Abstract
Ubiquitination has long been recognised as a key determinator of protein fate by tagging proteins for proteasomal degradation. Most recently, the ability of conjugated ubiquitin chains to confer selectivity to autophagy was demonstrated. Although autophagy was first believed to be a bulk, non-selective ‘self-eating' degradative process, the molecular mechanisms of selectivity are now starting to emerge. With the discovery of autophagy receptors – which bind both ubiquitinated substrates and autophagy specific light chain 3 (LC3) modifier on the inner sheath of autophagosomes – a new pathway of selective autophagy is being unravelled. In this review, we focus on the special role of ubiquitin signals and selective autophagy receptors in sorting a variety of autophagic cargos.
Keywords: ubiquitin, autophagy, mitophagy, Optineurin, phosphorylation
Facts
The ubiquitin–proteasomal system (UPS) and the ubiquitin-like system (UBL) have functional similarity.
Ubiquitination is frequently a prerequisite for substrate recognition and determines selectivity in autophagy in higher eukaryotes.
Crosstalk between ubiquitination and autophagy is provided by autophagic adaptor proteins (or autophagy receptors), which bind both ubiquitin and autophagy-specific UBL modifiers (LC3 (light chain 3)/GABARAP (gamma-aminobutyric acid receptor-associated protein)).
To date, seven autophagy receptors have been identified, which specifically regulate the selective autophagosomal degradation of large protein aggregates, mitochondria and bacterial pathogens.
Open Questions
What are the spatio-temporal mechanisms governing the dynamics of selective autophagosomal degradation?
Are there regulatory feedback loops, whereby autophagy activity affects ubiquitination?
What are the molecular basics that regulate selective autophagy under certain conditions?
Are there germline or somatic mutations in key autophagy regulators that are causative for disease?
What is the interplay between selective autophagy pathways and macroautophagy during starvation?
Ubiquitin, a small protein consisting of 76 amino acids, is found in all tissues of eukaryotic organisms. With only three amino-acid differences between mammals, yeast and plants, ubiquitin displays a remarkable evolutionary conservation.1 It acts as a modifier by covalent attachment to cellular proteins through an enzymatic cascade, which involves three classes of enzymes termed E1 (activation), E2 (conjugation) and E3 (ligation).1 The process of tagging a protein with ubiquitin is called ubiquitination and is one of the most versatile known cellular regulatory mechanisms for controlling physiological and pathological cellular events. Discovered in the 1970s it took several decades to elucidate that ubiquitination serves as the ‘kiss of death' signal for protein turnover. More recently, it has been shown that ubiquitination also regulates key cellular processes including gene transcription, cell cycle progression, DNA repair, apoptosis, virus budding and receptor endocytosis. Among the three major protein degradation pathways in eukaryotes, ubiquitination constitutes the main denominator by targeting substrates to (i) the proteasome, (ii) the lysosome and as a part of the lysosomal system, (iii) the autophagosome.
Autophagy is a catabolic degradative process delivering cytoplasmic components to the lysosome.2 This ancient and highly conserved pathway occurs through one of the three different routes, that is, macroautophagy, microautophagy and chaperone-mediated autophagy (CMA). Macroautophagy describes the process of forming first a lunate-shaped membrane structure (the phagophore) leading to a LC3-containing vacuole (the autophagosome). Subsequently, the cargo is sequestered before being delivered for lysosomal degradation (Figure 1). By contrast, in microautophagy and CMA cytosolic components are directly incorporated into the lysosomes. This review focuses mainly on macroautophagy and – unless specified otherwise – is referred to as autophagy.
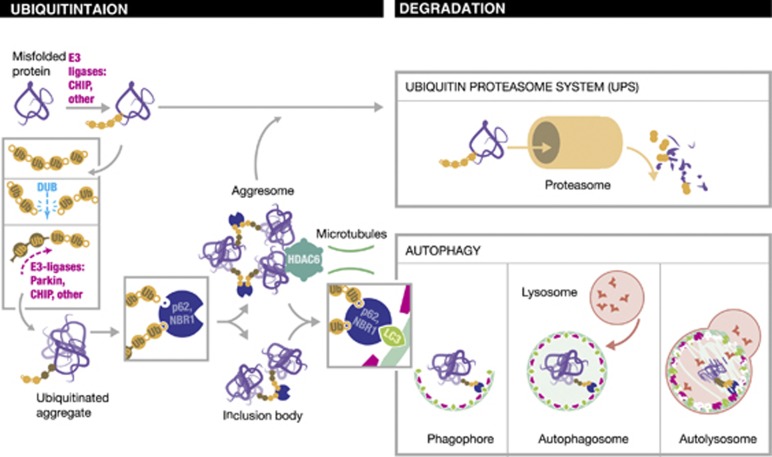
Figure 1.
The Ubiquitin code links between proteasomal and lysosomal degradation. A model for different degradation routes of a misfolded protein. As illustrated, a misfolded protein can be degraded by proteasome or lysosomal system. Chaperone-dependent E3-ligase CHIP or other related E3-ligases leads to ubiquitination of the misfolded protein, predominantly in a K48-linked mode, which targets for the proteasome. Under certain conditions i.e., if the capacity of the chaperone-mediated refolding machinery and the UPS is overloaded, protein aggregation occurs, which are then targeted for autophagic clearance. Thereby, ubiquitin chains on misfolded proteins can undergo remodelling by combined activity of deubiquitinating enzyme (DUB) and E3-ligases. Newly formed ubiquitin chains, i.e., K63-linked chains are then recognised by the UBD of p62, NBR1 to form inclusion bodies or by the corresponding UBD of HDAC6, which direct protein aggregates to the aggresome. Aggresomes can be degraded via the proteasome or via autophagy pathway. If degradation occurs via autophagy, targeting of the protein aggregates are determined by the LIR motif of p62 and NBR1. Autophagy takes part in distinct steps. Cytosolic components are enclosed by an isolation membrane so-called phagophore, which leads to the formation of the autophagosomes. Thereby, LC3 and other ATG proteins such as the ATG 5/12/16L complex are recruited to autophagosome. Finally, the outer membrane of the autophagosome fuses with the lysosome and the internal material is degraded in the autolysosome. LC3, mammalian LC3 modifier including all LC3 and GABARAP family protein; Ub, ubiquitin
When autophagy was discovered over 50 years ago it was assumed to represent a general non-selective degradative pathway activated by nutrient limitation.3 Since then autophagy has been linked to human pathophysiology, including cancer, neurodegeneration, immune response and ageing.4, 5 Through ‘self-digestion' of its own cytoplasmic contents, the cell recycles all nutrients during limited energy supply. In addition, under normal (nutrient-rich) conditions basal autophagy serves as an intracellular quality control system: protein aggregates including damaged or redundant organelles are selectively eliminated. Compared with classical starvation-induced autophagy, this form of autophagy needs to distinguish between normal and ‘abnormal' cell content. Autophagy is considered selective when a precise cargo is specifically and exclusively targeted into autophagosomes. The exact mechanism of cargo recognition remains obscure. However, this process clearly involves ubiquitination. The molecular characterisation of ubiquitin binding proteins such as p62 and neighbour of breast cancer 1 (NBR1)6, 7 has demonstrated that, analogous to the proteasome where ubiquitinated cargos are delivered by ubiquitin receptors,8 a ubiquitin-dependent sensor system is responsible for substrate specificity. Before autophagic clearance, these receptors need to ‘tether' the ubiquitinated cargo to the nascent autophagosome, which carries UBL proteins known as LC3 or GABARAP proteins on its surface.9 Thus, autophagy receptors binding to both ubiquitin and LC3 or GABARAP proteins are able to control protein degradation by selective autophagy. In this review, we will concentrate on the aspect of substrate modification by ubiquitin and its effect on cargo turnover, particularly in selective autophagy.
Ubiquitin Degrades Proteins in Two Ways: via UPS and through Selective Autophagy
All cells monitor misfolded proteins resulting from translation errors, impaired folding or damages due to harsh conditions (i.e., oxidative stress, heat shock).10 Misfolded proteins severely impair cellular physiology, as exposed hydrophobic residues lead to formation of insoluble oligomers and larger aggregates alter essential protein–protein interactions, thereby causing cellular toxicity.11 Accumulation of toxic protein aggregates is a hallmark of several common human diseases, so-called protein misfolding disorders or proteinopathies.
The critical factors that direct a specific substrate to one degradation route or the other is incompletely understood, but is partially encoded in how ubiquitin is attached to its substrates: conjugation of a single ubiquitin monomer (monoubiquitination) or sequential conjugation of several ubiquitin moieties (polyubiquitination) of variable length. Dependent on which of its seven lysine (K) residues is linked to the ubiquitin monomer, the chains are called K6, K11, K27, K29, K33, K48 or K63 ubiquitin chains.12 Conjugation on its amino terminal methionine (M1) moieties leads to the formation of linear chains.13 ‘Classical' K48 ubiquitin chains were originally identified as the canonical signal to target proteins for proteasomal degradation.1 In contrast, non-classical linkage types such as K63, K11, M1-linked chains or single ubiquitin moieties (monoubiquitination) were thought to signal mainly for non-proteolytical functions.14, 15 These chain types are involved in controlling several processes such as receptor transport, DNA repair and signalling.14, 15, 16 However, recent reports have demonstrated that regulatory proteins of the cell cycle modified with K11 chains are also targeted for the proteasomal pathway,17 whereas K63 chains can target substrates for degradation via autophagy.18
Molecular chaperones of the heat-shock protein (HSP) family initiate the first protective mechanism to prevent toxicity and ensure intracellular quality control-mediated clearance. On the one hand HSP assist folding of de novo synthesised protein while they traffic into cellular compartments and promote refolding of denatured or damaged proteins. On the other hand protein aggregation is prevented by shielding hydrophobic surfaces of folding intermediates from the cytosol.11 If refolding of the protein is not successful, E3 ubiquitin ligases, such as the co-chaperone carboxyl terminus of heat-shock cognate70 (HSC70)-interacting protein (CHIP), induce ubiquitination and thereby select unfolded or damaged proteins for degradation preferentially by the UPS (Figure 1).11 The UPS has a major role in cellular quality, which unlike bulk degrades substrates in a highly selective manner via covalent attachment of ubiquitin chains. These chains serve as recognition motifs for delivery to 26S proteasomes. If the capacity of the chaperone-mediated refolding machinery and the UPS is overloaded, protein aggregation can occur. Accumulation of misfolded proteins leads to the formation of so-called inclusion bodies (also called aggresomes), which are then delivered to the lysosome for autophagic degradation (Figure 1). The term ‘inclusion bodies' has been referred to the intracellular foci into which protein aggregates are sequestered. Aggresomes are inclusion bodies, which require a microtubule-based apparatus involving the ubiquitin-binding histone deacetylase 6 (HDAC6) (Figure 1). Importantly, ubiquitinated aggresomes can also be degraded by the UPS (Figure 1). The biological relevance of aggregate formation remains obscure, although it may have a protective role by actively removing toxic misfolded monomers and preventing widespread intracellular toxicity.19 Conversely, it may block proteasomal degradation and increase cytotoxicity.20
Molecular Links between the Proteasomal and the Autophagic Machinery
Although it has long been assumed that the proteasomal and the autophagosomal machinery are two complementary (but strictly separate) degradative systems with distinct clients, increasing evidence suggests that there are numerous intersections. The way how a misfolded protein is degraded is partly determined by the relative activity of each degradation systems (Figure 1).21 For instance, recent studies demonstrate that activation of the forkhead transcription factor O 3 induces both autophagosomal and proteasomal degradation.22, 23 It remains unclear whether autophagy is strictly a parallel or a compensatory degradation system when the UPS is impaired. A molecular link between these systems could be HDAC6, a microtubule-associated deacetylase that interacts with polyubiquitinated proteins and can rescue defects of the UPS by increasing autophagy.24 Moreover, long-term inhibition of autophagy can increases proteasomal substrates level by compromising the UPS as a consequence of p62 accumulation.25 Finally, chaperones and co-chaperones such as BCL-2-associated athanogene 1 (Bag1) and Bag3 have a very important regulatory role in recognising misfolded proteins for degradation.26 For instance, as a consequence of cell ageing, the amount of misfolded proteins gradually increases. To cope with this situation, ageing cells switch co-chaperone expression from Bag1 to Bag3 to control recognition of misfolded proteins and gather them into p62-containing aggregates.27 Bag1, highly expressed in young cells, promotes high use of proteasomal degradation, whereas Bag3 was upregulated in ageing cells correlating with increased autophagic activity and enhanced p62 formation, suggesting that the Bag1/Bag3 ratio decides between proteasomal and autophagic degradation. Bag3 was found in the same complex with p62 and interacts with Hsp/Hsc70.27 Also p97/VCP, a HDAC6-interacting chaperone, is linked to both degradation systems by disassembling protein complexes for degradation to the UPS28, 29 but also interacting with protein aggregates destined for autophagy.30, 31 Importantly, mutations in p97 cause an autophagosome maturation defect by preventing autophagosome–lysosome fusion (reviewed in Ju and Weihl32). This leads to the development of the proteinopathy inclusion body myopathy associated with Paget's disease of the bone and fronto-temporal dementia.32
Another kind of crosstalk between proteasomal and lysosomal degradation takes place at the substrate level. The ubiquitin code predicts that the respective degradation system is selected upon specific types of ubiquitin chains, as described above. Main factors for this process are the ubiquitin E3-ligases, as they, together with specific E2 enzyme, are responsible for the conjugation of different ubiquitin chains. Misfolded proteins resistant to refolding by chaperones are ubiquitinated by the E3-ligase CHIP with K48-linked chains and degraded by the proteasome (Figure 1). In combination with the E2 enzyme ubiquitin-conjugating enzyme, CHIP is able to make K63 chains33 and target the substrate to the autophagic machinery. As an example, α-synuclein, is degraded by both proteasomal and autophagic after CHIP ubiquitination.34 A recent study demonstrated that coexpression of aggregation-prone proteins with different ubiquitin mutants in cultured cells leads to formation of inclusion bodies. Thereby, K48- and K63-linked polyubiquitination, as well as monoubiquitin modifications, all contributed to the biogenesis of these inclusions, but only K63-linked chains induced autophagic degradation of the inclusion bodies.18 Also, the E3-ligase Parkin is able to ligate several ubiquitin linkages (Figure 1), though the mechanism remains elusive. Numerous proteins have been shown to undergo Parkin-dependent K48-linked ubiquitination followed by proteasomal degradation.35 Others have identified that Parkin forms K63-linked polyubiquitin chains on misfolded DJ-1 protein, which leads to recruitment into aggresome via HDAC6.36 In addition, recent studies showed that Parkin promotes selective autophagy of dysfunctional mitochondria (mitophagy, see below).37, 38 By interacting with different sets of cofactors under specific conditions it may be conceivable that CHIP, Parkin and other related E3-ligases act like dual-function ubiquitin ligases targeting substrates to either proteasomal or autophagic degradation pathways (Figure 1).
The Autophagy Modifiers have UBL Functions
The process of autophagy is divided mechanistically into distinct steps: (1) the initiation starts with the formation of a cup-shaped isolation membrane, the so-called phagophore (Figure 1);39, 40 (2) the elongation of the phagophore engulfs cytoplasmic components, including membrane bound organelles and finally closes to form a double membrane vacuole, the autophagosome;40, 41 (3) fusion of the outer membrane of an autophagosome with a lysosome forms an autolysosome (Figure 1). Over 30 autophagy-related genes (ATG) involved in the process of autophagosome formation have been identified to date.42, 43 The ‘core' ATG proteins, which show high functional homology to mammalian genes, can be categorised into four subgroups: (1) Atg1/ULK1 kinase complex (ULK, ATG13, FIP200, ATG101); (2) PI3K complex (VPS34-BECN1), and two UBL systems comprised by the (3) ATG12-ATG5-ATG16L and (4) MAP1LC3/GABARAP modifiers.
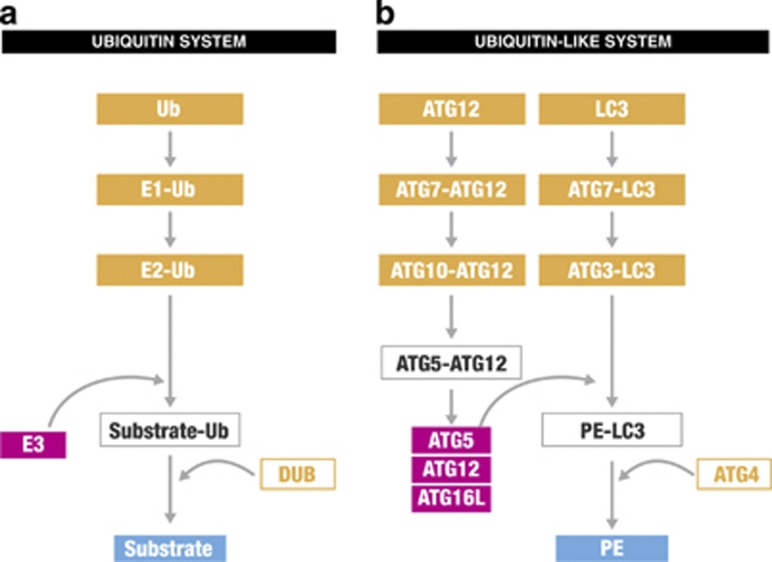
The two UBL conjugation systems ATG12 and LC3 are highly conserved from yeast to humans and function in both selective and non-selective autophagy during the autophagosome formation. Although Atg12 and LC3 modifiers (LC3 and GABARAP family proteins) do not have apparent sequence homology with ubiquitin from yeast to mammals, they share a common ubiquitin-like beta-grasp superfold and show similarities to the ubiquitin conjugation system (schematically summarised in Figure 2).44, 45 Both systems utilise an E1-like enzyme in the first activation step, called Atg7 in the UBL system. Then, the E2-like enzyme Atg10 conjugates Atg12 to Atg5 to form a complex with Atg16. Finally, similar to the E3 ubiquitin ligase, this complex assists in the ligation of LC3 to phosphatidylethanolamine (PE) by Atg3 (E2-like enzyme) (Figure 2).46 The cysteine protease Atg4 (corresponding to deubiquitinating enzymes) has a dual role: it processes LC3 modifiers before their activation and subsequently it removes LC3 from PE on the autophagosome after vesicle completion (Figure 2). Reactive oxygen species (ROS) seem to be involved in regulating Atg4 activation at this step.47 The exposed glycine residue of LC3 can then be conjugated to PE (i.e. formation of LC3-II). In the conjugated form, LC3 seems to be indispensable for hemifusion of lipid membranes, thereby driving expansion of the autophagosome.48 Furthermore, LC3 may also control the size of autophagosomes.49
Figure 2.
Two UBL are required for autophagy. (a) In the ubiquitin system, ubiquitination takes part in three steps. First, the enzymes E1 leads to activation of ubiquitin followed by a conjugation step, which is catalysed by E2 enzyme. Finally, a ubiquitin E3-ligase process tagging of ubiquitin to a protein. Removal of ubiquitin is performed by DUB. (b) The UBLs ATG12 and LC3 have functional similarities to the ubiquitin system. Both systems utilise a E1-like enzyme (Atg7) in the first activation step. Then, the E2-like enzyme Atg10 conjugates Atg12 to Atg5 to form a complex with Atg16L, which assists as a E3 ubiquitin ligase in the ligation of PE to LC3 by Atg3 (E2-like enzyme). The corresponding deubiquitinating enzyme (DUB) ATG4 removes LC3 from PE. LC3, mammalian LC3 modifier including all LC3 and GABARAP family protein; Ub, ubiquitin
The yeast Atg8 protein has eight orthologs in mammals, which are divided into two subfamilies according to their sequence homology: the microtubule-associated protein 1 LC3 (MAP1LC3) family with MAP1LC3A-C (MAP1LC3A has two splicing variants), and the GABARAP/GATE-16 (golgi-associated ATPase enhancer of 16 kDa) family with GABARAP, GABARAPL1, GATE-16 (also called GABARAPL2) and GABARAPL3.9 Among all Atg8 homologues LC3B has been extensively studied and is emerging as the main actor in starvation-induced autophagy.50 The exact function of either LC3 or GABARAP/GATE-16 in the autophagic process remains yet unclear. It appears that both subfamilies are required for autophagy and that members of both subfamilies contribute to different aspects of autophagosome biogenesis.51
The Role of Ubiquitin in Different Types of Selective Autophagy and its Implications for Disease
As ubiquitin moieties are recognised by proteins containing ubiquitin binding domains (UBD),52 autophagic UBLs such as the LC3 modifiers, interact to a short hydrophobic sequence, termed LC3-interacting region (LIR) motif (named LC3 recognition sequence53; LRS54). LIR motifs were identified in adapter proteins, which have also the ability to bind ubiquitinated substrates simultaneously facilitating autophagic degradation.6, 7 These so-called autophagy receptors are involved in the elimination of various substrates such as protein aggregates, organelles and pathogens (Figure 3).
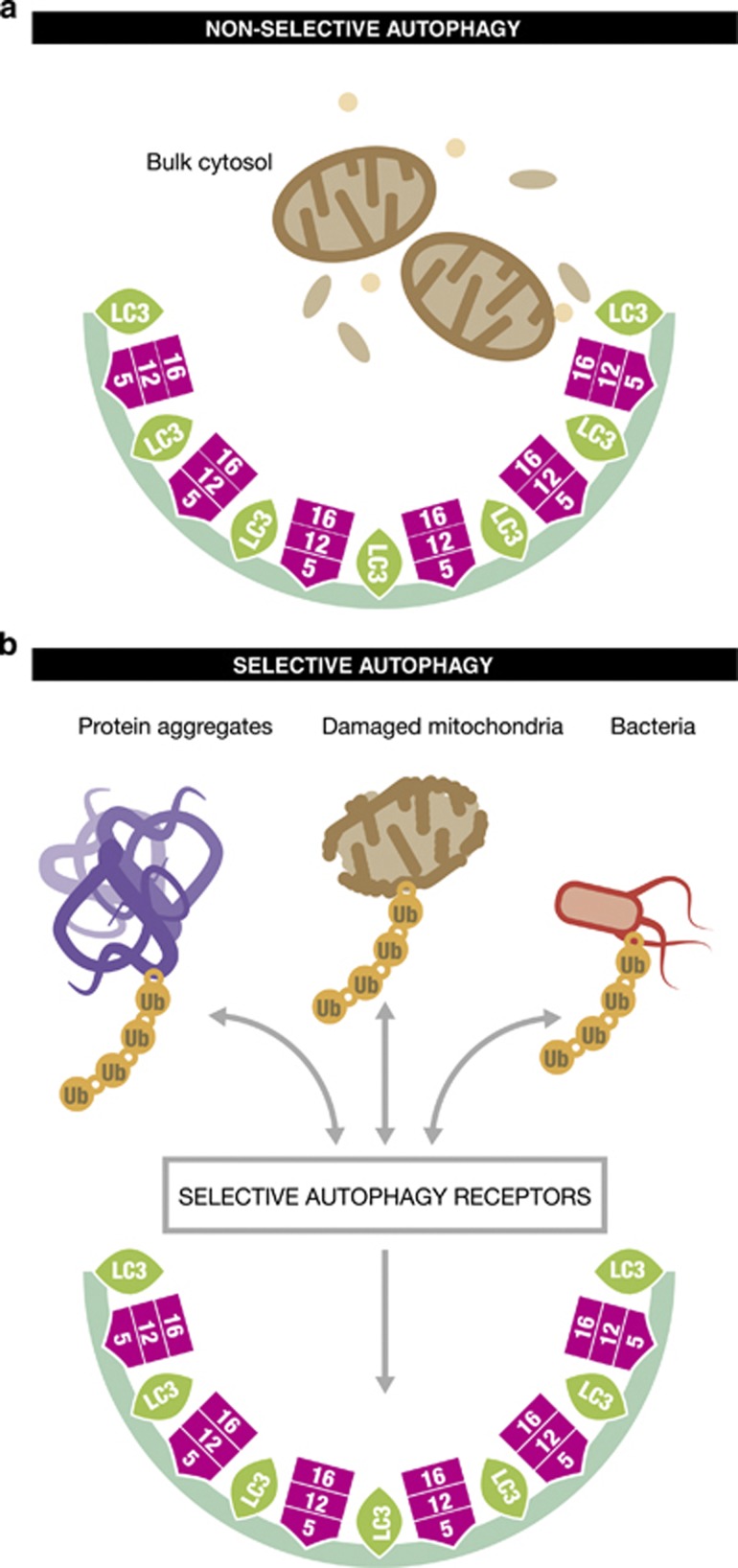
Figure 3.
Non-selective and selective Autophagy. (a) Upon nutrient deprivation, autophagy catabolizes cytoplasmic components nonselectively into autophagosome and mediates recycling and global turnover of cytoplasmic materials. (b) In selective autophagy particular substrate are targeted into the autophagosome by selective autophagy receptors. The targeted cargo includes protein aggregates, damaged mitochondria or pathogens such as bacteria. 16/12/5, ATG 16/12/5 complex; LC3, mammalian LC3 modifier including all LC3 and GABARAP family protein; Ub ubiquitin
Several types of selective autophagy can be distinguished, based on the remarkable substrate specificity of this process: aggrephagy, mitophagy, xenophagy, ribophagy and pexophagy.55 The following sections emphasise on (i) ubiquitination as a signal for substrate targeting and (ii) the crucial role of autophagy receptors (Figure 3). The rapidly growing number of autophagy receptors identified to date are listed in Figure 4.
Figure 4.
Hitherto identified proteins involved in selective autophagy and their domain architecture. (a) p62, NBR1, OPTN, NDP52 and c-Cbl are autophagic adaptor proteins. Excluding c-Cbl they all interact with both ubiquitin and LC3 to promote autophagic degradation. The UBD domain of c-Cbl instead seems not to be involved in autophagic degradation. (b) NIX and newly found FUNDC1 are mitochondrial membrane proteins, which bind LC3/GABARP via their LIR motifs. (c) HDAC6 has only a UBD corresponding called BUZ and binds to ubiquitin (but not to LC3), (d) whereas the proteins Alfy, Bag3 and Tecpr1 are indirectly associated with ubiquitinated proteins or with LC3. Although this may ultimately also result in bridging substrates to autophagosomes, these proteins are not referred to as autophagy receptors. Numbers indicates length of human proteins in amino acids. BEACH, BEACH domain; BH3, Bcl-2 homology 3 domain; BUZ, ubiquitin-binding zinc finger; CC, coiled coil domain; Dysf, Dysferlin domain; FYVE, Fab1,YOTB/ZK632.12, Vac1, and EEA1 domain; PH, Pleckstrin homology domain; TM, transmembrane domain; WD40, WD40 repeats; WW, WW domain; ZnF, Zinc-finger domain
Role of p62/SQSTM1 (sequestosome 1) and NBR1 in Selective Autophagy
p62/SQSTM1 is a common component of ubiquitinated inclusion bodies found in neurodegenerative and liver disease as well as in cultured cells. Accumulation of so-called p62 bodies is associated with autophagy induction or subjected to misfolded protein stresses.56 p62 as a multifunctional adaptor protein is involved in several biological processes such as cell signalling, differentiation and particularly in the removal of toxic protein aggregates.57 Through its Phox and Bem1p (PB1) domain, it undergoes oligomerisation and interaction with protein kinases (i.e MAP-kinases) and NBR1.58, 59, 60 Via its ubiquitin-associated (UBA) domain (Figure 4), p62 is able to bind mono- or poly-ubiquitinated proteins.61 Furthermore, p62 is required for the concentration of ubiquitinated proteins forming aggregates and crucial for their clearance.6, 62 Depletion of p62 in combination with inhibition of autophagy was shown to prevent accumulation of ubiquitin-positive protein aggregates, indicating that p62 is indispensable for basal autophagy.53 By directly binding to LC3 via its LIR motif, p62 becomes a selective autophagy receptor, bringing ubiquitinated protein aggregates to the emerging autophagosome.6, 53 The importance of p62 as a selective autophagy receptor for degradation of misfolded proteins or aggregates is reflected in the development of several human bone diseases. Thereby, the role of the UBA domain seems to be critical. Paget disease of the bone, a chronic and metabolic disorder characterised by increased bone turnover with lesion throughout the skeleton has been frequently associated with mutation of p62, predominantly in the UBA domain.63 Dominant-acting mutations in one of the p62 alleles were observed either leading to deletion of the UBA domain or the disability of binding ubiquitin.64 The loss of functional UBA causes increased osteoclastogenesis by activating TNF receptor-associated factor 6–NF-kB signalling,64 suggesting that regulation of p62 is important for specific biological settings such as bone formation. The efficiency of ubiquitin chain binding may constitute a platform for cellular signal pathways. For example, phosphorylation of serine 403 phosphorylation within the UBA domain of p62 can enhance the binding to ubiquitin chains and leads to increased autophagic clearance of ubiquitinated proteins.65 Moreover, it was shown that p62 can selectively regulate the turnover of signalling molecules: under starvation conditions, wingless-type MMTV integration site signalling is negatively regulated by dishevelled 2 (Dvl2) degradation by p62-mediated selective autophagy.66 Ubiquitination of Dvl2 by the E3-ligase Von Hippel–Lindau is recognised by p62, which in turn facilitates aggregation and LC3-mediated autophagosome recruitment.
NBR1 has a similar domain structure as p62 and binds directly to ubiquitin and LC3.7 Although NBR1 and p62 differ in sequence and size, they both share an N-terminal PB1 and a C-terminal UBA domain as well as a LIR sequence (Figure 4). NBR1 undergoes dimerisation via the coiled coil domain, whereas p62 dimerises through its PB1 domain. NBR1 can directly bind to p62, and together they act as cargo receptors for autophagic degradation of ubiquitinated substrates.6, 7, 53 In contrast, elimination of midbody derivatives through autophagic degradation is only dependent on NBR1 without any detectable involvement of p62.67 However, it had been previously reported that p62 has a role in midbody clerarance.68 Kuo et al.67 demonstrated that accumulation of midbody derivatives through evasion of autophagy is associated with cellular reprogramming of stem cells and enhanced tumorigenicity of potential cancer stem cell. Thus, NBR1 promotes cell differentiation and may act as a tumour suppressor in this case. Similar to p62, NBR1 is also involved in bone remodelling.69 It appears that NBR1 may also be involved in protein misfolding disorders such as sporadic inclusion body myositis and autophagic degradation may have role in the pathophysiology.70
Selective Autophagy of Mitochondria – Mitophagy
Mitophagy mediates the removal of mitochondria, either by selective elimination of damaged mitochondria as a form of quality control71 or by removal of surplus mitochondria for steady-state turnover to adapt either to metabolic requirements or during specialised development stages, such as red blood cell differentiation.72 Also, the role of ubiquitin in these two forms of mitophagy is distinct. During reticulocyte maturation it was shown that Nix (Bnip3L) acts as a mammalian mitophagy receptor and is important for the complete removal of mitochondria.71 In comparison with other autophagic adapter proteins, Nix has a LIR motif but is lacking a UBD (Figure 4). Probably due to its physiological localisation within the mitochondrial outer membrane, ubiquitination might not be required for NIX-dependent delivery of damaged mitochondria to autophagosomes.
During fertilisation, mitochondrial clearance has an important role as strictly maternal inheritance of mitochondria and its DNA in mammals derives from selective elimination of sperm mitochondria, which are tagged by ubiquitin.73 Later it was shown that ubiquitinated mitochondria in the midpiece of mouse sperms after fertilisation were also LC3-positive.74 Intriguingly, p62 and K63 ubiquitin chains were also detected in the midpiece of spermatozoa, suggesting that this selective autophagy receptor, might be also involved in the degradation process.74 Besides, it was demonstrated that in Caenorhabditis elegans paternal mitochondria are degraded by fertilisation-induced autophagy.74, 75 However, spermatozoan mitochondria were not ubiquitinated at all, neither before nor after entry into the ooplasm. Indeed, spermatozoan-specific membranous organelles (MO) were instead rapidly ubiquitinated after fertilisation. Despite these differential ubiquitination states, both mitochondria and MOs were degraded by autophagy. Overall, autophagy-mediated degradation of paternal mitochondria is an evolutionarily conserved process, though the dependency of this process on ubiquitin is not conserved.
Mitochondria form large cellular networks and serve as major sites of ATP production; their morphology also determine cellular response to autophagy as, elongated mitochondria are spared from autophagic degradation, have increased activity of ATP synthase, and maintain ATP production.76 At the same time dysfunctional mitochondria are a main source of ROS and cytochrome c, which cause cell damage and death. Accumulation of damaged mitochondria is accompanied with neurodegenerative diseases, hypoxia and cancer. Therefore, removal of depolarised mitochondria is essential and loss of membrane potential upon mitochondrial damage leads to autophagic degradation.77 This process is ubiquitin-dependent, especially in case of Parkin-mediated mitophagy. The E3-ligase Parkin is translocated to damaged mitochondria and mediates ubiquitination of voltage-dependent anion channel 1 (VDAC1).37 p62 is in turn recruited to ubiquitinated VDAC1 in a K63- and K27-dependent manner leading to clustering of mitochondria and autophagic degradation.78 Although it is clear that p62 can bind Parkin-ubiquitinated mitochondria and promotes mitochondrial ‘clumping', several conflicting reports show that p62 is not responsible for parkin-mediated mitophagy and that VDAC1 is dispensable for both.79 Concordantly, translocation of Parkin to uncoupled mitochondria and also its induction seems to be activated by PTEN-induced putative kinase 1.78, 80, 81 Recently, it was demonstrated that HDAC6 is recruited to mitochondria after Parkin-mediated ubiquitination and is needed for degradation. As HDAC6 binds to ubiquitin and microtubules, it potentially links transport of damaged mitochondria with mitophagy.82 In addition to K63-linked ubiquitination, other Parkin substrates relevant for mitophagy, such as mitofusin, can be ubiquitinated.83 Mitofusins are small transmembrane guanosine 5′-triphosphatases located in the outer membrane of mitochondria and mediate mitochondrial fusion. By selective degradation of mitofusin, Parkin can inhibit re-fusion of mitochondria.84 Recognition of damaged mitochondria for selective removal can be initiated by integral mitochondrial membrane proteins. For instance, Nix has been reported to promote Parkin translocation and Parkin-mediated mitophagy in mouse embryonic fibroblast85 and can recruit GABARAPL1 to depolarised mitochondria.71 Another mitochondrial membrane protein FUN14 domain containing 1 (FUNDC1) was identified as a receptor for hypoxia-induced mitophagy by interacting with LC3 through a LIR motif.86 Intriguingly, hypoxia-induced dephosphorylation of Tyr18 in the LIR motif (YxxL) of FUNDC1 (Figure 4C) enhances its interaction with LC3 for selective mitophagy. Thus, under normal physiological conditions FUNDC1-mediated mitophagy is inhibited by sarcoma viral oncogene homologue (src)-mediated LIR phosphorylation, whereas under hypoxia dephosphorylation takes over and increases LC3 binding and mitophagy.
Ubiquitination and Autophagy of Pathogens – Xenophagy
In xenophagy pathogen-containing phagosomes are exclusively targeted for autophagic degradation. The influence of ubiquitin in xenophagy within the context of bacterial invasion is clearly depicted in several studies.87, 88, 89 The association of ubiquitin with intracellular bacteria led to the hypothesis that antimicrobial autophagy requires a ubiquitin-dependent mechanism for cargo recognition and degradation, analogous to aggrephagy.87 In fact, at least three autophagic adaptor proteins are known – p62, nuclear domain 10 protein 52 (NDP52) and optineurin (OPTN) – which fulfil a dual role in recognising ubiquitinated cytosolic bacteria and bringing them into autophagosomes.90, 91, 92 p62, NDP52 and OPTN are not redundant, as depletion of either protein causes hyperproliferation of Salmonella typhimurium90, 91, 92 and all three proteins are independently recruited to the same bacterium.92, 93 NDP52 appears to function in innate immunity against cytosolic pathogens by linking the TANK binding kinase 1 (TBK1) signalling pathway to autophagic elimination. TBK1 is a serine/threonine kinase from the IKK family that has an antibacterial activity.91 Moreover, it was shown that TBK1 enhances LC3 binding to OPTN by phosphorylation of a conserved serine residue adjacent to the LIR motif of OPTN.92 Similar to NDP52 and TBK1 signalling, p62 interacts with atypical PKC, which promotes delivery of Salmonella-containing vacuoles for autophagy.89 TBK1 also forms a complex with autophagy-linked FYVE protein, which may facilitate formation of autophagosomal membranes around large ubiquitinated cargos, such as bacteria. Besides pathogen recognition, p62 mediates recruitment of antibacterial peptides derived from ribosomes and ubiquitinated proteins to Mycobacterium tuberculosis containing compartments, which is critical for antibacterial defence.94 Another explanation for the non-redundant function of p62, NDP52 and OPTN might rely on their UBD. The ability of UBDs to bind different ubiquitin chains may explain the differential localisation of autophagy receptors in distinct microdomains surrounding the bacterium. However, neither the identity of the E3 ubiquitin ligase nor the type of ubiquitin chains conjugated to the bacterial surface ise currently known. Also, modifications in binding affinity to ubiquitinated substrates may control pathogen elimination. The UBA domain of p62 is critical for the recruitment to ubiquitin-positive S. typhimurium. Similar to TBK1-mediated LIR phosphorylation in OPTN (resulting in stronger LC3 binding), phosphorylation of serine 403 in the UBA domain of p62 by casein kinase 2 was shown to enhance the binding to ubiquitin chains.65 Thus, recruitment of several kinases to the vicinity of bacteria targeted by autophagy might ensure coordinated regulation of pathogen sensing and efficient elimination of invading pathogens by autophagy.
Besides ubiquitin-dependent pathogen defence, subscribing a later time point against bacterial invasion, there is an early line of defence comprising accumulation of diacylglycerol on bacteria-containing vesicles.89 Subsequently, they are targeted selectively to autophagy. Bacteria escaping this early defence line expose host glycans on their damaged vacuoles. Recently, it was shown that the host glycans on the surface of Salmonella are targeted by galectin 8, a cytosolic lectin, which directly binds NDP52 promoting antibacterial autophagy in a ubiquitin-independent way.95 This interaction is transient and followed by a ubiquitin-dependent NDP52 recruitment, outlining the third layer of defence.
Ubiquitination and Other Forms of Selective Autophagy
Mounting evidence suggests that peroxisomes (pexophagy), ribosomes (ribophagy) and surplus endoplasmatic reticulum (ER) (reticulophagy) are also selectively degraded by autophagy. However, the mechanism of cargo recognition is poorly understood, but may require ubiquitination. For instance, a significant role of ubiquitin in pexophagy has been described: monoubiquitination of peroxisomes was sufficient for targeting them to autophagosomes in a p62-dependent manner.96 The deubiquitination activity of Ubp3/Bre5 is crucial for selective autophagy of excess ribosomes.97 Similarly, selective autophagy of zymogen granules is dependent on the activity of the ubiquitin-specific protease USP9x.98 It seems that USP9x regulates zymogen granule-selective engulfment presumably by deubiquitination of vacuole membrane protein 1, an autophagy-associated protein. Alternatively, both ubiquitination and deubiquitination of distinct critical molecules might be required for zymophagy; however, specific E3-ligases have not been yet identified. Another example of selective autophagy is described as ER-phagy or reticulophagy induced by UPR (unfolded protein response). Upon severe ER stress, ER-phagy may constitute a cell survival mechanism against UPR-induced ER expansion. As a consequence, reducing ER volume is followed by decreasing unfolded ER proteins, though the molecular mechanisms remain elusive.99, 100
Future Direction/Outlook
There is a growing body of evidence, suggesting that the specificity factor for selective autophagy is determined by ubiquitination and binding of this ubiquitin signal by autophagic adaptor proteins. Beside ‘classical' autophagy receptors such as p62 and NBR1 characterised by their UBD and LIR motifs, also ‘non-classical' autophagy receptors are appearing. Such adaptor proteins target substrates selectively for autophagy independently of classical UBD and LIR motifs. They are giving new molecular insights in selective autophagy. For instance, quite recently the relatively promiscuous c-Casitas B-lineage lymphoma (c-Cbl) was shown to act as an autophagic adapter protein under certain conditions.101 In the absence of focal adhesion kinase activity c-Cbl targets Src into autophagsosomes by binding LC3 via its LIR motif.101 Intriguingly, only the LIR motif and not the E3-ligase activity of c-Cbl is required for the autophagosomal targeting. Thus, the ubiquitin ligase activity of c-Cbl is essential for proteasomal degradation of Src but dispensable for Src degradation by autophagy. Furthermore, the role of ubiquitination in this model remains unclear, as binding of c-Cbl to Src is a ubiquitin-independent process. SMAD-specific E3 ubiquitin protein ligase 1 (SMURF1), another E3-ligase, was also shown to operate as an adaptor protein in selective autophagy independently of its catalytic activity.102 As a mediator of both viral autophagy and mitophagy, it was assumed that its C2 domain bridges autophagic substrates to the autophagosomes. Analogous to c-Cbl, SMURF1 may have dual functions in targeting substrates to either proteasomal degradation via its ubiquitination activity or in targeting to autophagic degradation via its C2 domain. In general, it will be interesting to clarify what exactly determines proteasomal or lysosomal degradation of the same protein and, in particular, which factors or signals can switch an E3-ligase from its ubiquitination function to its autophagic adaptor mode.
It will be important to understand how autophagy substrates are selected and labelled for degradation. Selection does not always mean ubiquitin labelling. As an example, Tachylectin-II-like beta-propeller domain (Tecpr1) was initially identified as an Atg5 binding partner.103 Later it was shown to colocalise with ATG5 at Shigella-containing autophagosomes promoting efficient autophagic degradation of bacteria.104 Importantly, targeting of bacterial pathogens for selective autophagy by Tecpr1 is ubiquitin-independent, as Shigella are able to avoid polyubiquitin tagging. Furthermore, Tecpr1 interacts with WIPI-2, a yeast Atg18 homologue and PI(3)P-interacting protein, which is required for LC3-positive autophagosomes formation. Hence, neither a LIR nor a UBD domain is required for autophagic degradation of Shigella by Tecpr1.104 In addition, Tecpr1 appears to have a more general role in autophagosome maturation in starvation-induced autophagy.105
The future challenge will be to delineate the spatio-temporal mechanisms by which ubiquitin signals control selective autophagy and its importance in physiological and pathophysiological processes. Among the questions to be addressed are how post-translational modifications including phosphorylation, acetylation and ubiquitination coordinate selective autophagy.106 Recent reports indicate an intricate network of regulatory steps, as exemplified by the phosphorylation of the LIR motif of OPTN that is crucial for xenophagy92 and phosphorylation of the UBA domain of p62 that mediates autophagy of protein aggregates.65 Lastly, it will be of great interest to elucidate the role of selective autophagy in different signalling pathways, especially to reveal how autophagy can control cellular homoeostasis by modulating the capacity of intracellular signalling networks.
Acknowledgments
We apologize to scientists whose important contribution was not referenced in this review owing to space limitations. We would like to thank Ligia Gomes and Christian Behrends for critical reading of the manuscript and also to Philipp Wild and Evgenij Fiškin for comments and discussions. Research in the I.D. laboratory is supported by the Deutsche Forschungsgemeinschaft, the Cluster of Excellence ‘Macromolecular Complexes' of the Goethe University Frankfurt (EXC115), LOEWE OSF and a European Research Council Advanced Grant and in the C.B. laboratory through a LOEWE Cell and Gene Therapy (CGT) grant, SFB834 funding, LOEWE OSF and Frankfurt Autophagy Network (FAN) funding. S.S. is the recipient of a fellowship from the Faculty of Medicine of the Goethe University, Frankfurt am Main.
Glossary
- ATG
autophagy-related gene
- BAG
BCL-2-associated athanogene
- BECN1
Beclin1
- c-CBL
c-Casitas B-lineage lymphoma
- CHIP
carboxyl terminus of HSC70-interacting protein
- CMA
chaperone-mediated autophagy
- Dvl2
dishevelled 2
- ER
endoplasmatic reticulum
- FIP200
FAK family kinase-interacting protein of 200 kDa
- FUNDC1
FUN14 domain containing 1
- GABARAP
gamma-aminobutyric acid receptor-associated protein
- GATE-16
golgi-associated ATPase enhancer of 16 kDa
- HDAC6
histone deacetylase 6
- Hsp
heat-shock protein
- Hsc70
heat-shock cognate70
- K
lysine
- LC3
light chain 3
- LIR
LC3-interacting region
- M
methionine
- MO
membranous organelle
- MAP1LC3
microtubule-associated protein 1 light chain 3
- NBR1
neighbour of breast cancer 1
- NDP52
nuclear domain 10 protein 52
- NF-kB
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1
- OPTN
optineurin
- p97/VCP
protein 97/valosin-containing protein
- PB1
Phox and Bem1p domain
- PE
phosphatidylethanolamine
- PI3K
1-phosphatidylinositol 3-kinase
- ROS
reactive oxygen species
- src
sarcoma viral oncogene homologue
- SMURF1
SMAD-specific E3 ubiquitin protein ligase 1
- p62/SQSTM1
sequestosome 1
- TBK1
TANK binding kinase 1
- Tecpr
Tachylectin-II-like beta-propeller domain
- UBA
ubiquitin-associated
- UBL
ubiquitin-like system
- UBD
ubiquitin binding domain
- Ubp3
ubiquitin carboxyl-terminal hydrolase 3
- ULK
Unc-51-like kinase
- UPS
ubiquitin–proteasomal system
- UPR
unfolded protein response
- USP9x
ubiquitin-specific peptidase 9, X-linked
- VDAC1
voltage-dependent anion channel 1
- VPS34
vacuolar protein sorting 34
The authors declare no conflict of interest.
Footnotes
Edited by M Piacentini
References
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- De Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Kubota H. Quality control against misfolded proteins in the cytosol: a network for cell survival. J Biochem. 2009;146:609–616. doi: 10.1093/jb/mvp139. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. Embo J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Dikic I. Ubiquitylation and cell signaling. Embo J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18:520–528. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- Walczak H. TNF and ubiquitin at the crossroads of gene activation, cell death, inflammation, and cancer. Immunol Rev. 2011;244:9–28. doi: 10.1111/j.1600-065X.2011.01066.x. [DOI] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JM, Wong ES, Kirkpatrick DS, Pletnikova O, Ko HS, Tay SP, et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet. 2008;17:431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettern N, Dreiseidler M, Tawo R, Hohfeld J. Chaperone-assisted degradation: multiple paths to destruction. Biol Chem. 2010;391:481–489. doi: 10.1515/BC.2010.058. [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. Embo J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik C, Yano M, DeMartino GN. RNA interference of valosin-containing protein (VCP/p97) reveals multiple cellular roles linked to ubiquitin/proteasome-dependent proteolysis. J Cell Sci. 2004;117:281–292. doi: 10.1242/jcs.00841. [DOI] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Mizuno Y, Hori S, Kakizuka A, Okamoto K. Vacuole-creating protein in neurodegenerative diseases in humans. Neurosci Lett. 2003;343:77–80. doi: 10.1016/s0304-3940(03)00280-5. [DOI] [PubMed] [Google Scholar]
- Hirabayashi M, Inoue K, Tanaka K, Nakadate K, Ohsawa Y, Kamei Y, et al. VCP/p97 in abnormal protein aggregates, cytoplasmic vacuoles, and cell death, phenotypes relevant to neurodegeneration. Cell Death Differ. 2001;8:977–984. doi: 10.1038/sj.cdd.4400907. [DOI] [PubMed] [Google Scholar]
- Ju JS, Weihl CC. Inclusion body myopathy, Paget's disease of the bone and fronto-temporal dementia: a disorder of autophagy. Hum Mol Genet. 2010;19:R38–R45. doi: 10.1093/hmg/ddq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, et al. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ. The co-chaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- Hattori N, Mizuno Y. Pathogenetic mechanisms of parkin in Parkinson's disease. Lancet. 2004;364:722–724. doi: 10.1016/S0140-6736(04)16901-8. [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, et al. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda NN, Ohsumi Y, Inagaki F. ATG systems from the protein structural point of view. Chem Rev. 2009;109:1587–1598. doi: 10.1021/cr800459r. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA, Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Paz Y, Elazar Z, Fass D. Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J Biol Chem. 2000;275:25445–25450. doi: 10.1074/jbc.C000307200. [DOI] [PubMed] [Google Scholar]
- Suzuki NN, Yoshimoto K, Fujioka Y, Ohsumi Y, Inagaki F. The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy. 2005;1:119–126. doi: 10.4161/auto.1.2.1859. [DOI] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. Embo J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. Embo J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Strnad P, Zatloukal K, Stumptner C, Kulaksiz H, Denk H. Mallory-Denk-bodies: lessons from keratin-containing hepatic inclusion bodies. Biochim Biophys Acta. 2008;1782:764–774. doi: 10.1016/j.bbadis.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. 2007;32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Wilson MI, Gill DJ, Perisic O, Quinn MT, Williams RL. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol Cell. 2003;12:39–50. doi: 10.1016/s1097-2765(03)00246-6. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Kimple AJ, Siderovski DP, Johnson GL. PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NF-kappaB activation. J Biol Chem. 2010;285:2077–2089. doi: 10.1074/jbc.M109.065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, et al. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Joung I, Strominger JL, Shin J. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J Biol Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode A, Layfield R. Recent advances in understanding the molecular basis of Paget disease of bone. J Clin Pathol. 2010;63:199–203. doi: 10.1136/jcp.2009.064428. [DOI] [PubMed] [Google Scholar]
- Cavey JR, Ralston SH, Hocking LJ, Sheppard PW, Ciani B, Searle MS, et al. Loss of ubiquitin-binding associated with Paget's disease of bone p62 (SQSTM1) mutations. J Bone Miner Res. 2005;20:619–624. doi: 10.1359/JBMR.041205. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–790. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- Kuo TC, Chen CT, Baron D, Onder TT, Loewer S, Almeida S, et al. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat Cell Biol. 2011;13:1214–1223. doi: 10.1038/ncb2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl C, Jentsch S. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat Cell Biol. 2009;11:65–70. doi: 10.1038/ncb1813. [DOI] [PubMed] [Google Scholar]
- Waters S, Marchbank K, Solomon E, Whitehouse CA. Autophagic receptors Nbr1 and p62 coregulate skeletal remodeling. Autophagy. 2010;6:981–983. doi: 10.4161/auto.6.7.13155. [DOI] [PubMed] [Google Scholar]
- D'Agostino C, Nogalska A, Cacciottolo M, Engel WK, Askanas V. Abnormalities of NBR1, a novel autophagy-associated protein, in muscle fibers of sporadic inclusion-body myositis. Acta Neuropathol. 2011;122:627–636. doi: 10.1007/s00401-011-0874-3. [DOI] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitin tag for sperm mitochondria. Nature. 1999;402:371–372. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation Autophagy 20106e-pub ahead of print 16 May 2010. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH. Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnazari S, Brumell JH. Mechanisms and consequences of bacterial targeting by the autophagy pathway. Curr Opin Microbiol. 2011;14:68–75. doi: 10.1016/j.mib.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cemma M, Kim PK, Brumell JH. The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target Salmonella to the autophagy pathway. Autophagy. 2011;7:341–345. doi: 10.4161/auto.7.3.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010;32:329–341. doi: 10.1016/j.immuni.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Grasso D, Ropolo A, Lo ReA, Boggio V, Molejon MI, Iovanna JL, et al. Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J Biol Chem. 2011;286:8308–8324. doi: 10.1074/jbc.M110.197301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:2311–2324. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilands E, Serrels B, McEwan DG, Morton JP, Macagno JP, McLeod K, et al. Autophagic targeting of Src promotes cancer cell survival following reduced FAK signalling. Nat Cell Biol. 2011;14:51–60. doi: 10.1038/ncb2386. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, Sumpter R, Xiao G, Ng A, Zou Z, Tang Y, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Yoshikawa Y, Kobayashi T, Mimuro H, Fukumatsu M, Kiga K, et al. A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host Microbe. 2011;9:376–389. doi: 10.1016/j.chom.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Chen D, Fan W, Lu Y, Ding X, Chen S, Zhong QA. Mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 Conjugate. Mol Cell. 2012;45:629–641. doi: 10.1016/j.molcel.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DG, Dikic I. The three musketeers of autophagy: phosphorylation, ubiquitylation and acetylation. Trends Cell Biol. 2011;21:195–201. doi: 10.1016/j.tcb.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]