Abstract
Autophagy is a bulk degradation system, widely conserved in eukaryotes. Upon starvation, autophagosomes enclose a portion of the cytoplasm and ultimately fuse with the vacuole. The contents of autophagosomes are degraded in the vacuole, and recycled to maintain the intracellular amino-acid pool required for protein synthesis and survival under starvation conditions. Previously, autophagy was thought to be an essentially nonselective pathway, but recent evidence suggests that autophagosomes carry selected cargoes. These studies have identified two categories of selective autophagy – one highly selective and dependent on autophagy-related 11 (Atg11); another, less selective, that is, independent of Atg11. The former, selective category comprises the Cvt pathway, mitophagy, pexophagy and piecemeal microautophagy of the nucleus; acetaldehyde dehydrogenase 6 degradation and ribophagy belong to the latter, less selective category. In this review, I focus on the mechanisms and the physiological roles of these selective types of autophagy.
Keywords: selective autophagy, autophagosome, ATG genes, yeast, Atg11
Facts
Autophagy is a bulk degradation system widely conserved in eukaryotes and especially induced upon starvation.
Under starvation conditions, autophagosomes enclose a portion of the cytoplasm and the contents of autophagosomes are degraded in the vacuole.
Autophagy is essentially a nonselective pathway but carries a number of selective cargoes.
There are two categories of selective autophagy – one highly selective and dependent on autophagy-related 11 (Atg11); another, less selective, that is independent of Atg11.
Atg11 functions by connecting cargo−receptor complexes and organelles, with Atg proteins essential for autophagosomal membrane biogenesis.
Open questions
Physiological roles of selective autophagy are not fully understood.
The mechanism of how each type of selective autophagy is induced is mostly unknown.
The molecular mechanisms underlying Atg11-independent selective autophagy remain to be addressed.
Autophagy is a bulk degradation system widely conserved in eukaryotes.1, 2 Under starvation conditions, autophagosomes enclose a portion of the cytoplasm and the contents of autophagosomes are degraded in the vacuole. Autophagy is essentially a nonselective pathway but carries a number of selective cargoes.
The first selective autophagy cargo to be identified was the vacuolar aminopeptidase I (Ape1). Ape1 is synthesized in precursor form (prApe1) and subsequently processed in the vacuole to its mature form (mApe1).3 This biosynthetic pathway, which occurs under nutrient-rich conditions, was named the cytoplasm-to-vacuole targeting (Cvt) pathway. Mutants defective in the maturation of Ape1 were screened to obtain cvt mutants.4 Around the same time, other groups identified mutants defective in starvation-induced autophagy, termed apg (autophagy) and aut (autophagocytosis).5, 6 apg and aut mutants have a phenotype similar to that of the cvt mutants,7 suggesting that the Cvt and autophagy pathways share some common machinery. Electron microscopy revealed that in the Cvt pathway, prApe1 is exclusively enclosed in double-membrane-bound organelles called Cvt vesicles, which are topologically similar to autophagosomes. However, the two compartments are of different sizes, ∼150 nm for Cvt vesicles and ∼500 nm for autophagosomes.8, 9
Peroxisome degradation mediated by autophagy has been described by several groups, and a number of the genes involved were named independently, for example, GSA,10 PAZ,11 and PDD.12 This list includes genes essential for peroxisome degradation as well as some required for bulk autophagy. To avoid confusion, the nomenclature was later consolidated; the genes are now collectively referred to as ATG genes.13
Atg11 as a Scaffold Protein for Selective Autophagy
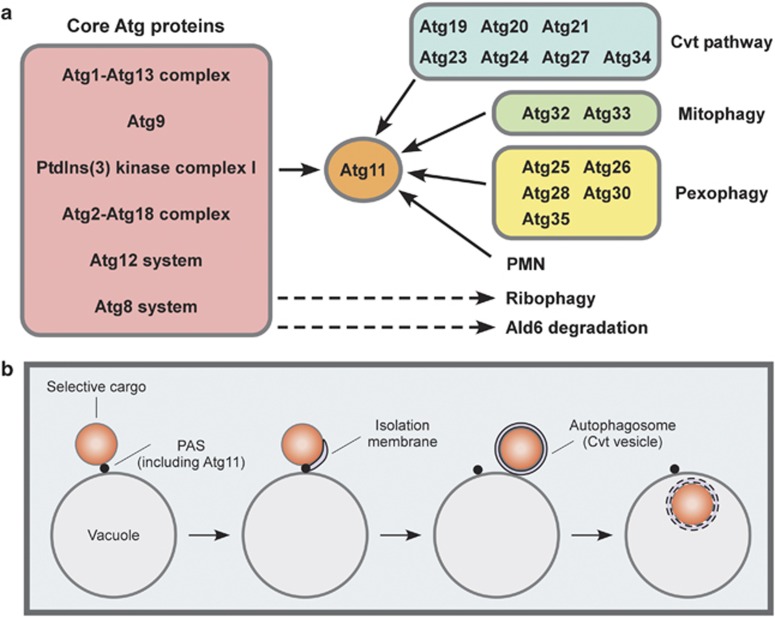
The pre-autophagosomal structure (PAS) mediates the membrane biogenesis of Cvt vesicles/autophagosomes.14, 15 In atg11Δ cells, the core Atg proteins, a subgroup of Atg proteins responsible for membrane biogenesis of Cvt vesicles/autophagosomes,16 are not targeted to the PAS under nutrient-rich conditions, leading to a defect in the Cvt pathway.17, 18 As PAS localization of the core Atg proteins is largely abolished in atg17Δ cells under starvation conditions, Atg17 is thought to function as a scaffold protein for bulk autophagy.19 In atg17Δ cells, Ape1 transport to the vacuole is normal under nutrient-rich conditions.20, 21 During starvation, Ape1 maturation is partially defective in atg11Δ and atg17Δ cells, indicating that Ape1 maturation depends on both Atg11 and Atg17 during autophagy.22 When ATG11 and ATG17 are both deleted, PAS formation is completely abolished, resulting in a total block in Ape1 maturation.22 Thus, Atg11 is involved in Ape1 maturation under both nutrient-rich and -starvation conditions, via its role in organization of the PAS. Moreover, Atg11 is important for other types of selective autophagy, such as mitophagy (selective degradation of mitochondria by autophagy), pexophagy (selective degradation of peroxisomes by autophagy) and piecemeal microautophagy of the nucleus.18, 23, 24 Cargoes degraded by Atg11-independent selective autophagy have also been reported.
Atg11-dependent Selective Autophagy
Atg11 is required for various types of selective autophagy. It functions by connecting cargo−receptor complexes and organelles with core Atg proteins essential for autophagosomal membrane biogenesis. In the following section, I provide an overview of Atg11-dependent selective autophagy.
The Cvt pathway
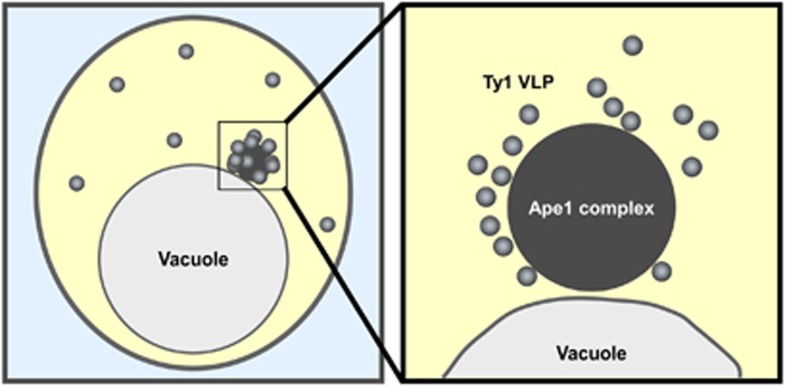
The Cvt pathway, a constitutive biosynthetic pathway mediated by Cvt vesicles, is responsible for the transport of the Cvt complex under nutrient-rich conditions.8 Similarly, under starvation conditions, autophagy facilitates transport of the Cvt complex. prApe1, the major component of the Cvt complex, is synthesized, oligomerized to a dodecamer, and assembled to form the Ape1 complex, which constitutes the core structure of the Cvt complex.9, 25, 26 The Cvt complex is morphologically defined by microscopy (Figure 1). In addition to the Ape1 complex, the Cvt complex contains vacuolar α-mannosidase (Ams1) and Ty1 virus-like particles (VLPs).27, 28, 29 Ty1 VLPs, which are produced by Ty1 retrotransposons in the yeast genome, can be observed by electron microscopy as particles surrounding the Ape1 complex.29 Atg19 and Atg34 receptors are also required for organization of the Cvt complex.30, 31 In the absence of Atg19, the Ape1 complex and the Ty1 VLPs are both localized to the cytoplasm, but are not associated with one another, preventing their selective delivery to the vacuole27, 28, 29 Atg19 is peripherally localized to the Ape1 complex,32 and may promote the association of Ty1 VLPs with the Ape1 complex as well as target the Cvt complex to the PAS. During starvation, either Atg19 or Atg34 is sufficient to target Ams1 to the Cvt complex.31 Thus, the Cvt complex is composed of cargo (prApe1, Ams1 and Ty1 VLP) and receptors (Atg19 and Atg34). The physiological role of Cvt complex transport to the vacuole is not well understood. The vacuolar enzymes may have a role in protein turnover. It has been reported that selective Ty1 VLP degradation is involved in maintaining genome integrity during starvation by decreasing the frequency of Ty1 transposition.29 Ape4 (aspartyl aminopeptidase) and Lap3 (leucine aminopeptidase) are also delivered to the vacuole by selective autophagy through association with the Cvt complex.33, 34
Figure 1.
Schematic of the Cvt complex. The Cvt complex is localized near the vacuole in S. cerevisiae (left panel). Ty1 VLPs (gray) are associated with the Ape1 complex, the main component of the Cvt complex (dark gray). These two structures can be observed by electron microscopy
Mitophagy
When yeast cells are cultured in media containing a nonfermentable carbon source, such as glycerol or lactate, they shift from anaerobic to aerobic respiration. As the latter condition places oxidative stress on the mitochondria, stress-related damage can occur. As a quality control step to eliminate damaged mitochondria in post-log phase cells, mitochondria-specific autophagy (mitophagy) is induced under these conditions. Mitophagy also occurs when cells grown under aerobic respiration conditions are transferred to nitrogen-starvation medium. In addition to the core Atg proteins, Atg11 and Atg32 are required for mitophagy (Figure 2).23 Atg32 is anchored to the outer mitochondrial membrane, and confers selectivity for mitochondrial sequestration by recruiting autophagic machinery through interactions with Atg8 and Atg11.35, 36, 37 When mitophagy is induced, Atg32 is phosphorylated; this modified form of Atg32 is able to bind Atg11. Hog1 and Pbs2, kinases involved in the osmoregulatory signal transduction cascade, have a role in Atg32 phosphorylation.38 Moreover, Atg33, another mitophagy-specific protein, has an important role in mitophagy in post-log phase cells.39
Figure 2.
Multiple pathways of selective autophagy. (a) Atg11 is a key component of a number of selective autophagy routes, including the Cvt pathway, mitophagy, pexophagy and PMN. By contrast, Atg11 is dispensable for ribophagy and preferential Ald6 degradation. ‘Core' Atg proteins function in the membrane biogenesis of Cvt vesicles/autophagosomes. (b) Schematic of Atg11-dependent selective autophagy. Atg11 functions as a scaffold protein for PAS formation by recruiting core Atg proteins. Autophagosomes are generated from the PAS and, subsequently, delivered to the vacuole, where the selected cargoes are degraded
Mitophagy maintains mitochondrial quality by eliminating damaged mitochondria. This physiological role of mitophagy was confirmed using atg11Δ and atg32Δ cells. When these mitophagy-deficient cells are faced with nutrient starvation, mitochondria damaged by exposure to reactive oxygen species (ROS) are not degraded. These damaged mitochondria lose their mitochondrial DNA, and host cells exhibit the ‘petite' phenotype, an indication that aerobic respiration has been compromised.40 Bulk autophagy is also important for maintenance of mitochondrial quality. When cells grown in fermentable medium with glucose as the sole carbon source are transferred to nitrogen-starvation medium, autophagy-defective cells mostly die within 5 days. In nitrogen-starvation medium that has been buffered at neutral pH, autophagy-defective cells can survive, but the majority demonstrate the ‘petite' phenotype.41 This may occur because autophagy-defective cells cannot scavenge ROS accumulated in the mitochondria. Autophagy-defective cells cannot produce ROS-scavenging enzymes, likely due to a shortage of free amino acids for de novo protein synthesis.42 In contrast, atg32Δ cells do not show the petite phenotype.41 Together, these different types of autophagy may have complementary roles in the maintenance of mitochondrial quality, for example, with bulk autophagy serving as a preventive measure to preserve mitochondrial activity by allowing synthesis of ROS-scavenging enzymes, while mitophagy eliminates damaged mitochondria to prevent their harmful effects.
Pexophagy
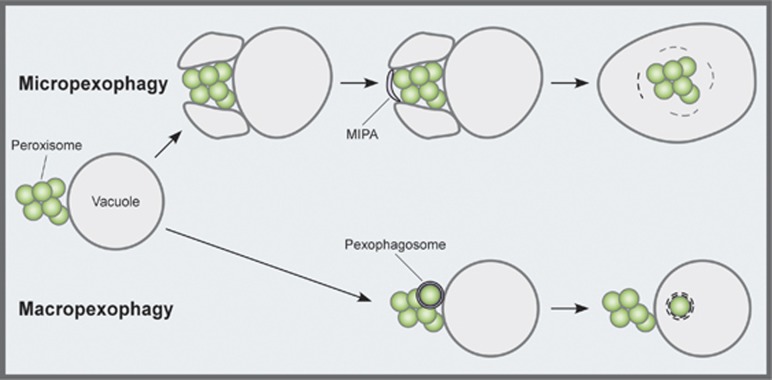
Peroxisome degradation by selective autophagy, called pexophagy, has been studied in several methylotrophic yeasts, such as Pichia pastoris, Hansenula polymorpha and Yarrowia lipolitica.43, 44 When these cells are grown in a methanol medium, they synthesize peroxisomes, which form clusters. Subsequent transfer of the cells to glucose medium induces a process called micropexophagy (Figure 3): first, the vacuolar membrane begins to enwrap the cluster; next, a cup-shaped membrane structure termed the micropexophagic membrane apparatus (MIPA) emerges on the cluster's open surface.45 Enclosure of the peroxisome cluster is completed by fusion between the MIPA and the vacuolar membrane, resulting in transport of the peroxisomes into the vacuole. Conversely, when cells grown in methanol are transferred to an ethanol medium, individual peroxisomes are enclosed in special autophagosomes, termed pexophagosomes, and delivered to the vacuole one by one. This type of pexophagy is termed macropexophagy (Figure 3). Core Atg proteins are required for both micro- and macropexophagy. Several other factors are specifically required for both types of pexophagy, including PpAtg26, a sterol glucosyltransferase; PpAtg28, a coiled-coil protein; and PpAtg30, a receptor protein.46, 47, 48 Moreover, PpAtg35 and HpAtg25 are specifically required for micropexophagy and macropexophagy, respectively.49, 50
Figure 3.
Two modes of pexophagy. When methylotrophic yeasts are grown in a methanol-containing medium, peroxisomes develop and form a cluster. Transferring the cells to glucose medium induces a process called micropexophagy, whereby the vacuolar membrane begins to enwrap the whole cluster (top). Subsequently, a cup-shaped membrane structure, the MIPA, emerges on the open surface of the peroxisome cluster. Transfer from methanol to an ethanol-containing medium induces a process called macropexophagy, during which, peroxisomes are enclosed in autophagosomes, termed pexophagosomes, and delivered to the vacuole one by one (bottom)
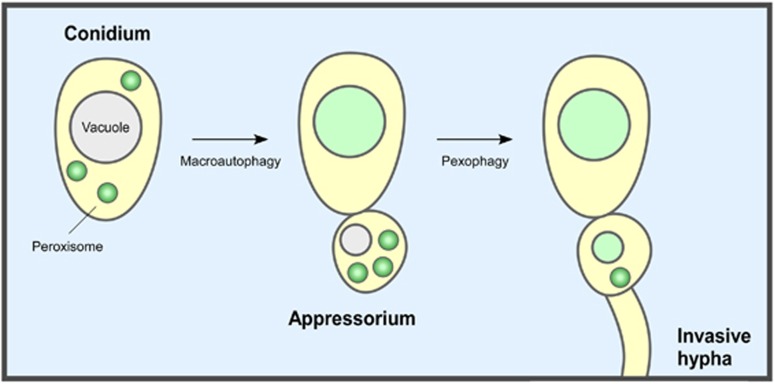
Peroxisome biogenesis is necessary for the pathogenicity of the plant fungus Colletotrichum orbiculare, which causes disease in cucumber plants.51 A recent study showed that pexophagy has an important role in pathogenicity (Figure 4).52 An insertional mutation library screen identified the CoATG26 gene as critical for pathogenicity. To invade host plants, C. orbiculare develops an infection structure termed the appressorium. In Coatg26 mutant cells, the biogenesis of peroxisomes is normal, but these structures accumulate in the appressoria, indicating that pexophagy is defective. Furthermore, Coatg26 mutants exhibit a specific defect in invasion. Upon infection, ring- or cup-shaped structures labeled with GFP-CoAtg8 are detected along peroxisomes, suggesting that macropexophagy has been induced in the appressoria. Domain and localization analyses of CoAtg26 show that both the phosphoinositide binding and sterol glucosyltransferase activities displayed by this enzyme are required for infection-related pexophagy. By contrast, normal appressoria do not differentiate in Coatg8 mutant cells, which are defective in both bulk autophagy and pexophagy. Together, these different autophagic pathways have complementary roles in the pathogenicity of the fungus: bulk autophagy is required for the early stages of infection-related C. orbiculare morphogenesis, and pexophagy is needed for later stages of infection occurring after development of appressoria.
Figure 4.
Differentiation of C. orbiculare during host invasion. Macroautophagy is required for germination and development of the appressorium from the conidium (asexual spore). Pexophagy in the appressorium is necessary for the subsequent development of the invasive hypha
Piecemeal microautophagy of the nucleus
When yeast cells are faced with nutrient limitations, a dispensable portion of the nucleus is protruded into the vacuolar lumen as a teardrop-shaped bleb and subsequently pinched off. The resultant vesicles are sequestered by three lipid bilayers and degraded by vacuolar hydrolases. This process is termed piecemeal microautophagy of the nucleus (PMN).24 PMN occurs at the nucleus−vacuole junction (NVJ) formed by interactions between Nvj1, localized to the outer nuclear membrane, and Vac8, localized to the vacuolar membrane.53 Nvj1 and Vac8 are required for PMN.24, 54 Moreover, an electrochemical gradient across the vacuolar membrane and lipid-modifying enzymes are necessary for PMN.55 Because Nvj1 is degraded in a PMN-dependent manner, progression of this phenomenon can be monitored by observing the degradation of Nvj1-GFP.54 As with mitophagy and pexophagy, Atg11 and core Atg proteins are essential for PMN.54 PMN also requires Atg17, Atg29 and Atg31, which are essential for starvation-induced autophagosome formation, but not for other types of selective autophagy.54 An understanding of the physiological roles of PMN in eukaryotic cells will hopefully emerge from future studies.
Atg11-independent Selective Autophagy
Previously, it was believed that abundant cytoplasmic components are enclosed in autophagosomes nonselectively. However, it has recently been reported that cytosolic acetaldehyde dehydrogenase (Ald6) and ribosomes are degraded by preferential autophagy, which is less selective autophagy than Atg11-dependent selective autophagy, in an Atg11-independent manner.
Acetaldehyde dehydrogenase 6
Ald6, a soluble cytoplasmic enzyme, was identified in a systematic differential proteomic screen of wild-type and autophagy-deficient yeast cells as a protein that is degraded by autophagy after nitrogen starvation for 24 h.56 This degradation depends on core Atg proteins and active vacuolar proteases, but not on known selective autophagic factors such as Atg11. Ald6 is preferentially enclosed in autophagosomes, then delivered to the vacuole for degradation. When active Ald6 accumulates in the cytosol, the viability of nitrogen-starved cells decreases. Accumulation of inactive Ald6 has little effect on viability; therefore, it may be that the enzymatic activity of Ald6 negatively impacts survival under starvation conditions, leading to rapid death of autophagy-defective cells. The molecular mechanisms underlying this preferential autophagy remain to be addressed.
Ribophagy
The observation of ribosomal degradation by autophagy was considered to support the hypothesis that autophagy is nonselective. Nevertheless, a recent report indicates that ribosomes are preferentially degraded by autophagy.57 Upon nutrient starvation, ribosomes are degraded along with other cytoplasmic components. In Saccharomyces cerevisiae, this degradation involves a novel type of selective autophagy termed ribophagy. As ribophagy is independent of Atg19, it is unlikely that Atg11 is important for this pathway. A genome-wide screen of a set of nonessential gene disruptants demonstrated that the Ubp3/Bre5 deubiquitination complex is involved in ribophagy.57 Moreover, this complex interacts with the Cdc48/Ufd3 complex, which has an important role in the ubiquitin-proteasome system.58 However, defects in proteasomal degradation do not greatly impact ribophagy. In cells defective in ribophagy, autophagic pathways other than ribophagy appear to be normal. A functional relationship between ubiquitination and ribophagy may exist, but the molecular mechanisms of ribophagy remain unknown.
Conclusion and Perspectives
Selective autophagy provides cells with multiple means to protect themselves against severe environmental conditions. Atg11 is a protein that functions as a scaffold for a group of selective autophagy pathways, including the Cvt pathway, mitophagy, pexophagy and PMN. Atg11 seems to be involved in the selective transport of protein aggregates and organelles. During Atg11-dependent selective autophagy, targets are associated with Atg11, which recruits autophagic machinery to the cargo by interacting with a variety of receptor proteins, for example, Atg19 and Atg34 (Cvt pathway); Atg32 (mitophagy); and PpAtg30 (pexophagy).31, 35, 36, 48, 59 These receptor proteins confer selectivity on the Atg11-dependent targets. By contrast, Ald6 and ribosomes are selectively degraded by Atg11-independent mechanisms, which exhibit weaker selectivity than Atg11-dependent mechanisms. Receptors for Ald6 and ribosomes have not yet been identified. Other mechanisms may confer selectivity to these cargoes. For instance, Ald6 and ribosomes might interact with the inner surface of the isolation membrane, or form a loose complex at the PAS.
Recently, autophagosome-associated proteins have been identified from human breast cancer cells.60 Future analysis of autophagosomal cargoes in yeast cells will hopefully elucidate the mechanisms of selective autophagy in eukaryotic cells.
Acknowledgments
I thank Dr. Yoshinori Ohsumi for helpful comments on this manuscript. This work was supported by the Hamaguchi Foundation for the Advancement of Biochemistry, the NOVARTIS Foundation (Japan) for the Promotion of Science and Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Glossary
- ATG genes
autophagy-related genes
- Ape1
aminopeptidase I
- prApe1
Ape1 precursor form
- mApe1
mature Ape1
- Cvt
cytoplasm-to-vacuole targeting
- PAS
pre-autophagosomal structure
- VLP
virus-like particles
- ROS
reactive oxygen species
- MIPA
micropexophagic membrane apparatus
- PMN
piecemeal microautophagy of the nucleus
- NVJ
nucleus−vacuole junction
The author declares no conflict of interest.
Footnotes
Edited by M Piacentini
References
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Tuttle DL, Shi YJ, Ralph GS, Dunn JrWA. Glucose-induced microautophagy in Pichia pastoris requires the alpha-subunit of phosphofructokinase. J Cell Sci. 1997;110:1935–1945. doi: 10.1242/jcs.110.16.1935. [DOI] [PubMed] [Google Scholar]
- Mukaiyama H, Oku M, Baba M, Samizo T, Hammond AT, Glick BS, et al. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells. 2002;7:75–90. doi: 10.1046/j.1356-9597.2001.00499.x. [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Keizer I, Harder W, Veenhuis M. Isolation and characterization of mutants impaired in the selective degradation of peroxisomes in the yeast Hansenula polymorpha. J Bacteriol. 1995;177:357–363. doi: 10.1128/jb.177.2.357-363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn JrWA, Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y. Current knowledge of the pre-autophagosomal structure (PAS) FEBS Lett. 2010;584:1280–1286. doi: 10.1016/j.febslet.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang CW, Klionsky DJ. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P, Moshitch-Moshkovitz S, Kvam E, O'Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda MN, Scott SV, Hefner-Gravink A, Caffarelli AD, Klionsky DJ. Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J Cell Biol. 1996;132:999–1010. doi: 10.1083/jcb.132.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Scott SV, Oda MN, Klionsky DJ. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kamada Y, Ohsumi Y. Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev Cell. 2002;3:815–824. doi: 10.1016/s1534-5807(02)00359-3. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Morimoto M, Kondo C, Ohsumi Y. Selective autophagy regulates insertional mutagenesis by the Ty1 retrotransposon in Saccharomyces cerevisiae. Dev Cell. 2011;21:358–365. doi: 10.1016/j.devcel.2011.06.023. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Noda NN, Kumeta H, Suzuki K, Ohsumi Y, Inagaki F. Selective transport of α-mannosidase by autophagic pathways: structural basis for cargo recognition by ATG19 and ATG34. J. Biol Chem. 2010;285:30026–30033. doi: 10.1074/jbc.M110.143545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kondo C, Morimoto M, Ohsumi Y. Selective transport of α-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J Biol Chem. 2010;285:30019–30025. doi: 10.1074/jbc.M110.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales Quinones M, Stromhaug PE. The propeptide of Aminopeptidase 1 mediates aggregation and vesicle formation in the Cytoplasm-to-vacuole targeting pathway. J Biol Chem. 2012;287:10121–10133. doi: 10.1074/jbc.M111.311696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuga M, Gomi K, Klionsky DJ, Shintani T. Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. J Biol Chem. 2011;286:13704–13713. doi: 10.1074/jbc.M110.173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama T, Suzuki K, Ohsumi Y. Lap3 is a selective target of autophagy in yeast, Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;378:551–557. doi: 10.1016/j.bbrc.2008.11.084. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo-Okamoto N, Noda NN, Suzuki SW, Nakatogawa H, Takahashi I, Matsunami M, et al. Autophagy-related protein 32 acts as autophagic degron and directly initiates mitophagy. J Biol Chem. 2012;287:10631–10638. doi: 10.1074/jbc.M111.299917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Kanki T, Hirota Y, Kurihara Y, Saigusa T, Uchiumi T, et al. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol Biol Cell. 22:3206–3217. doi: 10.1091/mbc.E11-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Kanki T, Aoki Y, Hirota Y, Saigusa T, Uchiumi T, et al. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J Biol Chem. 2012;287:3265–3272. doi: 10.1074/jbc.M111.280156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SW, Onodera J, Ohsumi Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS One. 2011;6:e17412. doi: 10.1371/journal.pone.0017412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- Veenhuis M, van der Klei IJ, Titorenko V, Harder W. Hansenula polymorpha: an attractive model organism for molecular studies of peroxisome biogenesis and function. FEMS Microbiol Lett. 1992;79:393–403. doi: 10.1111/j.1574-6968.1992.tb14068.x. [DOI] [PubMed] [Google Scholar]
- Dunn JrWA, Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, et al. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Mukaiyama H, Baba M, Osumi M, Aoyagi S, Kato N, Ohsumi Y, et al. Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol Biol Cell. 2003;15:58–70. doi: 10.1091/mbc.E03-05-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku M, Warnecke D, Noda T, Muller F, Heinz E, Mukaiyama H, et al. Peroxisome degradation requires catalytically active sterol glucosyltransferase with a GRAM domain. EMBO J. 2003;22:3231–3241. doi: 10.1093/emboj/cdg331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasyk OV, Stasyk OG, Mathewson RD, Farre JC, Nazarko VY, Krasovska OS, et al. Atg28, a novel coiled-coil protein involved in autophagic degradation of peroxisomes in the methylotrophic yeast Pichia pastoris. Autophagy. 2006;2:30–38. doi: 10.4161/auto.2226. [DOI] [PubMed] [Google Scholar]
- Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarko VY, Nazarko TY, Farre JC, Stasyk OV, Warnecke D, Ulaszewski S, et al. Atg35, a micropexophagy-specific protein that regulates micropexophagic apparatus formation in Pichia pastoris. Autophagy. 2011;7:375–385. doi: 10.4161/auto.7.4.14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrska I, Kiel JA, Krikken AM, Komduur JA, Veenhuis M, van der Klei IJ. The Hansenula polymorpha ATG25 gene encodes a novel coiled-coil protein that is required for macropexophagy. Autophagy. 2005;1:92–100. doi: 10.4161/auto.1.2.1832. [DOI] [PubMed] [Google Scholar]
- Kimura A, Takano Y, Furusawa I, Okuno T. Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell. 2001;13:1945–1957. doi: 10.1105/TPC.010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura M, Ninomiya S, Sugimoto M, Oku M, Yamashita S, Okuno T, et al. Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Plant Cell. 2009;21:1291–1304. doi: 10.1105/tpc.108.060996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Roberts P, Chen Y, Kvam E, Shulga N, Huang K, et al. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell. 2000;11:2445–2457. doi: 10.1091/mbc.11.7.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R, Muehe Y, Prick T, Bremer S, Schlotterhose P, Eskelinen EL, et al. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell. 2008;19:4492–4505. doi: 10.1091/mbc.E08-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawaliby R, Mayer A. Microautophagy of the nucleus coincides with a vacuolar diffusion barrier at nuclear-vacuolar junctions. Mol Biol Cell. 21:4173–4183. doi: 10.1091/mbc.E09-09-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y. Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae. J Biol Chem. 2004;279:16071–16076. doi: 10.1074/jbc.M312706200. [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bonizec M, Cohen M, Dokudovskaya S, Delalande F, Schaeffer C, et al. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep. 2010;11:548–554. doi: 10.1038/embor.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengjel J, Hoyer-Hansen M, Nielsen MO, Eisenberg T, Harder LM, Schandorff S, et al. Identification of autophagosome-associated proteins and regulators by quantitative proteomic analysis and genetic screens. Mol Cell Proteomics. 2012;11:014035. doi: 10.1074/mcp.M111.014035. [DOI] [PMC free article] [PubMed] [Google Scholar]