Abstract
Intracellular lipids are stored in lipid droplets (LDs) and metabolized by cytoplasmic neutral hydrolases to supply lipids for cell use. Recently, an alternative pathway of lipid metabolism through the lysosomal degradative pathway of autophagy has been described and termed lipophagy. In this form of lipid metabolism, LD triglycerides (TGs) and cholesterol are taken up by autophagosomes and delivered to lysosomes for degradation by acidic hydrolases. Free fatty acids generated by lipophagy from the breakdown of TGs fuel cellular rates of mitochondrial β-oxidation. Lipophagy therefore functions to regulate intracellular lipid stores, cellular levels of free lipids such as fatty acids and energy homeostasis. The amount of lipid metabolized by lipophagy varies in response to the extracellular supply of nutrients. The ability of the cell to alter the amount of lipid targeted for autophagic degradation depending on nutritional status demonstrates that this process is selective. Intracellular lipids themselves regulate levels of autophagy by unclear mechanisms. Impaired lipophagy can lead to excessive tissue lipid accumulation such as hepatic steatosis, alter hypothalamic neuropeptide release to affect body mass, block cellular transdifferentiation and sensitize cells to death stimuli. Future studies will likely identify additional mechanisms by which lipophagy regulates cellular physiology, making this pathway a potential therapeutic target in a variety of diseases.
Keywords: apoptosis, autophagy, cholesterol, hypothalamus, triglycerides
Facts
Cells break down triglycerides (TGs) and cholesterol stored in lipid droplets (LDs) by a form of the lysosomal degradative pathway of macroautophagy termed lipophagy.
The quantity of lipid undergoing autophagic degradation relative to the amounts of other cellular constituents can vary with nutritional status indicating that lipophagy is a selective form of autophagy.
Lipophagy regulates intracellular lipid content.
TG breakdown by lipophagy supplies free fatty acids (FFAs) needed to sustain cellular rates of mitochondrial β-oxidation and levels of ATP.
The intracellular content of FFAs and cholesterol regulates levels of autophagy.
Open Questions
How does the autophagic machinery selectively target stored lipids for degradation in response to changes in nutrient levels?
What are the relative contributions of cytosolic lipolysis and lipophagy to lipid metabolism in different cell types, and is there cross-talk between the two pathways?
What are the mechanisms by which intracellular lipids and other factors regulate levels of lipophagy?
Do defects in lipophagy underlie human disease and can lipophagy be a therapeutic target?
Intracellular lipids are essential to cells as an energy source, structural components for membranes, synthetic building blocks for other molecules, such as hormones, and as mediators of cell signaling. The ability to safely store adequate, but not excessive, amounts of lipids and to metabolize them when needed is critical for cell function and survival. The recent finding that lipids can be selectively degraded by the lysosomal pathway of macroautophagy through a process termed lipophagy1 has opened up a new understanding of how lipid metabolism regulates cellular physiology and pathophysiology. Many new functions for autophagic lipid metabolism have now been defined in diverse cellular processes ranging from transdifferentiation to resistance to death.
Intracellular Lipids are Degraded by Lipophagy
TGs and cholesterol are safely stored as neutral lipids in specialized cellular organelles called LDs.2, 3 Until recently, the breakdown of LD-stored TG and cholesterol was attributed exclusively to the actions of cytosolic hydrolytic enzymes or lipases. The role of lysosomes in lipid metabolism was previously thought to be confined to the breakdown of extracellular lipoproteins after endocytosis. Now for the first time a function has been demonstrated for the lysosomal degradative pathway of macroautophagy in the breakdown of intracellular LD stores (Figure 1).1 The recognition of this process of lipophagy has significantly altered our view of lipid metabolism by providing an alternative mechanism by which cells mobilize fat in response to cellular needs and external stimuli.4
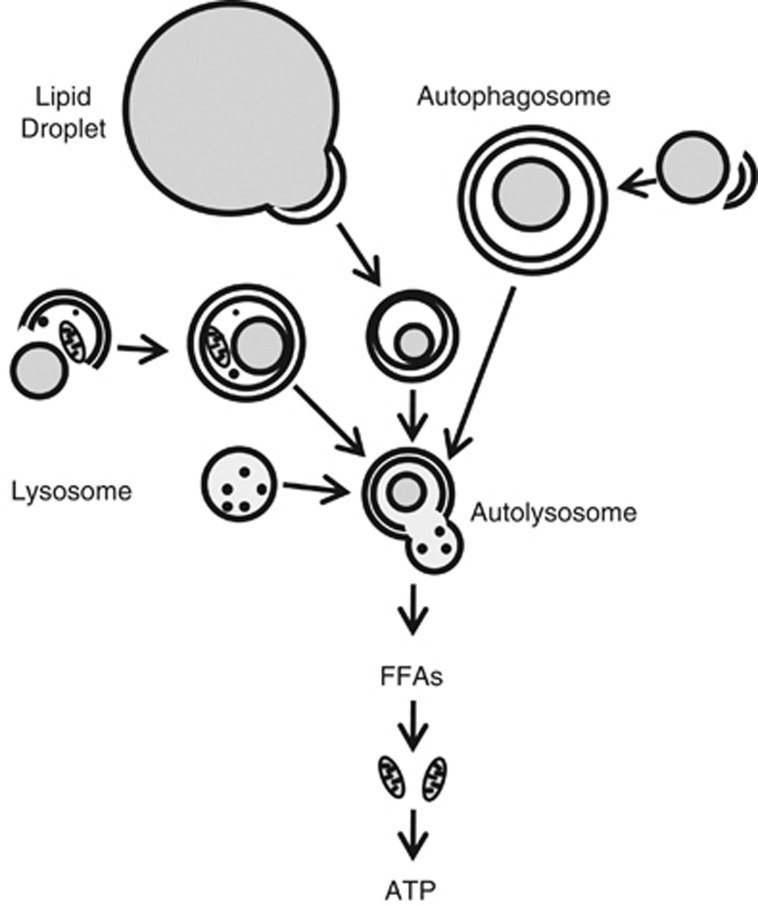
Figure 1.
Process of LD breakdown by lipophagy. Portions of large LDs or entire small LDs are sequestered by a double-membrane autophagosome. Lipids may be sequestered in autophagosomes in combination with other cellular constituents or as the only cargo. Autophagosomes fuse with lysosomes to form autolysosomes in which the substrates of the autophagosome and the hydrolytic enzymes of the lysosome are mixed for cargo degradation. Lipid breakdown leads to release into the cytoplasm of degradation products such as FFAs. FFAs serve to sustain rates of mitochondrial β-oxidation for the generation of ATP to maintain cellular energy homeostasis
Lipophagy was initially described in hepatocytes, which become a major site of excessive lipid accumulation in obesity and the metabolic syndrome.5, 6 A pharmacological or genetic inhibition of autophagy in cultured hepatocytes increased cellular TG content and LD number and size in response to a lipid challenge.1 As a consequence, mitochondrial β-oxidation is impaired due to decreased FFA generation from LD breakdown. Through the supply of FFAs to alter ATP generation by β-oxidation, lipophagy has the potential to regulate cellular energy homeostasis as well as lipid content. Fluorescence and electron microscopy demonstrated that intracellular lipids and LD proteins traffic through autophagosomes and lysosomes, and this flow is increased with lipid supplementation. These findings demonstrated for the first time that autophagy-mediated lysosomal degradation participates in the cellular mobilization and metabolism of intracellular lipids from LDs.
These in vitro findings were confirmed in mice with a hepatocyte-specific knockout of a gene essential for autophagosome formation, atg7.7 Deletion of atg7 led to a dramatic increase in hepatic TG and cholesterol content, demonstrating a function for autophagy in lipid metabolism in vivo.1 Biochemical and electron microscopic studies showed increased association of the autophagosomal protein microtubule-associated protein light chain 3 (LC3) with LDs in response to starvation. In addition, a remarkable switch of cargo selection occurred in response to nutritional status, as the percentage of autophagosomes containing lipid increased markedly with increasing time of starvation. These findings indicate that lipophagy, like many other subtypes of autophagy,8 is selectively upregulated in response to specific cellular needs. A switch in cargo selection to lipids enables the cell to quickly adapt to nutrient deprivation by generating energy from increased oxidation of FFAs.
Lipophagy is ubiquitous as it functions in other cells that do not store lipids in as large quantities as hepatocytes including fibroblasts,1 neurons,9, 10 and stellate cells.11 Questions remain as to how autophagy targets LDs and how lipophagy is selectively regulated in response to environmental stimuli. Possible candidates to mediate LD targeting are the soluble NSF attachment protein receptors (SNAREs). Long implicated in LD fusion,12 SNAREs have recently been described to mediate autophagosome biogenesis.13, 14 Another possibility is LC3, which is a protein critical for autophagosome membrane formation.7 The finding that LC3 associates with LDs in the apparent absence of an autophagosomal membrane1, 15 suggests that this protein may function in LD recognition. Another important remaining question is what are the relative contributions of the cytosolic neutral lipase and lipophagy pathways to total lipid breakdown, and is there cross-talk between these two lipolytic mechanisms. Do they work together, or function independently, and can each pathway be upregulated to compensate for the loss of the other? These questions must be addressed for different cell types as cytosolic lipases differ markedly among the cells, and the contributions of lipophagy to overall lipid metabolism may vary as well.16
Autophagy Regulates Cellular Differentiation and Phenotype
The finding that autophagy regulates lipid metabolism suggested a role for lipophagy in modulating lipid stores in the major fat-storing cell in the body, the adipocyte. Two types of differentiated adipocytes comprise either white adipose tissue (WAT) or brown adipose tissue (BAT).17 White adipocytes function as a lipid storage depot to compartmentalize FFAs into LDs to prevent lipotoxicity, and as a store of lipids for release to other cells for energy generation in times of nutrient deprivation.18, 19, 20 In contrast, brown adipocytes have a reduced capacity for lipid storage but a high rate of lipid metabolism through β-oxidation. Although various transcription factors and signaling pathways have been shown to modulate the differentiation of adipocytes into WAT and BAT,21, 22, 23 the mechanism of this process remains poorly understood.
Recent investigations have demonstrated a critical function for autophagy in regulating lipid accumulation during adipocyte differentiation. Studies in 3T3-L1 pre-adipocytes, a widely used in vitro model of white adipocyte differentiation,24 have demonstrated that in contrast to hepatocytes and fibroblasts, ablation of autophagy blocked the massive accumulation of TGs that normally occurs during chemically induced differentiation of 3T3-L1 cells into white adipocytes.25 Inhibition of autophagy reduced expression of master regulators of adipogenesis such as peroxisome proliferator-activated receptor gamma (PPARγ) and CCAAT-enhancer-binding protein.21, 22, 23 Protein markers of mature white adipocytes were also decreased in cells lacking autophagy, indicating that the failure to accumulate TGs was secondary to impaired differentiation.25 Similarly, deletion of the autophagy genes atg5 or atg7 blocked mouse embryonic fibroblast differentiation into adipocytes.26, 27
Adipocyte-specific atg7 knockout mice are leaner than control mice and resistant to high-fat diet (HFD)-induced obesity.25, 26 The knockouts have substantially diminished WAT weight (∼60%) in parallel with a significant gain in BAT mass (∼25%).25 Further investigations suggested that this phenotype resulted from enhanced transdifferentiation of WAT to BAT in the knockout mice, as fat depots that were normally composed of white adipocytes had increased morphological and molecular features of BAT.25 Consequently, the rate of FFA β-oxidation was increased 30-fold in WAT and 2-fold in BAT. Collectively, the transformation of WAT from an energy-storing tissue into an energy-expending tissue, together with increased BAT mass, resulted in a lean, obesity-resistant mouse.
Despite of the clear evidence that autophagy regulates adipocyte differentiation, the molecular mechanisms underlying autophagy-mediated adipogenesis remain to be defined. In this instance, there does not seem to be a direct effect of autophagy on lipid metabolism, but rather a primary effect on differentiation likely unrelated to lipophagy that leads to secondary alterations in cellular lipid storage and metabolism. It is possible that autophagy directly regulates expression of one or more of the transcriptional regulators of adipogenesis. Alternatively, autophagy may promote adipogenesis through cytoplasmic remodeling. With differentiation, the cytoplasm of white adipocytes becomes almost completely occupied by a unilocular LD, and autophagy may function by removing cellular organelles and other cytosolic components to allow sufficient space for the LD. WAT was previously thought to experience minimal cell turnover, but recent findings indicate that 10% of human adult adipocytes are replaced annually.28 The recently reported increase in autophagy in the adipose tissue of obese humans, particularly with insulin resistance,29 suggests that increased autophagy might fuel WAT enlargement in obesity. However, studies to date have not determined the effects of altered levels of autophagy on differentiated WAT or BAT. Specifically, does lipophagy function in the conventional way in these cells to regulate lipid storage? Investigations with an inducible adipocyte-specific knockout of autophagy are needed to address this question in differentiated adipose tissue.
A more definitive role for lipophagy in determining cellular phenotype has been recently demonstrated in stellate cell transdifferentiation or activation. Hepatic stellate cells are normally in a quiescent state marked by their storage of lipid in the form of retinoid esters and TGs in LDs and their lack of matrix gene expression.30 With a fibrogenic stimulus, these cells lose their lipid stores and transdifferentiate into proliferative myofibroblasts to produce the extracellular matrix that leads to hepatic fibrosis.30 Although the loss of LDs is a characteristic feature of this transdifferentiation, the role of lipid degradation in the activation process has been unclear. Studies have now established a critical function for lipid removal by lipophagy during activation. Hepatic stellate cells activated in vitro or isolated in the activated state from fibrotic mouse livers had increased levels of autophagy as compared with quiescent cells.11, 31 A pharmacological or genetic inhibition of autophagy blocked this transdifferentiation into myofibroblasts.11, 31 A stellate cell-specific mouse knockdown of atg7 decreased hepatic fibrosis induced by two stimuli in parallel with decreased stellate cell activation.11 The specific role of lipophagy in this effect was demonstrated by the fact that autophagic inhibition increased stellate cell LD and TG content and decreased ATP levels. This lipid breakdown promoted transdifferentiation as chemical inhibition of β-oxidation blocked cell activation, and supplementation with the FFA oleate partially reversed the inhibitory effect of the loss of autophagy.11 These findings suggest that stellate cells increase autophagy in order to break down lipids to meet increased energy demands for activation. However, there may be other mechanisms by which autophagy promotes activation. How diverse fibrogenic stimuli increase stellate cell autophagy has not been addressed. The activation of tissue-specific stellate cells is a central mechanism of fibrosis in many organs in addition to the liver, and fibroblasts from fibrotic human lungs also have increased autophagy, which when inhibited resulted in a reversal of their fibrotic phenotype.11 Thus, the ability of lipophagy to promote stellate cell/fibroblast transdifferentiation might be a central mechanism for the development of fibrosis in many tissues.
Autophagy Functions in Hypothalamic Neurons to Regulate Whole-body Energy Stores
The hypothalamus is an important regulator of whole-body energy homeostasis and lipid stores through its integration of various nutrient signals to determine food intake.32, 33 Hypothalamic lipid metabolism serves as a sensor of nutrient levels by altering intracellular malonyl-CoA and long-chain fatty-acid levels.34, 35, 36 The importance of intracellular lipid metabolism in hypothalamic nutrient sensing, together with the ability of nutrient levels to regulate autophagy and of autophagy to degrade lipids, suggested that lipophagy may mediate hypothalamic regulation of food intake. Two recent studies have confirmed an important function for autophagy in hypothalamic neurons in the control of food intake,10, 37 however, opposing effects have been reported and the function of autophagy in these cells remains unclear (Figure 2).
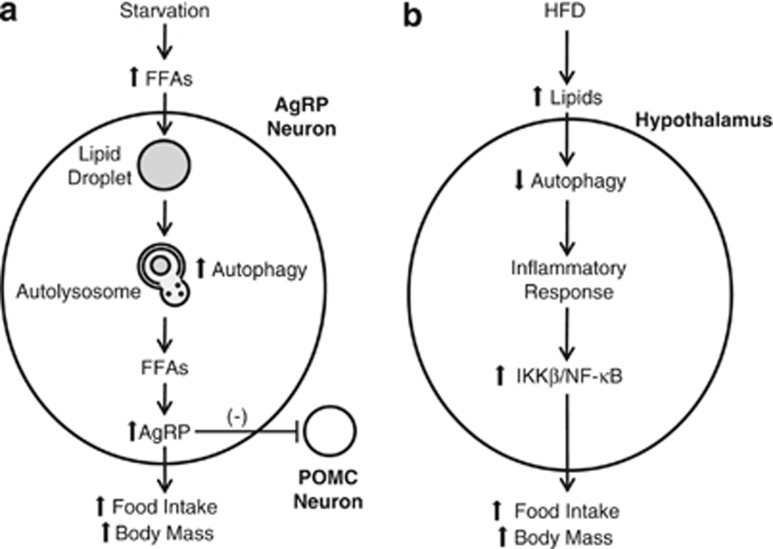
Figure 2.
Opposing views of the effects of autophagy on hypothalamic food sensing. (a) With acute starvation, the increased serum FFAs released from adipocyte lipid stores result in increased FFA levels in AgRP neurons and FFA incorporation into LDs. Starvation-stimulated autophagy breaks down LDs into FFAs that induce AgRP production. AgRP increases food intake both directly and indirectly through inhibitory effects on POMC neurons. An increase in autophagy in hypothalamic AgRP neurons therefore translates into increased food intake and body mass.10 (b) With chronic HFD feeding, the hypothalamic levels of autophagy are decreased. Reduced autophagy leads to an inflammatory response, which includes activation of the IκB kinase β (IKKβ)/NF-κB signaling pathway, that increases food intake. In this proposed model, a decrease in hypothalamic neuron autophagy increases food intake and body mass37
The arcuate nucleus of the mediobasal hypothalamus regulates food intake through the opposing actions of two populations of neurons. The anabolic agouti-related peptide (AgRP) neurons co-express AgRP and neuropeptide Y that stimulate food intake and reduce energy expenditure.32 The catabolic proopiomelanocortin (POMC) neurons express POMC, which is processed to α-melanocyte-stimulating hormone to suppress feeding.32 Studies in serum-starved AgRP-expressing GT1-7 cells and primary hypothalamic neurons from fasted mice have shown that autophagy is induced in these cells by nutrient deprivation.10 ATG7F/F-AgRP-cre-knockout mice with an inhibition of autophagy in AgRP neurons are leaner, more active and consume less food after starvation.10 These effects were attributed to decreased AgRP expression and a concomitant increase in POMC levels. AgRP decreased with the loss of autophagy because the AgRP neurons relied on lipophagy-mediated lipid metabolism to fuel production of this neuropeptide, although the mechanism of this effect was not defined. One possibility is through effects of autophagy on PPARγ signaling. PPARγ activates AgRP and suppresses POMC neurons by downregulating intracellular levels of reactive oxygen species.38 FFAs are a well-characterized ligand for PPARγ,39 and their generation by autophagy-mediated lipid metabolism may activate hypothalamic PPARγ to induce AgRP expression. The findings also suggest a possible new mechanism for the inhibition of food intake by the hormone leptin. Leptin activates hypothalamic mTOR signaling,40 which is the primary inhibitory pathway for autophagy,41 suggesting that leptin may induce an mTOR-mediated decrease in autophagy in AgRP neurons that reduces food intake. It would be interesting to see whether a loss of autophagy in AgRP neurons can prevent the effects of leptin deficiency by crossing ATG7F/F-AgRP-cre mice with ob/ob mice and examining the effects on food intake.
A second study employed an alternative experimental design to examine autophagic function in the arcuate nucleus of the hypothalamus. Based on the findings of decreased mediobasal hypothalamic autophagy in obese mice, Meng et al.37 delivered a lentiviral atg7 shRNA into this region in the brain in normal mice to inhibit autophagy in the arcuate nucleus. In contrast to the previous study,10 this approach presumably decreased autophagy in both AgRP and POMC neurons, and these findings reflect the impact of a generalized defect on hypothalamic autophagy. The knockdown mice had increased food intake and body weights and reduced energy expenditure, findings opposite to those in the AgRP cell-specific knockouts.37 Based on the known ability of a loss of autophagy to promote inflammation in non-neuronal cells,42, 43 the pro-inflammatory NF-κB pathway was examined and found to be activated in the hypothalamus of the mice with inhibited autophagy.37 Although evidence of inflammation was not provided in this study, subsequent investigations by others have confirmed that hypothalamic inflammation occurs in obese mice.44, 45 NF-κB activation presumably mediated the effects of the loss of autophagy on body mass and food intake as these effects were reversed in mice with a knockout of the NF-κB-activating IκB kinase β.37 The opposing findings in the two studies of hypothalamic autophagy may have resulted from the different experimental designs of a selective versus generalized inhibition of hypothalamic autophagy. Which approach is more physiologically relevant needs to be determined by technically difficult cell-specific studies of the levels of autophagy in the different neuronal cell types in vivo in an obese animal. An additional difference between the two investigations that may have led to the divergent results is that Kaushik et al.10 employed the rapid and transient lipid stimulus of starvation, whereas Meng et al.37 examined the chronic model by HFD feeding that might better reflect the hypothalamic changes in obese humans.
Lipophagy Regulates Macrophage Cholesterol Efflux
Atherosclerosis leads to coronary artery and cerebral vascular disease.46, 47 Many factors promote atherosclerosis, but prominent among them is dysregulated lipid metabolism in macrophages leading to their conversion into lipid-laden foam cells that contribute to the development of atherosclerotic plaques.48 The predominant lipid in foam cells is cholesterol in its free or esterified form. Pivotal to the mobilization of cholesteryl esters (CEs) from macrophages is their breakdown from LDs by neutral CE hydrolases.49 Enhancing the efflux of CE from macrophages is a potential approach to preventing foam cell formation and attenuating atherosclerotic lesions.50, 51
The discovery of lipophagy suggested that autophagy may contribute to macrophage cholesterol efflux. Ouimet et al.,52 examined this question in cultured macrophages preloaded with acetylated low-density lipoproteins (AcLDLs) that were rapidly hydrolyzed and reesterified into LDs.53 Cellular CE levels increased with inhibitors of either neutral CE hydrolases or lysosomal function. In parallel, efflux of free cholesterol to ApoA-1, a major protein component of high-density lipoprotein, was impaired with either inhibitor. These findings indicate that besides mobilization by neutral CE hydrolases, CEs in LDs can be degraded in lysosomes for efflux. Lysosomal inhibition only affected efflux from lipid-loaded, but not normal cells, suggesting a specific role for lysosomal degradation in foam cells. Lysosomal acid lipase (LAL) was identified as the enzyme-hydrolyzing CEs in lysosomes. In lipid-loaded atg5, knockout macrophage cholesterol efflux was decreased, but LAL inhibition had no additional effect on cholesterol efflux in these cells, indicating that autophagy trafficked CEs to lysosomes for degradation by LAL. Studies in vivo demonstrated that AcLDL-pretreated, autophagy-deficient macrophages injected into normal mice had decreased cholesterol efflux.
Although these findings are intriguing, further studies are needed to confirm the atheroprotective effects of autophagy in vivo. The conclusions of this study were largely based on investigations with AcLDL, which is not the most physiologically relevant lipid molecule.54 Other modified LDLs, such as oxidized low-density lipoprotein (OxLDL) and LDL aggregates, are more important in the development of atherosclerosis.55 OxLDL tends to be trapped in lysosomes,56 suggesting that this lipoprotein does not enter LDs to be a substrate for lipophagy. Important to the consideration of autophagy as a therapeutic target is whether levels of autophagy are altered in macrophages as they progress to foam cells. Ouimet et al.52 concluded that lipid loading increased macrophage autophagy but a careful quantification of autophagic function was not performed. In addition, this process might not be ameliorable to drug therapy, as agents to augment autophagy could trigger a macrophage inflammatory response that would promote formation of atherosclerotic lesions.57 Increased autophagic cholesterol breakdown would raise free cholesterol levels, which also might have an adverse cellular effect. For example, free cholesterol induces the unfolded protein response in the endoplasmic reticulum and triggers apoptosis in macrophages.58
Viruses Utilize Lipophagy for Replication
The ability of autophagy to eliminate unwanted cellular components suggests an important role for this degradative pathway in the elimination of intracellular pathogens.59 Conversely, infectious agents may subvert the autophagic pathway to promote their own survival. The two major hepatitis viruses, both of which are significant causes of chronic human liver disease, alter host cell autophagic function by unknown mechanisms. The RNA hepatitis C virus (HCV)60, 61 and the DNA hepatitis B virus62, 63 upregulate autophagy, which acts to promote rather than to block viral replication. HCV has a well-established association with LDs and the core protein of HCV attaches to LD for the apparent purpose of promoting viral assembly through a mechanism that is unknown.64 Studies with another RNA flavivirus family member, dengue virus, have for the first time demonstrated that lipophagy is one mechanism by which augmented autophagy fuels viral replication.
The site of dengue virus assembly is less well-defined than for HCV, but similar to HCV evidence for LD-associated replication exists.65 A recent study indicates that this upregulation of autophagy serves the specific purpose of promoting lipid breakdown to supply energy for viral replication.66 In hepatoma cells, dengue virus infection promoted the formation of autophagosomes that associated with LDs. As a result, lipophagy was stimulated in infected cells as reflected by a significant reduction in cellular TGs and LD volume in concert with an increase in FFA β-oxidation. This increase in cellular β-oxidation was blocked by an inhibition of autophagy, which also prevented viral replication. Supplementation with FFAs prevented the block in replication induced by an inhibition of autophagy, indicating that autophagy degrades LDs into FFAs that are metabolized by β-oxidation to support viral replication. Preliminary studies with HCV indicate that this virus is also dependent on β-oxidation.66 The ability of flaviviruses to induce lipophagy may therefore be a critical part of the viral life cycle in hepatocytes and a potential target for new therapeutic agents. The mechanism of viral induction is unknown but has been linked to a specific viral gene, NS4A.67 The function of virus-induced autophagy in other cell types may differ, however, as autophagy has been reported to limit dengue replication in monocytes.68 Whether other viruses such as hepatitis B, which alter autophagic function, utilize lipophagy to support replication also remains to be determined.
Lipophagy Mediates Resistance to Cell Death
Cell death and autophagy have long been considered interrelated, although the nature of their relationship has been controversial.69 Although early evidence suggested that autophagy is a form of cell death, more recent investigations support a predominant if not exclusive function for autophagy as a pro-survival pathway.70, 71 The metabolism of lipids by lipophagy suggests several new mechanisms by which autophagy may act to prevent cell death. Potential cell-protective effects of lipophagy include the breakdown of lipids to supply energy to maintain cell function and prevent ATP depletion that leads to necrosis or apoptosis from mitochondrial dysfunction and the ability to promote the metabolism of potentially toxic lipid molecules (Figure 3).
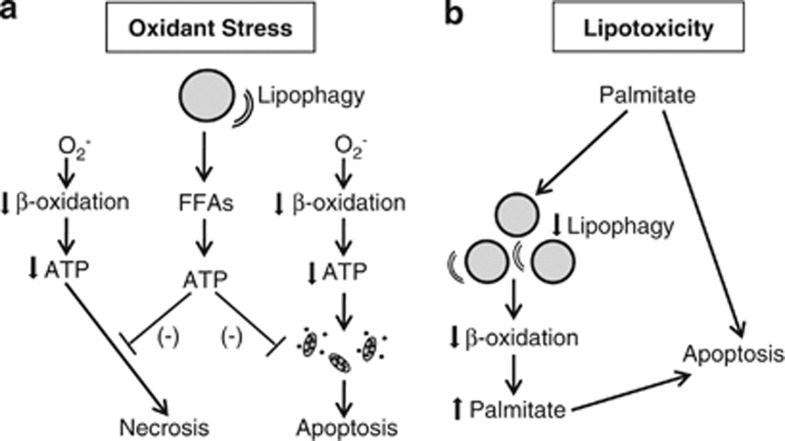
Figure 3.
Two mechanisms for the alteration of cell death responses by lipophagy. (a) Superoxide-induced oxidant stress decreases rates of mitochondrial β-oxidation, leading to decreased cellular ATP content. A profound decrease in ATP leads to cell death from necrosis. A more modest decrease in ATP promotes mitochondrial dysfunction and activation of the mitochondrial death pathway with cytochrome-c release that triggers cell death from apoptosis. Lipophagy mediates resistance to both of these forms of cell death by breaking down LDs into FFAs that can be utilized to make ATP. (b) Lipotoxicity from the saturated FFA palmitate may be mediated in part by its ability to inhibit lipophagy, resulting in decreased levels of β-oxidation and palmitate metabolism, which increase cellular levels of palmitate to trigger apoptosis
Lipophagy has been demonstrated to mediate hepatocyte resistance to death from oxidant stress. A genetic knockdown of atg5 sensitized hepatocytes to death from nontoxic levels of oxidative stress from the superoxide generator menadione and increased cell death from toxic levels of oxidant stress.72 In the absence of autophagy, menadione-induced decreases in cellular rates of β-oxidation and ATP content were amplified and sustained rather than transient. These findings suggest that lipophagy-generated FFAs are critical to sustain rates of β-oxidation sufficient to maintain cellular energy homeostasis under oxidant stress. This conclusion was further supported by the fact that an inhibition of β-oxidation sensitized cells to death from menadione-induced oxidant stress and that supplementation with the FFA oleate blocked death from menadione in cells lacking autophagy. How widespread this function of lipophagy is in other cell types under oxidant stress, or in cellular responses to other death stimuli, needs to be established.
Cell death can result from excessive organ lipid accumulation as occurs in obesity and the metabolic syndrome. One mechanism of cellular injury from lipid overload can be through the toxic effects of saturated FFAs or lipotoxicity. Two of the most prevalent human liver diseases, nonalcoholic fatty liver disease and alcoholic liver disease, are associated with increased hepatic lipid accumulation or steatosis.73 Excessive intracellular lipids can be a source of oxidative stress or toxic FFAs that trigger hepatocyte injury and cell death.73 Lipophagy has the potential to prevent liver injury by limiting steatosis, metabolizing toxic FFAs or maintaining energy homeostasis.74 Autophagy has been shown to protect against ethanol-induced fatty liver injury.75 Autophagy was found to be increased both in the livers of binge alcohol-fed mice and cultured hepatocytes treated with ethanol. Autophagic degradation of long-lived proteins was not increased, but findings of increased numbers of autophagosomes containing LDs and mitochondria suggested that selective increases in lipophagy and mitophagy occurred in response to alcohol. Autophagy protected against alcohol-induced injury as a pharmacological or genetic inhibition of autophagy increased cell death in ethanol-treated cultured hepatocytes and mouse liver. Increasing autophagy with rapamycin decreased alcohol-induced lipid accumulation in vivo, and autophagic inhibition worsened steatosis. These studies indicate that lipophagy functions to block the excessive lipid accumulation induced by alcohol, but do not directly prove that the protective effects of autophagy on liver injury are the result of lipophagy. The mechanisms by which autophagy ameliorates alcoholic liver disease need to be further examined, preferably in rodents with chronic alcohol ingestion that are a better model of human alcoholic liver disease. Whether lipophagy protects other tissues from injury from excessive lipid accumulation must also be investigated.
Levels of Autophagy are Altered by Cellular Lipid Content
Several studies have suggested that in addition to the ability of autophagy to mediate lipid metabolism, lipids regulate cellular levels of autophagy. Initial evidence for this relationship was provided from studies in which HFD feeding suppressed autophagy in mouse liver.1, 76 Mice fed a HFD have decreased trafficking of lipids through the autophagic pathway.1 Decreased numbers of autophagosomes by electron microscopy and protein levels of autophagic factors have been reported in genetically obese and HFD-fed mice, although in the HFD-fed animals the protein changes occurred only after prolonged feeding.76 Adenoviral expression of the autophagy gene atg7 increased autophagic function in obese mice suggesting that the decrease in autophagic proteins reduced levels of autophagy by decreasing autophagosome formation.76 In contrast, other investigations of obese ob/ob mouse livers have demonstrated increased autophagosome number, normal fusion of autophagosomes to lysosomes, but decreased autophagic function due to a defect in lysosomal acidification and reduction in cathepsin L that impaired substrate degradation in autolysosomes.77 Other studies using an in vitro assay to measure autophagosome–lysosome fusion have reported a fusion defect in the organelles from the livers of HFD-fed mice.78 This defect was attributed to HFD-induced changes in membrane lipid composition. Thus, there is agreement among studies that hepatic autophagy is impaired with increased body mass, but defects at the levels of autophagosome formation, fusion and substrate degradation have all been reported as mechanisms of this effect (Figure 4). Further studies to clarify the effects of a chronic lipid overload on autophagic function in the liver and other organs are needed to resolve these differences as the site of impairment may be an important therapeutic target for diseases that complicate obesity.
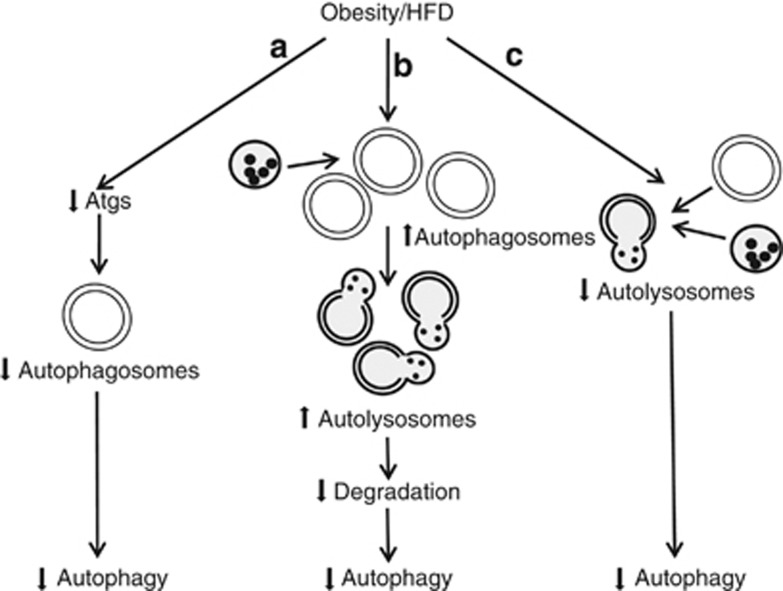
Figure 4.
Potential mechanisms implicated in the decreased hepatic autophagy that occurs with obesity or HFD feeding. (a) Obesity or HFD may reduce levels of various autophagy factors (Atgs), causing decreased autophagosome formation and levels of autophagy.76 (b) The liver may form more autophagosomes and autolysosomes, but levels of autophagy decrease nonetheless due to lysosomal defects that preclude degradation.77 (c) Impaired fusion of autophagosomes and lysosomes may also decrease autophagy78
A potential mediator of decreased hepatic autophagic function with HFD feeding is increased FFA levels. Studies have focused on the selective ability of toxic saturated FFAs such as palmitate, but not nontoxic, unsaturated FFAs, such as oleate, to inhibit autophagy. In hepatoma cells and primary mouse hepatocytes, palmitate decreased and oleate increased autophagy.79 Pharmacological inhibition of autophagy sensitized these cells to death from palmitate or oleate, indicating that autophagy was essential for resistance to FFA toxicity. The differential effects of the two FFAs on autophagy may in part explain the selective toxicity of palmitate. Neither the mechanism of autophagic inhibition or cell death was defined. Similarly autophagy was inhibited by palmitate in a pancreatic β-cell line and primary β cells.80 Autophagic inhibition was secondary to decreased lysosomal acidification that led to reduced hydrolase activity. Reversal of the inhibition of autophagy by rapamycin decreased palmitate toxicity in these cells. These findings suggest that autophagy is a critical mechanism of resistance against FFA toxicity, and lipotoxicity may be the mechanism by which an inhibition of autophagy leads to spontaneous pancreatic β cell death in vivo.81, 82 In contrast to hepatocytes,79 oleate also inhibited β cell autophagy as measured by long-lived protein degradation, although some of this effect was mediated by effects on proteasomal rather than lysosomal degradation.80 There may therefore be cell-type specificity to the effects of FFAs on autophagy, and these studies need to be extended to other cell types. Factors such as the concentration of FFAs and time of exposure may also affect cellular responses and complicate comparisons between studies.
Variable effects on autophagy have been reported for cholesterol. An acute cholesterol stimulus such as AcLDL loading in macrophages,52 or free cholesterol treatment of smooth muscle cells,83 increased autophagy. Alternatively, cholesterol84 and sterol85 depletion have also been reported to increase autophagy. Potentially, autophagy functions to both rid the cell of excessive cholesterol and break down LD-stored cholesterol in times of deprivation. One mechanism for the effects of cholesterol on autophagy was demonstrated by a genome-wide DNA-binding study of sterol regulatory element-binding protein 2 (SREBP-2),85 a transcription factor that regulates cholesterol metabolism,86 which showed that autophagy genes were enriched as SREBP-2 targets. This in silico result was confirmed in cell culture studies, which demonstrated that inhibition of srebp-2 dramatically diminished autophagosome formation during sterol or nutrient depletion, indicating that SREBP-2 can upregulate autophagy under certain circumstances. A further delineation of the mechanism by which cholesterol regulates autophagy may facilitate an understanding of the pathogenesis of many neurodegenerative diseases, as altered cholesterol homeostasis and autophagic function are commonly present in these diseases.87, 88
In complex metabolic settings such as obesity and diabetes, many factors likely regulate autophagic function. Increased glucose amplified the inhibition of autophagy in pancreatic β cells by palmitate, suggesting that hyperglycemia from diabetes could worsen the effects of FFAs on autophagy.80 Future investigations will likely determine that autophagy in metabolically active cells, such as hepatocytes and pancreatic β cells, is regulated by multiple serum factors that are altered with obesity and insulin resistance such as glucagon-like peptide 1.89
Conclusions
Studies have demonstrated a second fundamental mechanism for the metabolism of LD-stored lipids through the lysosomal pathway of autophagy. Lipophagy has now been demonstrated to function in a number of different cell types, indicating that this form of autophagy is a common pathway of cellular lipid metabolism. Lipophagy is a selective form of autophagy, but the mechanism by which LDs are recognized as substrate and how the relative amounts of lipids targeted for degradation are regulated by nutritional status remain unknown. Studies to date have begun to reveal a number of disparate functions for lipophagy in cellular physiology and pathophysiology. The most obvious function for lipophagy is as a regulator of cellular lipid content. In the liver, it has been clearly demonstrated that a defect in autophagy promotes hepatic steatosis and lipophagy might have a similar role in other organs. The second function is that breakdown products from this pathway such as FFAs affect cell physiology. The most important role of lipophagy in this regard is the maintenance of adequate levels of β-oxidation to supply ATP. This function of lipophagy has been implicated in the regulation of cell resistance to death stimuli and in cellular transdifferentiation. Likely, the ability of lipophagy to break down lipids serves other as-yet undelineated functions such as to supply lipids for structural elements of the cell or to regulate lipid-dependent cell signaling.
Considerable interest exists in whether defects in autophagy mediate human diseases. The existence of lipophagy suggests that impaired autophagy may be a fundamental mechanism of disorders of lipid metabolism such as obesity and the metabolic syndrome. Individual variation in autophagic function may determine the development or outcome from these human diseases. Autophagy therefore has the potential to serve as an important therapeutic target for the treatment of these disorders.
Acknowledgments
This work was supported in part by the National Institutes of Health grants DK044234, DK061498 and AG031782 to MJ Czaja and a training grant DK007218 for K Liu.
Glossary
- TG
triglyceride
- LD
lipid droplet
- FFA
free fatty acid
- LC3
microtubule-associated protein light chain 3
- SNARE
soluble NSF attachment protein receptor
- WAT
white adipose tissue
- BAT
brown adipose tissue
- PPARγ
peroxisome proliferator-activated receptor gamma
- HFD
high-fat diet
- AgRP
agouti-related peptide
- POMC
proopiomelanocortin
- CE
cholesteryl ester
- AcLDL
acetylated low-density lipoprotein
- LAL
lysosomal acid lipase
- OxLDL
oxidized low-density lipoprotein
- SREBP-2
sterol regulatory element-binding protein 2
The authors declare no conflict of interest.
Footnotes
Edited by M Piacentini
References
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- Thiele C, Spandl J. Cell biology of lipid droplets. Curr Opin Cell Biol. 2008;20:378–385. doi: 10.1016/j.ceb.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Zechner R, Madeo F. Cell biology: another way to get rid of fat. Nature. 2009;458:1118–1119. doi: 10.1038/4581118a. [DOI] [PubMed] [Google Scholar]
- Marchesini G, Brizi M, Bianchi G, Tomassetti S, Bugianesi E, Lenzi M, et al. Nonalcoholic fatty liver disease: a feature of the metabolic syndrome. Diabetes. 2001;50:1844–1850. doi: 10.2337/diabetes.50.8.1844. [DOI] [PubMed] [Google Scholar]
- Sanyal AJ, Campbell-Sargent C, Mirshahi F, Rizzo WB, Contos MJ, Sterling RK, et al. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 2001;120:1183–1192. doi: 10.1053/gast.2001.23256. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Rodriguez-Navarro JA, Arias E, Kiffin R, Sahu S, Schwartz GJ, et al. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metab. 2011;14:173–183. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom P, Andersson L, Rutberg M, Perman J, Lidberg U, Johansson BR, et al. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat Cell Biol. 2007;9:1286–1293. doi: 10.1038/ncb1648. [DOI] [PubMed] [Google Scholar]
- Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Yoshimura K, Furuya N, Koike M, Ueno T, Komatsu M, et al. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun. 2009;382:419–423. doi: 10.1016/j.bbrc.2009.03.039. [DOI] [PubMed] [Google Scholar]
- Czaja MJ. Autophagy in health and disease. 2. Regulation of lipid metabolism and storage by autophagy: pathophysiological implications. Am J Physiol Cell Physiol. 2010;298:C973–C978. doi: 10.1152/ajpcell.00527.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, et al. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest. 2011;121:2102–2110. doi: 10.1172/JCI46069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger RH. Minireview: weapons of lean body mass destruction: the role of ectopic lipids in the metabolic syndrome. Endocrinology. 2003;144:5159–5165. doi: 10.1210/en.2003-0870. [DOI] [PubMed] [Google Scholar]
- Unger RH, Orci L. Diseases of liporegulation: new perspective on obesity and related disorders. FASEB J. 2001;15:312–321. doi: 10.1096/fj.00-0590. [DOI] [PubMed] [Google Scholar]
- Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Ann Rev Cell Dev Biol. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci USA. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerga R, Zhang Y, Chen PH, Goldman S, Jin S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5:1118–1130. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding KL, Arner E, Westermark PO, Bernard S, Buchholz BA, Bergmann O, et al. Dynamics of fat cell turnover in humans. Nature. 2008;453:783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- Kovsan J, Bluher M, Tarnovscki T, Kloting N, Kirshtein B, Madar L, et al. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metabol. 2011;96:E268–E277. doi: 10.1210/jc.2010-1681. [DOI] [PubMed] [Google Scholar]
- Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoen LF, Guimaraes EL, Dolle L, Mannaerts I, Najimi M, Sokal E, et al. A role for autophagy during hepatic stellate cell activation. J Hepatol. 2011;55:1353–1360. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Sandoval D, Cota D, Seeley RJ. The integrative role of CNS fuel-sensing mechanisms in energy balance and glucose regulation. Annu Rev Physiol. 2008;70:513–535. doi: 10.1146/annurev.physiol.70.120806.095256. [DOI] [PubMed] [Google Scholar]
- Schwartz MW, Porte D. Diabetes, obesity, and the brain. Science. 2005;307:375–379. doi: 10.1126/science.1104344. [DOI] [PubMed] [Google Scholar]
- Lam TK, Schwartz GJ, Rossetti L. Hypothalamic sensing of fatty acids. Nat Neurosci. 2005;8:579–584. doi: 10.1038/nn1456. [DOI] [PubMed] [Google Scholar]
- He W, Lam TK, Obici S, Rossetti L. Molecular disruption of hypothalamic nutrient sensing induces obesity. Nat Neurosci. 2006;9:227–233. doi: 10.1038/nn1626. [DOI] [PubMed] [Google Scholar]
- Hu Z, Cha SH, Chohnan S, Lane MD. Hypothalamic malonyl-CoA as a mediator of feeding behavior. Proc Natl Acad Sci USA. 2003;100:12624–12629. doi: 10.1073/pnas.1834402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Cai D. Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IκB kinase β (IKKβ)/NF-κB pathway. J Biol Chem. 2011;286:32324–32332. doi: 10.1074/jbc.M111.254417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diano S, Liu ZW, Jeong JK, Dietrich MO, Ruan HB, Kim E, et al. Peroxisome proliferation-associated control of reactive oxygen species sets melanocortin tone and feeding in diet-induced obesity. Nat Med. 2011;17:1121–1127. doi: 10.1038/nm.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARγ 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Cota D, Proulx K, Smith KA, Kozma SC, Thomas G, Woods SC, et al. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- McNay DE, Briancon N, Kokoeva MV, Maratos-Flier E, Flier JS. Remodeling of the arcuate nucleus energy-balance circuit is inhibited in obese mice. J Clin Invest. 2012;122:142–152. doi: 10.1172/JCI43134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JP, Yi CX, Schur EA, Guyenet SJ, Hwang BH, Dietrich MO, et al. Obesity is associated with hypothalamic injury in rodents and humans. J Clin Invest. 2012;122:153–162. doi: 10.1172/JCI59660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- Weber C, Noels H. Atherosclerosis: current pathogenesis and therapeutic options. Nat Med. 2011;17:1410–1422. doi: 10.1038/nm.2538. [DOI] [PubMed] [Google Scholar]
- Moore KJ, Tabas I. Macrophages in the pathogenesis of atherosclerosis. Cell. 2011;145:341–355. doi: 10.1016/j.cell.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Zhao B, Bie J, Song J. Macrophage cholesteryl ester mobilization and atherosclerosis. Vascular Pharmaco. 2010;52:1–10. doi: 10.1016/j.vph.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brufau G, Groen AK, Kuipers F. Reverse cholesterol transport revisited: contribution of biliary versus intestinal cholesterol excretion. Arterioscler Thromb Vasc Biol. 2011;31:1726–1733. doi: 10.1161/ATVBAHA.108.181206. [DOI] [PubMed] [Google Scholar]
- Khera AV, Rader DJ. Future therapeutic directions in reverse cholesterol transport. Curr Atheroscler Rep. 2010;12:73–81. doi: 10.1007/s11883-009-0080-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BE, Griffin EE, Ullery JC, Jerome WG. Effects of cellular cholesterol loading on macrophage foam cell lysosome acidification. J Lipid Res. 2007;48:1012–1021. doi: 10.1194/jlr.M600390-JLR200. [DOI] [PubMed] [Google Scholar]
- Steinberg D. The LDL modification hypothesis of atherogenesis: an update. J Lipid Res. 2009;50:S376–S381. doi: 10.1194/jlr.R800087-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin EE, Ullery JC, Cox BE, Jerome WG. Aggregated LDL and lipid dispersions induce lysosomal cholesteryl ester accumulation in macrophage foam cells. J Lipid Res. 2005;46:2052–2060. doi: 10.1194/jlr.M500059-JLR200. [DOI] [PubMed] [Google Scholar]
- Yancey PG, Jerome WG. Lysosomal cholesterol derived from mildly oxidized low density lipoprotein is resistant to efflux. J Lipid Res. 2001;42:317–327. [PubMed] [Google Scholar]
- Schrijvers DM, De Meyer GR, Martinet W. Autophagy in atherosclerosis: a potential drug target for plaque stabilization. Arterioscler Thrombosis Vasc Biol. 2011;31:2787–2791. doi: 10.1161/ATVBAHA.111.224899. [DOI] [PubMed] [Google Scholar]
- Feng B, Yao PM, Li Y, Devlin CM, Zhang D, Harding HP, et al. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5:781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Akira S. Regulation of innate immune responses by autophagy-related proteins. J Cell Biol. 2010;189:925–935. doi: 10.1083/jcb.201002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci USA. 2010;107:4383–4388. doi: 10.1073/pnas.0911373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60–71. doi: 10.1002/hep.22581. [DOI] [PubMed] [Google Scholar]
- McLauchlan J. Lipid droplets and hepatitis C virus infection. Biochim Biophys Acta. 2009;1791:552–559. doi: 10.1016/j.bbalip.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, et al. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JE, Wudzinska A, Datan E, Quaglino D, Zakeri Z. Flavivirus NS4A-induced autophagy protects cells against death and enhances virus replication. J Biol Chem. 2011;286:22147–22159. doi: 10.1074/jbc.M110.192500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyasrivanit M, Greenwood MP, Murphy D, Isidoro C, Auewarakul P, Smith DR. Induced autophagy reduces virus output in dengue infected monocytic cells. Virology. 2011;418:74–84. doi: 10.1016/j.virol.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Singh R, Massey AC, Kane SS, Kaushik S, Grant T, et al. Loss of macroautophagy promotes or prevents fibroblast apoptosis depending on the death stimulus. J Biol Chem. 2008;283:4766–4777. doi: 10.1074/jbc.M706666200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Kepp O, Michaud M, Martins I, Minoux H, Metivier D, et al. Association and dissociation of autophagy, apoptosis and necrosis by systematic chemical study. Oncogene. 2011;30:4544–4556. doi: 10.1038/onc.2011.168. [DOI] [PubMed] [Google Scholar]
- Wang Y, Singh R, Xiang Y, Czaja MJ. Macroautophagy and chaperone-mediated autophagy are required for hepatocyte resistance to oxidant stress. Hepatology. 2010;52:266–277. doi: 10.1002/hep.23645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy JK, Rao MS. Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am J Physiol Gastrointest Liver Physiol. 2006;290:G852–G858. doi: 10.1152/ajpgi.00521.2005. [DOI] [PubMed] [Google Scholar]
- Amir M, Czaja MJ. Autophagy in nonalcoholic steatohepatitis. Expert Rev Gastroenterol Hepatol. 2011;5:159–166. doi: 10.1586/egh.11.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inami Y, Yamashina S, Izumi K, Ueno T, Tanida I, Ikejima K, et al. Hepatic steatosis inhibits autophagic proteolysis via impairment of autophagosomal acidification and cathepsin expression. Biochem Biophys Res Commun. 2011;412:618–625. doi: 10.1016/j.bbrc.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei S, Ni HM, Manley S, Bockus A, Kassel KM, Luyendyk JP, et al. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J Pharmacol Exp Ther. 2011;339:487–498. doi: 10.1124/jpet.111.184341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Las G, Serada SB, Wikstrom JD, Twig G, Shirihai OS. Fatty acids suppress autophagic turnover in β-cells. J Biol Chem. 2011;286:42534–42544. doi: 10.1074/jbc.M111.242412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, et al. Autophagy is important in islet homeostasis and compensatory increase of β cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, et al. Loss of autophagy diminishes pancreatic β cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Xu K, Yang Y, Yan M, Zhan J, Fu X, Zheng X. Autophagy plays a protective role in free cholesterol overload-induced death of smooth muscle cells. J Lipid Res. 2010;51:2581–2590. doi: 10.1194/jlr.M005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Ohsaki Y, Tauchi-Sato K, Fujita A, Fujimoto T. Cholesterol depletion induces autophagy. Biochem Biophys Res Commun. 2006;351:246–252. doi: 10.1016/j.bbrc.2006.10.042. [DOI] [PubMed] [Google Scholar]
- Seo YK, Jeon TI, Chong HK, Biesinger J, Xie X, Osborne TF. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 2011;13:367–375. doi: 10.1016/j.cmet.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenza M, Cattaneo E. Emerging roles for cholesterol in Huntington's disease. Trends Neurosci. 2011;34:474–486. doi: 10.1016/j.tins.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Mells JE, Fu PP, Saxena NK, Anania FA. GLP-1 analogs reduce hepatocyte steatosis and improve survival by enhancing the unfolded protein response and promoting macroautophagy. PLoS One. 2011;6:e25269. doi: 10.1371/journal.pone.0025269. [DOI] [PMC free article] [PubMed] [Google Scholar]