Abstract
In this study, combinatorial libraries were used in conjunction with ultra-high throughput sequencing to comprehensively determine the impact of each of the 19 possible amino acid substitutions at each residue position in the TEM-1β-lactamase enzyme. The libraries were introduced into E. coli and mutants were selected for ampicillin resistance. The selected colonies were pooled and subjected to ultra-high throughput sequencing to reveal the sequence preferences at each position. The depth of sequencing provided a clear, statistically significant picture of what amino acids are favored for ampicillin hydrolysis for all 263 positions of the enzyme in one experiment. Although the enzyme is generally tolerant of amino acid substitutions, several surface positions far from the active site are sensitive to substitutions suggesting a role for these residues in enzyme stability, solubility or catalysis. In addition, information on the frequency of substitutions was used to identify mutations that increase enzyme thermodynamic stability. Finally, a comparison of sequence requirements based on the mutagenesis results versus those inferred from sequence conservation in an alignment of 156 class A β-lactamases reveals significant differences in that several residues in TEM-1 do not tolerate substitutions and yet extensive variation is observed in the alignment, and vice versa. An analysis of the TEM-1 and other class A structures suggests residues that vary in the alignment may nevertheless make unique, but important, interactions within individual enzymes.
Introduction
Enzymes have long been the subject of structure-function studies to determine the amino acid sequence requirements for folding, stability and catalysis. These studies often utilize site directed mutagenesis to alter amino acid residues that are hypothesized to play a key role in an aspect of catalysis or folding followed by biochemical and biophysical characterization of the altered enzyme to test the hypothesized role1. Another site-directed mutagenesis approach is a systems level, unbiased strategy to systematically alter each position in an enzyme and assess the importance of the position for the structure and function of the enzyme. Those positions that have stringent sequence requirements, that is, those positions that do not tolerate amino acid substitutions without disruption of stability, solubility or catalytic activity are inferred to be critical for enzyme function. Subsequent biochemical studies of non-functional mutants at these critical positions can be performed to infer a role for the residue in enzyme structure and function.
Several proteins have been the subject of systematic amino acid substitution studies including HIV protease, CcdB protein2, T4 lysozyme3 and lac repressor4. These studies have shown that proteins are, in general, accepting of substitutions with approximately 80% of the positions tolerant of some substitutions while retaining function and buried positions are less tolerant of substitutions than surface positions. In addition, we have previously performed systematic mutagenesis studies on the TEM-1 β-lactamase that have yielded information on which residues are critical for stability, solubility and catalysis as well as which residues control the substrate specificity of the enzyme with regard to various β-lactam antibiotics5.
β-lactamases catalyze the hydrolysis of β-lactam antibiotics and thereby provide for bacterial resistance to these drugs. The TEM-1 β-lactamase efficiently hydrolyzes pencillins and many cephalosporins and is a common plasmid-encoded β-lactamase in Gram negative bacteria6.
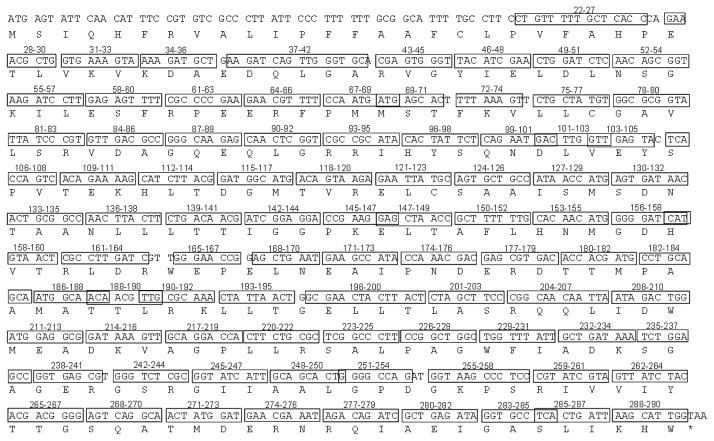
The approach taken to study the determinants of structure and function for the TEM-1 β-lactamase was to use site directed mutagenesis to randomize codons within the blaTEM-1gene to create libraries of all possible amino acid substitutions for the region randomized 5. The majority of the libraries were created by randomization of three contiguous codons to contain all 8000 (203) possible amino acid combinations for the positions randomized. This process was repeated to create 88 random libraries that encompassed the entire 263 amino acid coding region of the mature portion of TEM-1 β-lactamase (Fig. 1) 5. Each library was then introduced into E. coli and cells were spread on agar plates containing ampicillin to select for mutants with wild type levels of β-lactamase function. An average of 10 functional, ampicillin-resistant clones for each library was chosen and DNA sequencing was performed to examine the spectrum of allowable substitutions at each position 5. In this way it was possible to systematically determine the amino acid sequence requirements for TEM-1 β-lactamase folding, stability and ampicillin hydrolysis.
Figure 1.
Position of random libraries on TEM-1 β-lactamase sequence. The nucleotides randomized for each library are boxed. The amino acids randomized for each library are indicated above each box.
A key element of codon randomization and selection studies is obtaining DNA sequence information on enough functional clones to make robust conclusions about what types of amino acid replacements are functional (and which are not) for a given antibiotic selection. As more sequences are accumulated the power of the approach increases. For example, an average of 10 clones was sequenced for each library, which provides a first approximation of sequence requirements but does not allow robust statistics or a ranking of residue types. In this regard, the recent development of ultra-high-throughput sequencing technologies provides a means of obtaining orders of magnitude increases in the number of sequences for a fraction of the effort expended using standard sequencing technologies 7.
In this study, ultra-high throughput sequencing was used to sequence en masse functional clones that were selected from the 88 β-lactamase random libraries. This resulted in hundreds to thousands of sequences of functional clones from each of the libraries and thereby provided comprehensive information on the tolerance of each position in β-lactamase to substitution as well as a robust ranking of the amino acid sequence preferences at each position. The results indicate that TEM-1 β-lactamase is generally tolerant of amino acid substitutions. However, several surface positions far from the active site are sensitive to substitutions suggesting a role for these residues in enzyme stability, solubility, or catalysis. The findings also revealed a number of previously unidentified amino acid substitutions that act to increase the thermodynamic stability of TEM-1. Finally, a comparison of the mutagenesis results with sequence variability observed in an alignment of β-lactamases indicates a significant but relatively weak correlation due to many positions that do not tolerate substitutions in the mutagenesis experiments but vary in the alignment and vice versa. Taken together, the findings demonstrate that large-scale saturation mutagenesis in combination with ultra-high throughput sequencing is a powerful approach to study amino acid structure-activity relationships across the entire sequence of a protein.
Results and Discussion
Selection of functional β-lactamase sequences from random libraries
In order to make use of high throughput sequencing to study β-lactamase, functional mutants were isolated from each of the 88 β-lactamase random libraries by selecting for growth of E. coli containing the library clones on LB agar plates containing 1 mg/ml ampicillin, as was done previously (Fig. 1). This concentration of ampicillin selects for mutants with, on average, 85% of wild type β-lactamase function 5.
In this study, 454 ultra-high throughput sequencing was used for analysis of functional clones from the 88 TEM-1 random libraries 8. However, rather than sequencing individual ampicillin resistant clones from each library, approximately 1000 ampicillin resistant mutants were pooled for each of the 88 libraries. PCR was used to amplify the pooled clones in DNA fragments of the appropriate size for 454 sequencing and the PCR fragments for all of the pools were collected in three sets. DNA sequencing of the pooled sets was performed to obtain sequencing data from all of the 88 libraries (Materials and Methods)(Table S1). Approximately 700,000 sequencing reads were obtained and mutant sequences for each library were extracted from the large collection of pooled sequencing reads using custom computer programs developed to recognize the sequences from each library (Materials and Methods). After extraction and analysis, an average of 5,878 ampicillin resistant mutant sequences were obtained from each library and each library contained an average of 431 unique sequences. The maximum number of unique sequences that could be obtained for each library is approximately 1,000, i.e., the number of ampicillin resistant clones pooled for each library. The number of unique sequences for each library will depend on the stringency of sequence requirements and codon usage for the amino acids that are consistent with function. The total number of sequences as well as the number of unique sequences for each library is provided in Table S1. The error rate associated with the 454 sequencing of library clones was estimated to be 0.0237 (2.37%) as described in Materials and Methods.
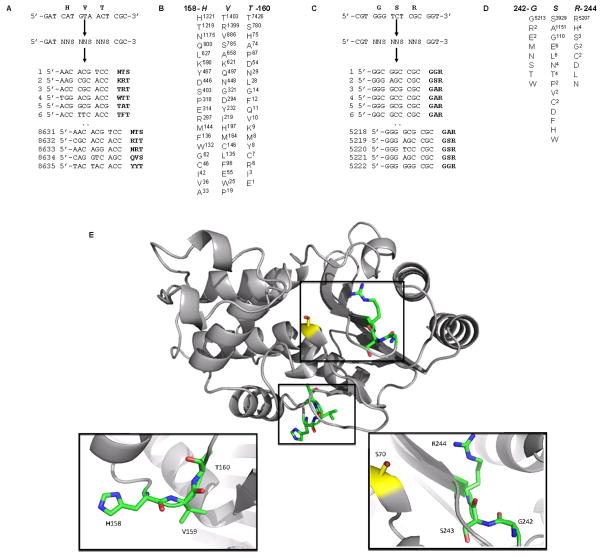
The results obtained for the two libraries encompassing residues 158-HVT-160 and 242-GSR-244 are described as examples of the data that has been obtained from the 88 libraries (Fig. 2). Residues 158-160 are far from the active site and 158 and 159 are largely surface exposed. Thus, these positions would be expected to contribute largely to protein stability and/or solubility rather than catalysis. Residues 242-244 are near the active site and Arg244 plays a role in binding the carboxylate group present on β-lactam substrates9. The selection and 454 DNA sequencing procedure resulted in data for 8635 ampicillin resistant clone sequences from the 158-160 library and 5222 from the 242-244 library. The results are summarized in Fig. 2, where the amino acids found among the functional mutants from 454 sequencing are shown below the wild type sequence. It is apparent that obtaining the sequences of thousands of functional clones for each library provides very detailed information on which residues are preferred at a position. For example, positions His158 and Val159 can be substituted by other amino acids and the β-lactamase retains high level function. In contrast, there is a strong preference for the wild type Thr at position 160 with Thr occurring 7426 times while the next most frequent amino acid, Ser, occurs 780 times among functional mutants (Fig. 2). The strong preference for Thr at position 160 can be rationalized based on the fact that Thr160 participates in a buried hydrogen bond network, the disruption of which is likely to destabilize the protein. Positions 158 and 159 are substantially surface exposed and previous results suggest surface exposed residues are relatively tolerant of amino acid substitutions 2. The data in Figure 2 also indicates that the Arg at position 244 is very strongly preferred, presumably due to its role in substrate binding. Gly at position 242 is also strongly preferred and, consistent with this finding; this residue is largely buried and is part of a β-turn structure. The wild type serine at position 243 is also preferred (3929 occurrences) but a number of alanine replacements (1151) are also found while glycine occurred 110 times. All other residue types were found less than 10 times among the functional clones (Fig. 2). This may be due to the fact that the Ser243 side chain is completely buried in the structure and its side chain forms an H-bond with that of Thr266.
Figure 2.
Ultra-high throughput sequencing of TEM-1 β-lactamase random libraries. A. 158-HVT-160 random library and list of sequences of ampicillin resistant clones obtained by 454 DNA sequencing. A total of 8635 sequence reads were obtained. B. Summary of 158-160 library sequencing results. The wild type sequence is at top and the amino acids found among ampicillin resistant clones are listed below. The number of times an amino acid type occurred is indicated by the superscript number. C. 242-GSR-244 library and list of ampicillin resistant clone sequences. D. Summary of 242-244 library sequencing results. E. Location of amino acid residues 158-160 and 242-244 on the TEM-1 β-lactamase structure (PDB code: 1BTL). A ribbon diagram of the TEM-1 structure is shown with the highlighted boxes containing a side chain view of the regions. The active site residue Ser70 is indicated in yellow.
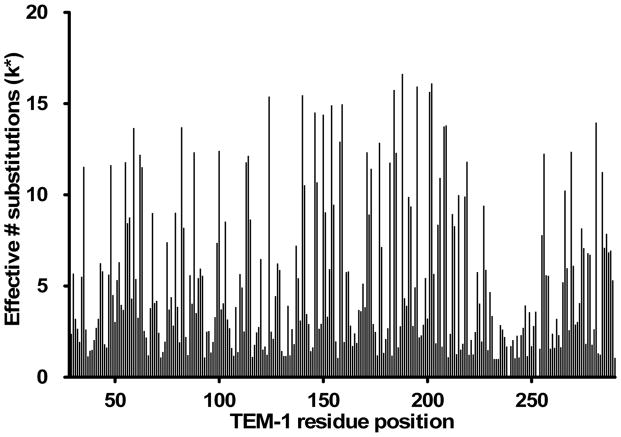
The important point from these examples is that the depth of sequencing provides a clear picture of what amino acids are favored at a position for ampicillin hydrolysis. In addition, data for all 263 positions was obtained from the sequencing of the pooled clones. The results of DNA sequencing of functional mutants selected from random libraries such as that illustrated by the 158-160 and 242-244 libraries provide a qualitative indication of the tolerance of each position to amino acid substitutions. A quantitative assessment of the sequencing data can be accomplished by calculating the effective number of amino acid types that appear at a position (k*)5,10. It is calculated from the information-theoretical entropy, S, where S is the entropy and pi is the fraction of times the ith type appears at a position and k is the number of different amino acid residue types that appear at a position10. The number of times an amino acid type appeared at a position was normalized for the number of codons encoding that particular amino acid type for the k*calculations (Materials and Methods). A k* value of one indicates complete conservation at a position, i.e., only one residue type is functional, while a value of 20 indicates all 20 amino acids occur at equal frequency. This statistic is useful in that it distinguishes between positions where multiple amino acid substitution types appear but at different frequencies. Because the sequences of functional clones from all 88 random libraries were obtained using the pooling and 454 sequencing strategy described above, the effective number of substitutions (k*) has been determined for all positions randomized in TEM-1 β-lactamase based on the ampicillin resistance selection (Fig. 3).
Figure 3.
Graph indicating the effective number of amino acid substitutions (k*) at each residue position in TEM-1 β-lactamase that are consistent with high levels of ampicillin resistance. Positions with low k* values do not tolerate substitutions while positions with high k* values can accept many different substitutions and retain high levels of function.
The quantitative assessment of the tolerance of the β-lactamase residue positions to substitutions in the form of the effective number of substitutions allows a comparison of the properties of a position with the ability to accept amino acid substitutions. Several previous studies have demonstrated that surface exposed residues in a protein are more tolerant of substitutions than buried positions2,3,5,11,12. A plot of the effective number of substitutions versus the solvent accessible surface area for TEM-1 β-lactamase reveals a weak correlation (r2=0.22, P value <0.0001) between accessible surface area and tolerance to amino acid substitutions in that surface positions generally have higher k* values, as was observed previously when only 10 clones per library were examined (Supplementary information, Fig. S1)5. The low correlation is partially due to residues in and near the active site that are solvent exposed but do not tolerate substitutions due to their role in substrate binding and catalysis.
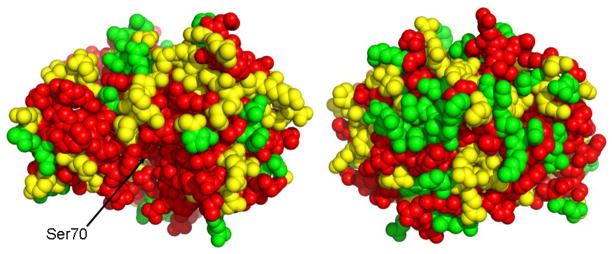
The effective number of substitutions of each position is shown mapped onto the structure of TEM-1 β-lactamase in Figure 4. It is apparent that the region in and around the active site exhibits k* values less than 5 and therefore is not tolerant of many amino acid types. This result is consistent with the important functional role of active site residues and with the fact that ampicillin is an excellent substrate and so the active site sequence is optimized for hydrolysis of this antibiotic.
Figure 4.
Summary of the effective number of amino acid substitutions (k*) on the structure of TEM-1 β-lactamase. At left is a view of enzyme with the active site serine 70 indicated with an arrow. At right is a 180o rotation of the structure. The color of the amino acid residues is based on the observed k* values. Red, k*<5; yellow, k*<10; green, k*>10.
Surface exposed residues with stringent sequence requirements
Examination of Figure 4 reveals that many surface positions outside the active site can tolerate multiple amino acid substitutions, which is consistent with previous reports2. However, it is also observed that several surface positions are not tolerant of multiple substitutions. Those positions where the effective number of substitutions does not correlate with solvent accessible surface area includes positions that are solvent accessible (>40% SAS) and yet tolerate few substitutions (k*<5) as listed in Table 1. None of these residues is directly involved in catalysis. However, several positions including D101, E104, P174, N175, E240, T271, M272, and D273 are near the active site and changes at these positions could influence substrate binding and catalysis. The remaining positions are surface exposed and are located quite distant from the active site suggesting that substitutions at these positions would impact stability or solubility, although it is possible substitutions could communicate a negative effect to the active site and reduce hydrolysis as well. Regardless of the mechanism, the results indicate that several surface positions far from the active site have stringent sequence requirements associated with their role in the structure and function of the enzyme.
Table 1.
List of TEM-1 β-lactamase residue positions that are on the surface of the enzyme (>40% solvent accessible) and not tolerant of amino acid substitutions (k*<5).
| Residue position | Effective number of substitutions (k*) | Solvent accessible surface |
|---|---|---|
| 31 | 3.20 | 80.3 |
| 53 | 3.97 | 55.7 |
| 87 | 4.03 | 79.0 |
| 93 | 1.09 | 43.4 |
| 94 | 2.5 | 46.8 |
| 96 | 1.36 | 71.6 |
| 98 | 3.30 | 54.6 |
| 101 | 3.72 | 41.9 |
| 104 | 3.17 | 73.0 |
| 111 | 4.92 | 74.0 |
| 112 | 2.51 | 45.2 |
| 121 | 1.52 | 49.2 |
| 143 | 2.92 | 58.3 |
| 156 | 1.97 | 57.3 |
| 174 | 2.92 | 63.2 |
| 175 | 2.49 | 95.8 |
| 223 | 2.48 | 42.1 |
| 226 | 1.96 | 41.3 |
| 240 | 1.71 | 68.9 |
| 252 | 3.60 | 42.7 |
| 271 | 2.89 | 75.3 |
| 272 | 3.03 | 53.4 |
| 273 | 4.07 | 80.3 |
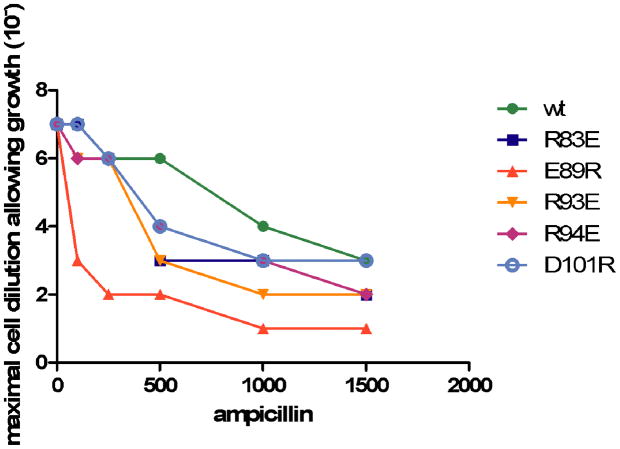
Several of the surface positions with low k* values are charged residues. The effect of substitutions at surface exposed, charged residues that are distant from the active site was investigated further by introducing single amino acid substitutions at several of these positions and measuring the effect on the ability of the mutant to confer ampicillin resistance. For this purpose, the R93E, R94E, and D101R mutants were constructed by site directed mutagenesis. The R83E and E89R substitutions were also tested, although the exposed surface area of Arg83 is slightly lower than 40% (k*= 8.2; SAS= 31.6%) and Glu89 is largely buried (k*= 3.5; SAS= 4.4%). Each of the substitutions occurs at a frequency significantly lower than wild type in the sequencing data and thus these substitutions are predicted to decrease ampicillin resistance levels of E. coli containing the mutants versus wild type. The ampicillin resistance level of E. coli containing each mutant was measured and each of the mutants retains significant ampicillin resistance but, consistent with the predictions based on substitution frequencies, each exhibits less resistance than wild type (Fig. 5). Therefore, a number of surface positions in TEM-1 are sensitive to amino acid substitutions. Previous studies have shown that optimization of charge-charge interactions on the surface of a protein can act to stabilize the protein 13. By this view, the charged surface positions that have low k* values would be predicted to participate in optimal charge-charge interactions. Alternatively or additionally, the charged surface positions could be important for maintaining the solubility of the enzyme in the periplasm of E. coli.
Figure 5.
Determination of ampicillin resistance levels of wild type and substitutions of surface charged residues in TEM-1 β-lactamase. The ampicillin resistance level of E. coli containing each mutant was measured by spotting serial dilutions of cultures containing each mutant on agar plates containing increasing concentrations of ampicillin. The maximum dilution for which growth of colonies occurred is indicated on the y-axis versus the concentration of ampicillin in the agar plates on the x-axis.
Estimating the impact of substitutions by frequency of occurrence
Deep sequencing of the ampicillin resistant clones also allows an estimate of the impact on enzyme function of any type of amino acid substitution at a position by calculating the number of times that amino acid occurs compared to the number of occurrences of the wild type amino acid at the position randomized. It has been shown previously with combinatorial libraries examining the contributions of residue positions in a protein-protein interaction using phage display technology that the frequency with which a residue appears among mutant clones after selection relative to the frequency of wild type correlates with the change in free energy of binding (ΔΔG) for the mutant versus wild type protein14,15. This statistical "ΔΔGstat" value is calculated as ΔΔGstat= RT ln (p-wt/p-mut), where p-wt and p-mut are the frequencies of occurrence of the wild type and mutant amino acid, respectively, at the position being examined 14. Note that the frequencies of occurrence of wild type and mutant amino acids were normalized for the number of codons encoding a particular amino acid type for the ΔΔGstat calculations (Materials and Methods).
The model being used for this analysis is that the probability that a colony forms is related to the concentration of cells spread on agar plates and the total activity of β-lactamase in the clone, which is related to the catalytic efficiency of the enzyme for ampicillin turnover as well as the stability and solubility of the enzyme. Unstable β-lactamase is known to be rapidly proteolyzed in E. coli, which reduces the amount of active enzyme in the cell16–18. In addition, mutations that reduce solubility result in protein aggregation, which also reduces active enzyme in the cell. It has been shown that E. coli containing a β-lactamase mutant has a certain plating efficiency on agar containing ampicillin based on its total hydrolytic activity19–21. The number of colony forming units for a mutant decreases with increasing ampicillin concentration in an agar plate and the rate of decrease is related to the activity of the mutant. A mutant with low activity may have a probability of near zero of forming colonies at high ampicillin concentrations, i.e., at concentrations above the minimum inhibitory concentration (MIC) for the mutant. This is the rationale for the ΔΔGstat calculation and the idea that the frequency with which an amino acid type is present among functional mutants is related to the level of total β-lactamase enzyme activity conferred by that amino acid. The ΔΔGstat value is clearly not a thermodynamic parameter in that it is a composite of catalytic efficiency, stability and solubility; however, it does provide a quantitative estimate of the total β-lactamase activity conferred by an amino acid substitution versus wild type for any position. The deep sequencing data from functional clones for each residue position allows for the calculation of a ΔΔGstat value for each possible single amino acid substitution in TEM-1 β-lactamase. The ΔΔGstat values for each position are provided in Table S2 and a summary of the results is shown in the form of a heat map for the entire enzyme in Figure 6.
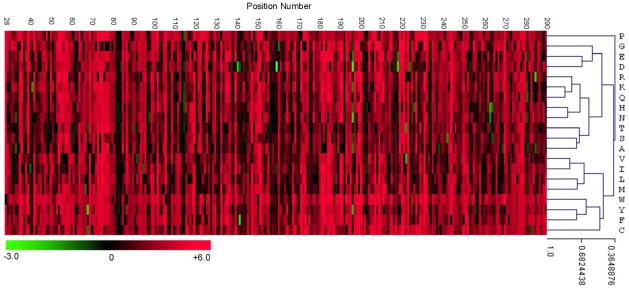
Figure 6.
Heat map representation of ΔΔGstat values for each randomized position in TEM-1 β-lactamase. The X-axis lists each residue position while the Y-axis indicates each of the two amino acid types. Each column above the residue position is therefore the set of ΔΔGstat values for each amino acid substitution. The correspondence between heat map color and ΔΔGstat is shown at lower right. Positive ΔΔGstat values indicate a residue occurs at a frequency lower than the frequency of the wild type residue (red) while a negative value indicates the residue occurs more frequently than the wild type (green) residue among the sequenced ampicillin resistant clones. The order of the amino acid rows (Y-axis) were clustered by comparison of the ΔΔGstat patterns for each amino acid type. Residues that are in the same branches of the tree exhibit similar effects on ΔΔGstat values.Pearson correlation coefficients are shown adjacent to the hierarchical tree of amino acid substitutions.
The data in the heat map in Fig. 6 were analyzed for correlations between amino acid types and their frequency of occurrence among the ampicillin resistance clones. A ΔΔGstat value is available for each amino acid type for each position in the enzyme. The correlation test asks if substitutions of certain amino acid types result in similar patterns of ΔΔGstat values across the enzyme, i.e., does the substitution of chemically similar amino acids result in similar effects on the enzyme. The similarities in patterns of ΔΔGstat values for each amino acid are indicated in the tree at the right in Fig. 6. It is apparent that amino acids with similar chemical properties exhibit similar patterns of ΔΔGstat values. For example, charged residues are clustered in the tree and within the charged cluster, arginine and lysine are in a sub-cluster and aspartate and glutamate are in a separate sub-cluster. In addition, hydrophobic residues are clustered in the tree and within the hydrophobic group the aromatic residues form a sub-cluster. Therefore, the results in Fig. 6 represent a systematic, experimental validation of the idea that conservative substitutions have a similar impact on the structure and function of an enzyme. It is interesting to note that cysteine is peripherally associated with the hydrophobic cluster while proline does not cluster with any other residues, as might be expected based on the special properties of these amino acids.
The average impact of each type of amino acid substitution on enzyme structure and function was also assessed by calculating the ΔΔGstat value for substitution of each amino acid type averaged from all residue positions in the enzyme (Supplementary information, Table S3). The results indicate that, on average, tryptophan (avg. ΔΔGstat = 3.31) and proline (3.04) are the least tolerated amino acid substitutions while threonine (2.10) and alanine (2.15) are the most tolerated substitutions. The negative impact of Trp and Pro substitutions may stem from large size of tryptophan resulting in steric clashes and the effect of proline on main chain conformation.
An interesting set of substitutions includes those where a non-wild type amino acid residue predominates compared to wild type among functional mutants in that these substitutions are predicted to result in increased levels of β-lactamase activity (Table 2). Because the wild type enzyme exhibits excellent catalytic efficiency for ampicillin hydrolysis, it seems unlikely that amino acid substitutions could improve hydrolysis rates. Alternatively, the changes could increase stability or solubility and thereby increase activity in E. coli. A genetic test was used to evaluate whether the high frequency substitutions listed in Table 2 can act to increase enzyme stability. In previous studies, we showed that an asparagine for leucine substitution (L76N) in the hydrophobic core of β-lactamase destabilizes and greatly reduces in vivo expression levels of the enzyme due to rapid proteolysis 16. Using this mutant it was possible to select a second site substitution (M182T) that stabilized the enzyme and thereby increased expression levels and ampicillin resistance levels. The M182T substitution was subsequently shown to increase the thermodynamic stability of the wild type enzyme 22. Stabilizing substitutions such as M182T in an otherwise wild type enzyme are difficult to detect genetically because they do not greatly increase the ampicillin resistance levels of the E. coli strain since the wild type enzyme is already stable and well expressed21,23. To circumvent this problem, the L76N substitution was used as a tester mutant for the ability of other substitutions to stabilize the enzyme. Because L76N is poorly expressed and provides low levels of ampicillin resistance, it is sensitive to small improvements in stability and expression levels which are reflected by easily measurable changes in ampicillin resistance levels for the E. coli strain harboring the enzyme16,19.
Table 2.
List of amino acid substitutions that occur at frequencies higher than the wild type residue among ampicillin resistant functional clones.
| Residue position | WT aa occur | WT aa adjusted occura | Non-wt aa occur | Non-wt aa adjusted occura | AMP MIC L76N double mutant (μg/ml) |
|---|---|---|---|---|---|
| 31 | V-69 | V-35 | R-2770 | R-923 | 128 |
| 33 | V-1120 | V-560 | C-2148 | C-2148 | 16 |
| 35 | D-88 | D-88 | Q-472 | Q-472 | 48 |
| 47 | I-782 | I-782 | A-4584 | A-2292 | 12 |
| 48 | E-845 | E-845 | L-3028 | L-1009 | 48 |
| 52 | N-164 | N-164 | S-1955 | S-652 | 24 |
| 53 | S-334 | S-111 | H-1705 | H-1705 | 4 |
| 60 | F-557 | F-557 | Y-1418 | Y-1418 | 128 |
| 63 | E-576 | E-576 | D-1174 | D-1174 | 12 |
| 74 | V-294 | S-3805 | 3 | ||
| 78 | G-1548 | G-774 | A-4694 | A-2347 | >256 |
| 82 | S-1497 | S-499 | H-1129 | H-1129 | 48 |
| 90 | Q-314 | Q-314 | H-1897 | H-1897 | 32 |
| 92 | G-567 | G-284 | D-1906 | D-1906 | >256 |
| 94 | R-258 | R-86 | V-2631 | V-1316 | 16 |
| 98 | S-390 | S-130 | D-4359 | D-4359 | 24 |
| 100 | N-670 | N-670 | D-1507 | D-1507 | 32 |
| 120 | R-182 | R-61 | D-3486 | D-3486 | 16 |
| 159 | V-886 | V-443 | T-1403 | T-701 | 8 |
| 174 | P-519 | P-260 | F-8088 | F-8088 | 4 |
| 175 | N-458 | N-458 | D-8268 | D-8268 | 24 |
| 201 | L-140 | L-47 | P-463 | P-232 | 64 |
| 218 | G-224 | G-112 | N-391 | N-391 | 8 |
| 219 | P-257 | P-129 | D-249 | D-249 | 24 |
| 221 | L-585 | L-195 | I-949 | I-949 | 16 |
| 225 | L-510 | L-170 | T-1114 | T-557 | 6 |
| 230 | F-611 | F-611 | Y-1466 | Y-1466 | 12 |
| 247 | I-419 | I-419 | V-1718 | V-859 | 8 |
| 249 | A-82 | A-41 | Y-1293 | Y-1293 | 2 |
| 250 | L-831 | L-277 | M-1511 | M-1511 | 4 |
| 278 | Q-335 | Q-335 | I-1464 | I-1464 | 16 |
| 285 | S-399 | S-133 | A-1243 | A-622 | 16 |
Occurences adjusted for codon usage based on the number of codons encoding each type of amino acid.
As seen in Table 2, a non-wild type residue occurred among TEM-1 β-lactamase functional mutants at significantly higher numbers than wild type at 32 positions. Each of the non-wild type substitutions from Table 2 was introduced by site-directed mutagenesis into the L76N enzyme encoded in the same plasmid as that used for the library selections and the ampicillin MIC of E. coli containing the β-lactamase double mutant was determined (Materials and Methods). Ten of the double mutants exhibited significantly higher ampicillin MICs (>24 μg/ml) than the L76N parent mutant (16 μg/ml) and thus are able to suppress the L76N stability defect, consistent with functioning as a stabilizing substitution. These substitutions include V31R, D35Q, E48L, F60Y, G78A, S82H, Q90H, G92D, N100D, and L201P. Interestingly, even though they occur at higher frequency than wild type, 22 of the substitutions did not substantially increase the ampicillin MIC of the L76N mutant and several had a negative effect on the mutant (Table 2). This result could indicate that the 22 substitutions do not act on protein stability and thus do not act to suppress that L76N stability defect. These substitutions could impact other aspects ofstructure and function such as enzyme solubility or catalytic efficiency. Alternatively, they may improve protein stability but do not enhance the stability of the L76N enzyme, i.e., they may act in an allele-specific manner. It has been previously shown that some stabilizing substitutions in β-lactamase are allele-specific in that they suppress some but not all destabilized primary mutants 21.
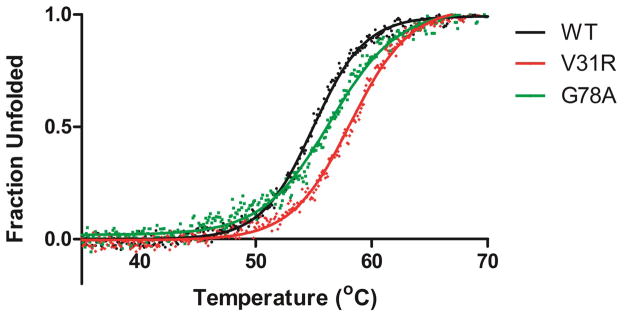
Among the 10 substitutions that do increase the ampicillin MIC of L76N, the L201P substitution has been shown to increase β-lactamase stability in several studies21,24,25. In addition, the G92D substitution was previously identified in a DNA shuffling/directed evolution experiment selecting for mutants with increased ceftazidime resistance26. Several of the remaining TEM-1 mutants including V31R, E48L, F60Y, G78A, S82H, as well as theG92D enzymewere constructed as single substitutions in the wild type enzyme and expressed from E. coli and purified for further characterization. The thermodynamic stability of each of the enzymes was determined by monitoring the folded state with increasing temperature using circular dichroism spectroscopy (Fig. 7, Table 3)19. It was found that all of the enzymes displayed increased thermal stability relative to the wild type enzyme, which is consistent with the substitutions serving as suppressors of the L76N mutant and is also in line with the hypothesis that the observed increased frequency of these substitutions relative to wild type is due to increased stability of the enzyme (Table 3).
Figure 7.
Thermal denaturation curves of wild type TEM-1 and selected β-lactamase variants obtained from circular dichroism measurements at increasing temperatures. Fractional changes in the CD signal are shown for the wild-type (black), V31R (red), and G78A (green).
Table 3.
Thermodynamic parameters for TEM-1 β-lactamase and substituted enzymes.
| Enzyme | Tm(°C) | ΔTma | ΔH (kcal/mol) | ΔS (kcal/mol*K) | ΔΔGu (kcal/mol) |
|---|---|---|---|---|---|
| WT | 54.9 ± 0.04 | --- | 87.2 ± 1.4 | 0.27 ± 0.004 | --- |
| V31R | 58.2 ± 0.07 | 3.2 | 91.1 ± 1.3 | 0.28 ± 0.004 | 0.85 |
| E48L | 55.3 ± 0.06 | 0.3 | 82.0 ± 0.9 | 0.25 ± 0.003 | 0.08 |
| F60Y | 57.5 ± 0.05 | 2.6 | 89.8 ± 1.2 | 0.27 ± 0.004 | 0.68 |
| G78A | 56.4 ± 0.06 | 1.5 | 75.7 ± 1.2 | 0.23 ± 0.004 | 0.39 |
| S82H | 57.1 ± 0.06 | 2.2 | 79.7 ± 0.9 | 0.24 ± 0.003 | 0.58 |
| G92D | 59.0 ± 0.08 | 4.1 | 71.3 ± 1.3 | 0.22 ± 0.004 | 1.09 |
Change relative to wild-type TEM-1
It is possible that altered catalytic parameters of the substituted enzymes could also influence the frequency at which mutants occurred relative to wild type. This possibility was tested by determining the kinetic parameters for hydrolysis of several β-lactam antibiotics for the V31R, E48L, F60Y, G78A, S82H and G92D enzymes to examine the impact of the substitutions on catalysis (Table 4). It was found that the kcat, Km and catalytic efficiency (kcat/Km) values for hydrolysis of ampicillin, nitrocefin and cephalosporin C were very similar to the wild type values for all enzymes tested. Therefore, consistent with their location far from the active site, the substitutions do not alter the catalytic activity of the enzymes. Taken together, the results suggest that the high frequency of the substitutions relative to wild type after the ampicillin selection is due to increased stability of the enzymes, which would be predicted to increase the half-life and expression levels of β-lactamase in the periplasm of E. coli.
Table 4.
Kinetic parameters of TEM-1 wild type and enzymes containing stabilizing substitutions.
| Substrates | Enzymes | k cat (s-1) a | km(μM) a | kcat /Km (s-1/μM) |
|---|---|---|---|---|
| Ampicillin | ||||
| WT | 1768 | 53 | 34 | |
| V31R | 1428 | 43 | 33 | |
| E48L | 1436 | 44 | 33 | |
| F60Y | 1845 | 46 | 40 | |
| G78A | 2114 | 61 | 34 | |
| S82H | 1473 | 37 | 40 | |
| G92D | 2519 | 51 | 50 | |
| Nitrocefin | ||||
| WT | 1493 | 69 | 22 | |
| V31R | 1538 | 84 | 18 | |
| E48L | 1309 | 87 | 15 | |
| F60Y | 1721 | 91 | 19 | |
| G78A | 1928 | 87 | 22 | |
| S82H | 1682 | 96 | 17 | |
| G92D | 2767 | 87 | 32 | |
| Cephalosporin C | ||||
| WT | 49 | 499 | 0.098 | |
| V31R | 53 | 518 | 0.10 | |
| E48L | 44 | 418 | 0.10 | |
| F60Y | 59 | 577 | 0.10 | |
| G78A | 48 | 402 | 0.12 | |
| S82H | 43 | 539 | 0.080 | |
| G92D | 66 | 542 | 0.12 | |
The standard errors of the kcat and Km values are ≤25%
The location of the ten substitutions (V31R, D35Q, E48L, F60Y, G78A, S82H, Q90H, G92D, N100D, and L201P) that were found to increase the ampicillin resistance levels of the TEM-1 L76N mutant are shown in Supplemental Fig. S2. They are dispersed widely on the structure of the enzyme and occur at various distances from L76N and also exhibit a range of values for solvent accessible surface area (Table S4). Thus, there is not an obvious trend in location or side chain characteristics that distinguishes the stabilizing substitutions.
The V31R and G92D stabilizer mutants are of particular interest with regard to the discussion above on the optimal distribution of charged surface residues. Both of the mutants involve the introduction of a new charged residue at the surface of the protein that results in stabilization of the protein. Molecular modeling of the substitutions suggests both the V31R and G92D substitutions would introduce favorable new charge-charge interactions on the surface of the enzyme. In the wild type enzyme Val31 is 80% solvent exposed and the V31R substitution could introduce a new salt bridge with the side chain of residue Glu28. In the wild type enzyme the Glu28 side chain does not interact with other TEM-1 residues. Gly92 is 55% solvent exposed and the Asp substitution would be highly solvent exposed. Depending on the side chain rotamer, the Asp carboxyl could interact with the guanidinium group of Arg94 or Arg120 and/or hydrogen bond with the side chain of Asn90. Thus, the V31R and G92D substitutions are predicted to optimize charge-charge and hydrogen bonding interactions on the surface of the enzyme.
The TEM-1 β-lactamase has been the subject of intense study with regard to protein structure, function and evolution and a number of substitutions have been identified that stabilize the enzyme including P62S, V80I, G92D, R120G, E147G, H153R,M182T, L201P, I208M, A184V, A224V, I247V, T265M, R275L/Q, and N276D19,24,25. With the exception of G92D and L201P, these mutants were not among the positions in Table 2 that contained substitutions that appeared at a higher frequency than wild type after the ampicillin resistance selection. There could be multiple reasons for this but one explanation lies in how the randomization experiments were performed. The libraries were constructed by randomizing three and in a few cases more than three, positions while none of the libraries were randomized at a single position (Fig. 1). Randomizing multiple positions could influence the frequency at which certain substitutions appear. In fact, randomizing multiple positions rather than a single position is likely what allowed the detection of any stabilizing substitutions. It is known that stabilizing substitutions such as M182T when introduced into the wild type enzyme do not greatly increase ampicillin resistance because the wild type enzyme is already very stable16,23. Thus, if a single position were randomized, there would be no ampicillin resistance advantage for mutants with increased stability and the frequency of these substitutions would not be greater than wild type. In contrast, when three positions are simultaneously randomized, a stabilizing substitution at one position can act as a suppressor of destabilizing substitutions at other positions to increase ampicillin resistance and therefore will be found at a higher frequency than wild type among the sequenced clones. Therefore, the observation that a substitution exists at a higher frequency than wild type is a good indication that it alters the properties of the enzyme such as increasing stability, but deep sequencing of the libraries will have false negatives, i.e., the failure to identify a stabilizing substitution such as M182T by the frequency of substitutions could be due to particular characteristics of the neighboring residues in the library.
It is also worth noting that, by the same argument as that above, the randomization of three codons could influence the frequency at which certain substitutions appear in this experiment in that if a substitutions at neighboring position in the same library can increase enzyme stability it could alter the spectrum of substitutions observed at a position compared to if that position were randomized alone.
Comparison of information from deep sequencing versus an alignment of class A β−lactamases
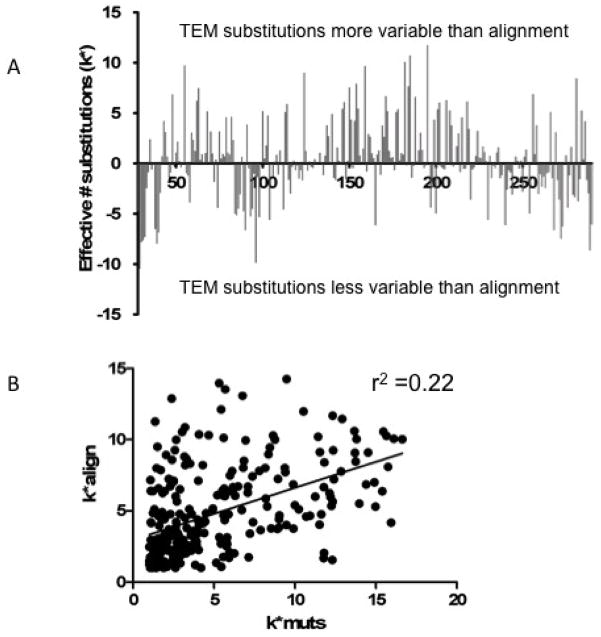
As indicated above, deep sequencing of functional clones from random libraries provides detailed information on sequence requirements at any given position. An interesting question is whether the same information is obtained from analysis of sequence conservation in an alignment of sequences from a protein family. We have previously assembled and aligned a collection of 156 class A β-lactamases including the TEM-1 enzyme27. This alignment was used to calculate the effective number of substitutions at each position (k*) as well as a ΔΔGstat value for each substitution at each position using TEM-1 as the reference sequence (Table S5). The results of the k* determinations revealed many positions in β-lactamase that differ substantially for tolerance to amino acid substitutions in the deep sequencing versus protein alignment based calculations as seen in Figure 8A. Thus, there are many positions that exhibit stringent sequence requirements (low k*) based on the deep sequencing and yet are not strongly conserved (high k*) in the alignment and vice versa (Fig. 8A). As a result, a plot of k* values obtained from deep sequencing versus alignment reveals a significant, but relatively weak correlation (r2=0.22, P value<0.0001) (Fig. 8B).
Figure 8.
Comparison of the effective number of substitutions (k*) determined from TEM-1 β-lactamase mutagenesis experiments versus an alignment of class A β-lactamase sequences. A. Bar graph indicated the difference in k* between the mutagenesis experiments and the class A enzyme alignment. The values shown are k* mutagenesis - k* alignment. B. Plot of k* values determined based on the class A alignment versus the random mutagenesis results. Analysis reveals a correlation coefficient r2 of 0.22 with a P value <0.0001.
An examination of positions where k* differs substantially between deep sequencing and alignment results reveals possible explanations for the observed differences in the mutants versus the family. A large percentage of the residues that exhibit more variation among the mutants than in the alignment are surface exposed positions that exhibit conservation of charge or hydrophilicity in the alignment. The average solvent accessible surface area of residues with a difference in k* values from mutagenesis versus the alignment of >7 is 50.6% (Table S5). Some examples that differ in k* by ≥9 are TEM-1 positions Lys55 (k*mut 11.8, k*align 2.0), Glu63 (k*mut 11.5, k*align 4.0), Ser124 (k*mut 15.4, k*align 6.4), and Thr195 (k*mut 15.9, k*align 4.2) (Tables S2, S5). Within the class A β-lactamase alignment, position 55 is most often deleted and among the remaining enzymes is largely Arg or Lys, which results in the low k* score for the alignment (Table S5). In the mutagenesis experiment, position 55 is often substituted by charged residues but also other hydrophilic residues which leads to a much higher k* score (Table S2). Similarly, Glu63 is on the surface of TEM-1 and is substituted by a number of residue types in the mutagenesis experiment (Table S2). In the class A family, however, the position is dominated by Asp, Glu, and Asn residues leading to a low k* value (Table S5). Ser124 is partially surface exposed in TEM-1 and the side chain is oriented so that substitutions will extend into solvent. The position is substituted most often by Asp or Asn but many other residues are observed among the mutants (Table S2). In contrast, position 124 is dominated by Ala or charged residues in the class A β-lactamase alignment which results in a low k* value (Table S5). Finally, Thr195 is largely surface exposed on TEM-1 and is substituted by a number of residues in the mutagenesis experiments while position 195 is often occupied by Leu or Val in the class A alignment resulting in a low k* value. Surface exposed residues that are far from the active site are often freely substituted in mutagenesis experiments. The low sequence variability observed for these positions in the class A alignment could reflect the sequence requirements to maintain solubility of the individual class A enzymes that differ from the requirements for TEM-1 β-lactamase.
The explanation for the positions where the variability of the sequences observed among the selected mutants is significantly lower (low k*) that the variability in the class A enzyme alignment (high k*) appears highly case-specific (Table S5). For example, position 31 is a surface exposed valine in TEM-1 and exhibits a k* value among mutants of 3.2 versus 10.9 in the class A alignment. The relatively low k* value for the mutants is due to the dominance of Arg31 mutants among the functional clones (Table 2, Table S2). The dominance of Arg31 is presumably due to the strong stabilizing effect of the arginine substitution (Fig. 7). Other substitutions at position 31 may be consistent with wild type levels of function but they are outcompeted by the Arg mutant.
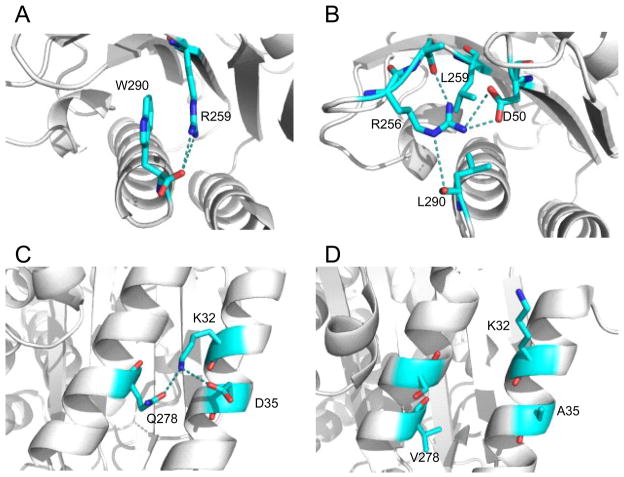
In other cases, it appears that the residue of interest occupies an environment that is unique to TEM-1 and makes an important contribution to enzyme structure and function. An example is the carboxy-terminal Trp290 residue in TEM-1 that fills a large hydrophobic cavity and is ideally positioned for a cation-pi interaction with Arg259. This position exhibits a low k* value (1.1) for the mutants but a high value (7.2) based on the class A alignment (Tables S2, S5). The Arg259 guanidinium group is also positioned to form a salt bridge with the terminal carboxylate of Trp290 (Fig. 9). Within the class A alignment position 290 is dominated by hydrophobic residues but many different types are observed including Leu, Ile, Val, Tyr, Ala and Met which results in a higher k* value. Arg259 also has a low k* value (1.6) in mutagenesis experiments and a modestly higher value of 4.6 for the alignment. Interestingly, Arg at 259 is rare in the alignment, which is dominated by Leu, Ile and other hydrophobics. An example of the structure of the position 259-290 environment in another class A enzyme (CTX-M-16) is shown in Fig. 9 where leucine is present at both 259 and 290 and the residue interactions are strikingly different than those observed in TEM-128. Thus, the environment of the Trp290 side chain in TEM-1 is unique in the class A family and explains the sensitivity of the positions to substitutions in the mutagenesis experiments but not in the alignment.
Figure 9.
Comparison of TEM-1 and CTX-M-16 β-lactamase structures at positions 32 and 290 showing the difference in residue environment in the different class A enzymes. A. Molecular environment of TEM-1 β-lactamase residues Trp290 and Arg259 (PBP ID 1BTL). B. Molecular environment of residue 290 in CTX-M-16 β-lactamase (PDB ID 1YLW). C. Environment of TEM-1 β-lactamase residue Lys32. D. Environment of CTX-M-16 β-lactamase residue 32.
Another example of a residue that is not substituted in the mutagenesis experiments but varies among class A enzymes involves Lys32 in the TEM-1 enzyme (Fig. 9). This position exhibits a low k* (2.7) in mutagenesis experiments but a higher value (10.0) in the class A alignment. This appears to be due to interactions that occur between Lys32 and Asp35 and Gln278 in TEM-1 that do not occur in other class A enzymes such as CTX-M-16 as shown in Fig. 928. Therefore, a unique environment in TEM-1 whereby residues that are not conserved in the class A family make important interactions for TEM-1 structure and function appears several times and may be a common reason for the difference between k* values in TEM-1 versus the gene family.
Statistical ΔΔG values were also calculated for each possible substitution at each residue position based on the alignment of 156 class A β-lactamases using the TEM-1 sequence as a reference (Table S5). The ΔΔGstat and k* values are related in that positions with low k* values exhibit high (unfavorable) ΔΔGstat for most substitutions and positions with high k* display ΔΔGstat values near zero or negative for individual substitutions. A comparison of ΔΔG values calculated from the mutagenesis versus the class A alignment reveals a significant but relatively weak correlation (r2=0.25, P value <0.0001) as observed for the correlation of k* values. The reason for the weak correlation is similar to that for k*, i.e., there are multiple residue positions that are more tolerant of substitutions in TEM-1 mutagenesis experiments versus conservation in the alignment and vice versa. The explanations provided for the k* observations also apply for ΔΔGstat values. For example, in the TEM-1 mutagenesis experiments, all substitutions are highly deleterious at Trp290 as indicated by ΔΔGstat values >4.0 and all substitutions except lysine at Arg259 exhibit ΔΔGstat values ≥3.0 (Table S2). In contrast, there is a wide range ΔΔGstat values for positions 259 and 290 in the class A alignment with Ile and Leu displaying negative (favorable) values for both 259 and 290. These observations are consistent with the unique environment at position 259-290 in the TEM-1 enzyme that, although not conserved in class A enzymes, is nevertheless important for the structure and function of the enzyme (Fig. 9). This observation may explain why the number of residues that do not tolerate amino acid substitutions (k*<2) is higher in the mutagenesis experiments (63) compared to those from the class A enzyme alignment (49).
Finally, it is worth noting that some differences in the variability of positions in the alignment versus the mutagenesis experiments could be that the natural sequences are phylogenetically related and so are not independent of each other while in the mutagenesis experiments the substitutions are independent of one another and an amino acid type will not appear frequently simply due to phylogenetic descent.
Conclusion
The use of deep sequencing of combinatorial libraries is a powerful method of exploring the structure-function and evolution of a protein. Fowler et al described a similar approach to study the structure and function of a human WW domain by selecting functional clones from large combinatorial libraries by phage display followed by ultra high throughput sequencing 29. In addition, the fitness effects of single amino acid substitutions over a nine residue region of Hsp90 in yeast have been examined using deep sequencing of random libraries 30. The ΔΔGstat values calculated in this study allow a quantitative comparison of the impact of each type of amino acid substitution at a given position with respect to ampicillin resistance, i.e., fitness of E. coli containing the mutant. Because the 454 sequencing experiment encompassed all 88 libraries, ΔΔGstat values are available for each of the 19 possible amino acid substitutions for each of the 263 positions in the mature TEM-1 β-lactamase (Fig. 6, Table S2). This provides a great deal of information about the effect of amino acid substitutions on TEM-1, and, more generally, on protein structure and function, which can be mined to explore questions of the impact of substitutions on stability, solubility and organismal fitness. For example, this study has shown that impact of substitutions on protein structure and function correlated with the chemical properties of the amino acids. The study has also provided the average impact of a substitution by each type of amino acid in a protein showing that tryptophan has the most deleterious and threonine the least deleterious effect when introduced into a protein.
Natural variants of TEM-1 β-lactamase that are capable of hydrolyzing extended spectrum cephalosporins such as ceftazidime or β-lactamase inhibitors such as clavulanic acid have emerged in the past twenty years and are a common source of drug resistance in Gram negative bacteria6. The random libraries and deep sequencing described here can be used to determine the sequence requirements and evolutionary potential of TEM-1 or other β-lactamases for hydrolysis of these drugs by replacing the ampicillin selection with a selection for ceftazidime or a β-lactam antibiotic-inhibitor combination followed by deep sequencing of the functional clones.
Materials and Methods
TEM-1 β-lactamase random libraries
The 88 β-lactamase random libraries used for this study were constructed previously5. The libraries were constructed in the pBG66 plasmid which encodes blaTEM-1 and cat and therefore provides resistance to ampicillin and chloramphenicol. Eleven of these libraries were constructed by random replacement mutagenesis and the relevant codons were replaced with NNN (where N is any of the 4 nucleotides A,C,G,T), while the remaining 77 libraries were constructed using oligonucleotide directed mutagenesis by the method of Kunkel and the codons were replaced with NNS, where S represents C or G5,31,32. The libraries constructed by random replacement mutagenesis include 22-27, 37-42, 69-71, 72-74, 103-105, 161-164, 165-167, 168-170, 196-200, 238-241, and 251-254 (Fig. 1) 32. Because the random replacement method results in the randomization of an even number of nucleotides, some codons in these libraries are not completely randomized. These include the codons for residues Pro22, Pro27, Glu37, Ala42, Thr71, Val74, Tyr105, Arg164, Trp165, Glu168, Gly196, Arg241, and Asp254.
Selection of functional mutants on ampicillin agar plates and pooling
To select for functional random mutants, each of the 88 random plasmid libraries contained in E. coli XL1-Blue was used to inoculate 5 ml of LB medium with 12.5 μg/ml chloramphenicol and was grown overnight at 37 °C. Then 1.2 ml of the overnight culture was diluted into different series: 1:10, 1:20, 1:40, 1:80, etc., and the diluted samples were spread on LB agar plates containing 1 mg/ml ampicillin incubated overnight at 37°C. Approximately 1000 clearly isolated single colonies for each library were then pooled together and plasmid DNA was isolated using ZyppyTM Plasmid Miniprep kit (Zymo Research) for further amplification and high-throughput sequencing.
The 454 GS FLX Titanium Series Amplicon Sequencing platform was used for high-throughput sequencing. The blaTEM gene region covered by 88 random libraries includes 800 bps, which were divided into three sections. Section 1 contains libraries 22-27 to 109-111 (269 bps, 29 libraries). Section 2 contains libraries from 112-114 to 196-200 (267 bps, 30 libraries) (Fig. 1). Section 3 contains libraries 201-203 – 288-290 (264 bps, 29 libraries). The plasmid DNA after selection from each library was PCR amplified according to sections. Libraries in the same section were amplified using the same PCR primers to amplify top and bottom strands of sequence plus the adapter tags for 454 Titanium sequencing (Supplementary information, Fig. S3). The primers used are listed in Supplementary Information, Table S6.
The 454 sequencing platform requires 5 – 10 μg DNA for a single run. To fulfill the requirement, 50 μL of PCR reaction for each section top and bottom strand were performed. The section 2 amplified libraries (from 112-114 to 196-200, 30 libraries including top and bottom strands) were mixed and 454 sequencing was performed at the Human Genome Sequencing Center (HGSC) at Baylor College of Medicine. The total DNA sample was approximately 10 μg. After obtaining long, highly accurate sequencing reads for the pooled section 2 PCR products, the process was repeated with section 1 and section 3 DNA combined in a single 454 run using the same methods.
Data processing and analysis of 454 sequencing reads
Because the libraries were pooled for sequencing, it was necessary to extract the mutant sequences for each library from the large collection of sequence reads. A custom Perl script was developed to extract the mutant sequences for each library by using matches to the sequences 5 base pairs (bp) upstream and downstream of the library as the frame. For example, the 5 upstream bases of the 220-222 library are GACCA, the 5 downstream bases are TCGGC and the length of the 220-222 library is 9 bps. Any read that contains a 9 bp sequence with the GACCA and TCGGC flanking the 9 bp is extracted by the script and translated into amino acid sequence. In the program, only the sequences in accordance with NNSNNSNNS (where N indicates A, T, C or G, and S indicates C or G) are extracted, i.e., the program also requires a C or G at the 3rd position in each codon. The NNS rule is enforced because NNS codons were designed into the mutagenic oligonucleotides for the randomization experiments5. Because this is required for the three consecutive codons in the library, it effectively eliminates the extraction of wild type, non-mutagenized sequences because the wild type sequences for the positions randomized in the libraries do not have C or G at the 3rd position for each codon.
Eleven libraries (22-27, 37-42, 69-71, 72-74, 103-105, 161-164, 165-167, 168-170, 196-200, 238-241, and 251-254) were constructed using a different randomization procedure in which the codons were randomized with NNN rather than NNS32. For these libraries, the program was adjusted to extract the mutated sequences while filtering the wild type sequences. For instance, 165-167 was randomized as the following, 5’-GAT CGT TNN NNN NNN GAG CTG. The program captured any reads that match TCGTT at the 5’ end and GAGCT at the 3’end while excluding wild type sequence GGGAACCG in the randomized region. It is worth noting that there are only 2400 possible amino acid combinations in 165-167 because the number of possible amino acids sequences for TNN is 6. In addition, because of the degeneracy of the genetic code, there are 7 nucleotide sequences consistent with a wild type amino acid sequence for this example and so the true wild type nucleotide sequence (which is filtered out by the program) represents only 1/7 of the wild type amino acid sequences present in the library.
Each round of 454 sequencing returned two files – the FASTA sequence file (.fna) and the quality score file (.qual). A total of 366,316 reads were obtained from the first round of 454 sequencing (section 2 libraries). The average length of the sequences was 407 bp. A total of 353,920 reads were obtained from the second round of sequencing (section 1 & 3 libraries combined together). The average length was 357 bp. The number of sequencing reads was limited by the usage of the 454 picotiter plate. For GS FLX Titanium Series, the PicoTiterPlate (PTP) Device contains 3.5 million wells (in practice, no more than one-half of the wells are occupied with beads). In our experiment, 1/8 of the 454 PTP was used in each round of sequencing.
The .qual file contained Phred equivalent quality scores associated with each base pair in the sequence file. Since the errors usually occur at the ends of the reads, the distribution of errors was not random. Most of the quality scores for the library sequencing were over 30 (meaning > 99.9% accuracy) while the scores dropped steadily near the ends of the reads. In order to compensate for the sequencing errors near the ends and to avoid bias, the complementary strand was also sequenced from 5’ end to 3’end. In addition, since the library mutant sequences were only extracted if they contained exact matches to the 5 base pairs flanking either side of the library, sequences from poor quality reads were largely eliminated.
DNA sequencing error rate
The error rate associated with 454 DNA sequencing of the pooled mutant libraries was estimated by taking advantage of the fact that the each mutant library represents a small window of sequence (9 bp) in a read that is otherwise wild type sequence. DNA sequencing errors were detected and the mutation rate was determined by first excluding the randomized region and the 5 bp upstream and 5 bp downstream to obtain the sequences outside the randomized window for each read. These sequences were then compared with the wild-type blaTEM-1 gene sequence template and the number unmatched base pairs and matched base pairs at each nucleotide position was determined and the frequency of mutations was calculated from number of unmatched bases divided by the total number of bases sequenced for each nucleotide position. This number provides an estimate of the probability of errors occurring at each position in the TEM-1 gene. The total number of unmatched bases divided by the total number sequenced was calculated to estimate the total error rate. The total error rate was estimated to be 0.0237 (2.37%). The regions making the largest contributions to error were homopolymeric tracts of nucleotides as well as the regions near the middle of each PCR section which contain relatively more ends of reads from the sequencing using primers from either end of the PCR fragment.
k-star calculations
The effective number of substitutions at each positions (k*) was calculated using the method of Shenkin using the equation below10:
where S is the entropy, pi stands for the fraction of times the ith type appears at a position and k is the number of different amino acid residue types that appear at a position. Note that for the k* calculations, the value for fraction of times an amino acid residue type appears at a position (pi) was adjusted for the number of codons encoding the amino acid type. For example, the large majority of the libraries were constructed using NNS codons which results in a 32 codon genetic code table. The number of occurrences of Arg, Leu, and Ser sequences was divided by three to account for three different codons for these amino acids. The number of Ala, Gly, Pro, Thr, and Val sequences was divided by two to account for two codons for these amino acids. The number of sequences for the remaining amino acids was not adjusted because there is only one codon for each when using NNS codons. For those eleven libraries using NNN codons (see above), the number of occurrences of each amino acid type was adjusted using the appropriate codon numbers.
Heat map construction and correlations of amino acid substitutions
Predicted ΔΔGstat values for each amino acid substitution for every position on TEM-1 were compiled into a two-color heat map using Multiexperiment Viewer33. The pairwise similarity of all each of the 20 amino acid substitutions was calculated as the Pearson correlation coefficient of these calculated ΔΔGstat values for all positions on TEM-1. Amino acid substitutions were grouped by hierarchical clustering using the average linkage method in Multiexperiment Viewer34.
Protein purification and kinetic analysis
The TEM-1 β-lactamase and its substituted variants were purified to >95% homogeneity and the kinetic parameters were determined as previously described19. Substrate hydrolysis was observed for ampicillin, nitrocefin, and cephalosporin C with a DU800 spectrophotometer at wavelengths 235nm, 482 nm and 280 nm, respectively. These experiments were performed in 50 mM sodium phosphate buffer at 30°C. Bovine serum albumin was added to the buffer at a concentration of 1 mg/mL for the nitrocefin substrate. The initial velocities were determined and fitted to the Michaelis-Menton equation using GraphPad Prism5. The initial velocities were measured in at least duplicate trials to determine the kinetic parameters.
Thermal denaturation
The thermostability of the β-lactamases was determined as previously described19. The β-lactamases were first buffer exchanged into 50 mM potassium phosphate. The far-UV CD signal at 223 nM was measured with a JASCO-810 CD spectrophotometer as the sample was increased in temperature from 35°C to 70°C at a rate of 2°C/min increments. The β-lactamases were shown to refold in that 95% of the signal was recovered when the sample was cooled back to 35°C at a protein concentration of 1.5μM. These experiments were performed in at least triplicate. The melting temperatures (Tm) were determined by fitting the fraction denaturation to a Boltzmann sigmoidal curve. The changes in enthalpy and entropy were determined by the fitting of the Van't Hoff equation, ln K = − H/RT + S/R, using GraphPad Prism5. The Becktel and Schellman method, ΔΔGu = ΔTmΔSWT, was used to calculate the ΔΔGu35.
Ampicillin resistance determinations
The ampicillin resistance levels of mutants was assessed by minimum inhibitory concentration determinations or by assaying the highest dilution at which colonies grew on ampicillin agar plates using a spot test. Ampicillin MIC measurements were performed using E-test strips or by ampicllin broth dilutions, as described previously. The spot test experiment was performed by serial dilution of cultures that had been grown overnight in LB medium at 37oC to saturation phase. The serial dilutions were done in a final volume of 200 μl in a 96-well microtiter plate. A total of 10 μl of each dilution was spotted onto agar plates containing increasing concentrations of ampicillin. This procedure follows that described by Foit et al20.
Supplementary Material
Acknowledgments
This research was supported by NIH grant AI32956 to T.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Benkovic SJ, Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj K, Chakrabarti P, Varadarajan R. Mutagenesis-based definitions and probes of residue burial in proteins. Proc Natl Acad Sci USA. 2005;102:16221–16226. doi: 10.1073/pnas.0505089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rennell D, Bouvier SE, Hardy LW, Poteete AR. Systematic mutation of bacteriophage T4 lysozyme. J Mol Biol. 1991;222:67–88. doi: 10.1016/0022-2836(91)90738-r. [DOI] [PubMed] [Google Scholar]
- 4.Suckow J, Markiewicz P, Kleina LG, Miller JH, Kisters-Woike B, Muller-Hill B. Genetic studies of the Lac repressor. XV: 4000 single amino acid substitutions and analysis of the resulting phenotypes on the basis of the protein structure. J Mol Biol. 1996;261:509–523. doi: 10.1006/jmbi.1996.0479. [DOI] [PubMed] [Google Scholar]
- 5.Huang W, Petrosino J, Hirsch M, Shenkin PS, Palzkill T. Amino acid sequence determinants of β-lactamase structure and activity. J Mol Biol. 1996;258:688–703. doi: 10.1006/jmbi.1996.0279. [DOI] [PubMed] [Google Scholar]
- 6.Perez F, Endimiani A, Hujer KM, Bonomo RA. The continuing challenge of ESBLs. Curr Opin Pharmacol. 2007;7:459–469. doi: 10.1016/j.coph.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Bubnoff A. Next-generation sequencing: The race is on. Cell. 2008;132:721–723. doi: 10.1016/j.cell.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 8.Mea Margulies. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zafaralla G, Manavathu S, Lerner A, Mobashery S. Elucidation of the role of arginine-244 in the turnover processes of class A beta-lactamases. Biochemistry. 1992;31:3847–3852. doi: 10.1021/bi00130a016. [DOI] [PubMed] [Google Scholar]
- 10.Shenkin PS, Erman B, Mastrandrea LD. Information-theoretical entropy as a measure of sequence variability. Proteins: Struc Funct Genet. 1991;11:297–313. doi: 10.1002/prot.340110408. [DOI] [PubMed] [Google Scholar]
- 11.Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS. The stability effects of protein mutations appear to be universally distributed. J Mol Biol. 2007;369:1318–1332. doi: 10.1016/j.jmb.2007.03.069. [DOI] [PubMed] [Google Scholar]
- 12.Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19:596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Gribenko AV, Patel MM, Liu J, McCallum SA, Wang C, Makhatadze GI. Rational stabilzation of enzymes by computational redesign of surface charge-charge interactions. Proc Natl Acad Sci USA. 2009;106:2601–2606. doi: 10.1073/pnas.0808220106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pal G, Kouadio J-LK, Artis DR, Kossiakoff AA, Sidhu SS. Comprehensive and quantitative mapping of energy landscapes for protein-protein interactions by rapid combinatorial scanning. J Biol Chem. 2006;281:22378–22385. doi: 10.1074/jbc.M603826200. [DOI] [PubMed] [Google Scholar]
- 15.Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci USA. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang W, Palzkill T. A natural polymorphism in β-lactamase is a global suppressor. Proc Natl Acad Sci USA. 1997;94:8801–8806. doi: 10.1073/pnas.94.16.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsell DA, Sauer RT. The structural stability of a protein is an important determinant of its proteolytic susceptibility in. Escherichia coli J Biol Chem. 1989;264:7590–7595. [PubMed] [Google Scholar]
- 18.Sideraki V, Huang W, Palzkill T, Gilbert HF. A secondary drug resistance mutation of TEM-1 beta-lactamase that suppresses misfolding and aggregation. Proc Natl Acad Sci USA. 2001;98:283–288. doi: 10.1073/pnas.011454198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown NG, Pennington JM, Huang W, Ayvaz T, Palzkill T. Multiple global suppressors of protein stability defects facilitate the evolution of extended-spectrum TEM β-lactamases. J Mol Biol. 2010;404:832–846. doi: 10.1016/j.jmb.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foit L, Morgan GJ, Kern MJ, Steimer LR, von Hacht AA, Titchmarsh J, Warriner SL, Radford SE, Bardwell JCA. Optimizing protein stability in vivo. Mol Cell. 2009;36:861–871. doi: 10.1016/j.molcel.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marciano DC, Pennington JM, Wang X, Wang J, Chen Y, Thomas VL, Shoichet BK, Palzkill T. Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 beta-lactamase. J Mol Biol. 2008;384:151–164. doi: 10.1016/j.jmb.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 2002;320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 23.Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- 24.Bershtein S, Goldin K, Tawfik DS. Intense neutral drifts yield robust and evolvable consensus proteins. J Mol Biol. 2008;379:1029–1044. doi: 10.1016/j.jmb.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 25.Kather I, Jakob RP, Dobbek H, Schmid FX. Increased folding stability of TEM-1 beta-lactamase by in vitro selection. J Mol Biol. 2008;383:238–251. doi: 10.1016/j.jmb.2008.07.082. [DOI] [PubMed] [Google Scholar]
- 26.Orencia MC, Yoon JS, Ness JE, Stemmer WP, Stevens RC. Predicting the emergence of antibiotic resistance by directed evolution and structural analysis. Nat Struct Biol. 2001;8:238–242. doi: 10.1038/84981. [DOI] [PubMed] [Google Scholar]
- 27.Marciano DC, Brown NG, Palzkill T. Analysis of the plasticity of location of positive charge within the active site of the TEM-1 β-lactamase. Protein Sci. 2009;18:2080–2089. doi: 10.1002/pro.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Delmas J, Sirot J, Shoichet BK, Bonnet R. Atomic resolution structures of CTX-M beta-lactamases: extended spectrum activities from increased mobility and decreased stability. J Mol Biol. 2005;348:349–362. doi: 10.1016/j.jmb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 29.Fowler DM, Araya CL, Fleishman SJ, Kellogg EH, Stephany JJ, Baker D, Fields S. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010;7:741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hietpas RT, Jensen JD, Bolon DN. Experimental illumination of a fitness landscape. Proc Natl Acad Sci U S A. 2011;108:7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 32.Palzkill T, Botstein D. Probing β-lactamase structure and function using random replacement mutagenesis. Proteins. 1992;14:29–44. doi: 10.1002/prot.340140106. [DOI] [PubMed] [Google Scholar]
- 33.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati NK, et al. TM4: An open source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 34.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Meth Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 35.Becktel WJ, Schellman JA. Protein stability curves. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.