-hydroxynonenal-induced cyclic 1,N2-propanodeoxyguanine DNA adducts by human cell nuclear extracts. Biochemistry. 43:7514–7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung FL, Young R, Hecht SS. 1984. Formation of cyclic 1,N2-propanodeoxyguanosine adducts in DNA upon reaction with acrolein or crotonaldehyde. Cancer Res. 44:990–995 [PubMed] [Google Scholar]
- Chung FL, Chen HJ, Nath RG. 1996. Lipid peroxidation as a potential endogenous source for the formation of exocyclic DNA adducts. Carcinogenesis. 17:2105–2111 [DOI] [PubMed] [Google Scholar]
- Emami A, Dyba M, Cheema AK, Pan J, Nath RG, Chung FL. 2008. Detection of the acrolein-derived cyclic DNA adduct by a quantitative 32P-postlabeling/solid-phase extraction/HPLC method: blocking its artifact formation with glutathione. Anal Biochem. 374:163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hu W, Hu Y, Tang MS. 2006. Acrolein is a major cigarette-related lung cancer agent: preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proc Natl Acad Sci U S A. 103:15404–15409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton F, Seto JT, North KN, Yang N. 2010. Validation of an automated computational method for skeletal muscle fibre morphometry analysis. Neuromuscul Disord. 20:540–547 [DOI] [PubMed] [Google Scholar]
- Maximova OA, Taffs RE, Pomeroy KL, Piccardo P, Asher DM. 2006. Computerized morphometric analysis of pathological prion protein deposition in scrapie-infected hamster brain. J Histochem Cytochem. 54:97–107 [DOI] [PubMed] [Google Scholar]
- Minko IG, Kozekov ID, Harris TM, Rizzo CJ, Lloyd RS, Stone MP. 2009. Chemistry and biology of DNA containing 1,N2-deoxyguanosine adducts of the α,β-unsaturated aldehydes acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem Res Toxicol. 22:759–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath RG, Ocando JE, Guttenplan JB, Chung FL. 1998. 1,N2- propanodeoxyguanosine adducts: potential new biomarkers of smoking-induced DNA damage in human oral tissue. Cancer Res. 58:581–584 [PubMed] [Google Scholar]
- Neville BW, Day TA. 2002. Oral cancer and precancerous lesions. CA Cancer J Clin. 52:195–215 [DOI] [PubMed] [Google Scholar]
- Onizawa K, Okamura N, Saginoya H, Yoshida H. 2003. Characterization of autofluorescence in oral squamous cell carcinoma. Oral Oncol. 39:150–156 [DOI] [PubMed] [Google Scholar]
- Pan J, Chung FL. 2002. Formation of cyclic deoxyguanosine adducts from ω-3 and ω-6 polyunsaturated fatty acids under oxidative conditions. Chem Res Toxicol. 15:367–372 [DOI] [PubMed] [Google Scholar]
- Pan J, Keffer J, Emami A, Ma X, Lan R, Goldman R, Chung FL. 2009. Acrolein-derived DNA adduct formation in human colon cancer cells: its role in apoptosis induction by docosahexaenoic acid. Chem Res Toxicol. 22:798–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Prabhu GK. 2012. Image analysis tools for evaluation of microscopic views of immunohistochemically stained specimen in medical research: a review. J Med Syst. 36:2621–2631 [DOI] [PubMed] [Google Scholar]
- Schwartz JL, Baker V, Larios E, Chung FL. 2005. Molecular and cellular effects of green tea on oral cells of smokers: a pilot study. Mol Nutr Food Res. 49:43–51 [DOI] [PubMed] [Google Scholar]
- Stevens JF, Maier CS. 2008. Acrolein: sources, metabolism, and biomolecular interactions relevant to human health and disease. Mol Nutr Food Res. 52:7–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CA, Burcham PC. 2008. Genome-wide transcriptional responses to acrolein. Chem Res Toxicol. 21:2245–2256 [DOI] [PubMed] [Google Scholar]
- Tsao AS, Liu D, Martin J, Tang XM, Lee JJ, El-Naggar AK, Wistuba I, Culotta KS, Mao L, Gillenwater A, et al. 2009. Phase II randomized, placebo-controlled trial of green tea extract in patients with high-risk oral premalignant lesions. Cancer Prev Res (Phila). 2:931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang IY, Chan G, Miller H, Huang Y, Torres MC, Johnson F, Moriya M. 2002. Mutagenesis by acrolein-derived propanodeoxyguanosine adducts in human cells. Biochemistry. 41:13826–13832 [DOI] [PubMed] [Google Scholar]
- Zhang S, Balbo S, Wang M, Hecht SS. 2011. Analysis of acrolein-derived 1,N2-propanodeoxyguanosine adducts in human leukocyte DNA from smokers and nonsmokers. Chem Res Toxicol. 24:119–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Villalta PW, Wang M, Hecht SS. 2007. Detection and quantitation of acrolein-derived 1,N2-propanodeoxyguanosine adducts in human lung by liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem Res Toxicol. 20:565–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Sun Z, Jiang Y, Chen F, Wang M. 2011. Acrolein scavengers: reactivity, mechanism and impact on health. Mol Nutr Food Res. 55:1375–1390 [DOI] [PubMed] [Google Scholar]