Abstract
Nonalcoholic fatty liver disease (NAFLD) is the most common liver disease worldwide. We previously showed that Perilipin 2 (Plin2), a member of lipid droplet protein family, is markedly increased in fatty liver, and its reduction in the liver of diet-induced obese mice by antisense oligonucleotide (ASO) decreased steatosis and enhanced insulin sensitivity. Plin2-ASO treatment markedly suppressed lipogenic gene expression. To gain a better understanding of the biological role of Plin2 in liver, we performed microarray analysis to determine genes differentially regulated by Plin2-ASO compared with a control (scrambled) oligonucleotide (Cont). Male C57BL/6J mice on a high-fat diet were treated with Plin2- or Cont-ASO for 4 wk. Plin2-ASO decreased hepatic triglycerides, and this was associated with changes in expression of 1,363 genes. We analyzed the data for functional clustering and validated the expression of representative genes using real-time PCR. On the high-fat diet, Plin2-ASO decreased the expression of enzymes involved in fatty acid metabolism (acsl1, lipe) and steroid metabolism (hmgcr, hsd3b5, hsd17b2), suggesting that Plin2 affects hepatic lipid metabolism at the transcriptional level. Plin2-ASO also increased the expression of genes involved in regulation of hepatocyte proliferation (afp, H19), mitosis (ccna2, incenp, sgol1), and extracellular matrix (col1a1, col3a1, mmp8). Plin2-ASO had similar effects on gene expression in chow-fed mice. Together, these results indicate that Plin2 has diverse metabolic and structural roles in the liver, and its downregulation promotes hepatic fibrosis and proliferation.
Keywords: perilipins, lipid, liver, microarray, antisense oligonucleotide
nonalcoholic fatty liver disease (NAFLD) is characterized by excessive accumulation of triglycerides and other lipids in the liver in the absence of alcohol consumption (8, 33). The incidence of NAFLD is closely associated with metabolic syndrome, insulin resistance, and Type 2 diabetes (18, 34) and has emerged as a major risk factor for end-stage liver disease and hepatocellular carcinoma (28, 30). Currently, there is no effective treatment to reverse hepatic steatosis or prevent its progression to nonalcoholic steatohepatitis. A better understanding of the pathogenesis of fatty liver may lead to the identification of therapeutic targets (2, 9, 21).
Hepatic triglycerides (TG) are stored within intracellular lipid droplets surrounded by a phospholipid monolayer (8). Structurally related perilipin proteins are found on the surface of lipid droplets and regulate the formation of lipid droplets and lipid metabolism in a wide range of cells (4, 12, 17, 27). Perilipin 2 (Plin2, ADFP) coats lipid droplets in hepatocytes, and its expression is increased with fatty liver in humans and rodents (6, 15, 23). Several lines of evidence indicate that Plin2 plays an active role in the development of fatty liver. Magnusson et al. (20) showed that a reduction in Plin2 decreased TG in hepatocytes, while induction of Plin2 increased TG in hepatocytes. Ablation of the plin2 gene or reduction of Plin2 in vivo by antisense oligonucleotide (ASO) treatment decreased fatty liver in diet-induced obese (DIO) and Lepob mice (6, 7, 15, 37). Plin2-ASO treatment also decreased hepatic VLDL secretion and improved insulin sensitivity (37). Thus, pathways regulated by Plin2 could potentially be targeted to prevent NAFLD and associated metabolic abnormalities.
Here, we performed an unbiased screening of genes whose expression is altered by Plin2-ASO treatment in C57BL/6J mice on a high-fat diet (HF). In agreement with our previous studies, Plin2-ASO treatment decreased hepatic and serum TG (37). Plin2-ASO differentially regulated genes involved in the regulation of fatty acids, TG, and steroid metabolism, as well as hepatocyte proliferation, mitosis, and extracellular matrix.
MATERIALS AND METHODS
Animal studies.
Experiments were performed in accordance with the Institutional Animal Care and Use Committee guidelines and approvals at the University of Pennsylvania, and Eastern Virginia Medical School. Eight-week-old wild-type male C57BL/6J mice (Jackson Laboratories or Harlan Laboratories) were housed n = 5/cage in 12 h light-dark cycle, at ambient temperature of 22°C, and allowed free access to water and an HF (45 kcal% fat; D124551, Research Diets). Another cohort of mice were fed regular rodent chow (5 kcal% fat). Mice were treated twice a week with intraperitoneal injection of Plin2-ASO or a control oligonucleotide (Cont), 25 mg/kg body wt, for 4 wk as described previously (n = 5/diet/treatment) (15, 37). We aimed to detect 45% difference between Plin2-ASO and Cont-ASO treatment. The power calculation predicted that five mice in each group were needed with alpha error of 0.05 and power of test of 0.8. A separate cohort of mice were fed regular rodent chow or HF ad libitum without ASO treatment (n = 4–5 per group). Body composition was measured with nuclear magnetic resonance spectroscopy (Echo Medical Systems, Houston, TX).
Tissue chemistry.
Body weight and tail blood glucose were measured at the end of treatment, after which the mice were euthanized and blood was obtained via cardiac puncture. Liver was rapidly dissected, frozen in liquid nitrogen, and stored at −80°C until further analysis. Serum TG, nonesterified fatty acids (NEFAs), and cholesterol were measured using enzymatic colorimetric assays (5, 37). Liver samples were processed for measurement of TG and cholesterol by enzymatic colorimetric assays as previously described (36). Lipids were also extracted from liver samples using a mixture of chloroform and methanol (2:1), and separated via thin-layer chromatography (TLC) on Silica Gel 60A TLC plates (Whatman), with the solvent system hexane-ether-acetic acid (80:20:1.5). TLC plates were exposed to iodine vapor and then charred with 8% phosphoric acid/3% cupric sulphate solution. Lipid species were quantified with NIH Image J software (10).
Hepatic RNA extraction and analysis of gene expression.
RNA was extracted from liver using TRIzol reagent (Invitrogen), and cDNA was prepared using iSCRPT (Invitrogen). Gene expression was analyzed by qPCR using commercial primers (Applied Biosystems) as described before (37). The level of mRNA expression was normalized to β-actin or GAPDH (37).
Microarray analyses.
Total RNA was extracted from livers, 50 ng of RNA was amplified with the Ovation aminoallyl RNA amplification and labeling system (NuGEN), and 2 μg was applied to whole mouse genome oligo microarray G4122A (Agilent Technologies). Array data were analyzed using GenePix 5.0 software (Axon Instruments). Significance Analysis of Microarrays (Stanford University) was used to define differentially expressed genes with a fold change cut-off of ≥1.5 (up or down) and a false discovery rate of 1%. The probe lists thus obtained (1,407 probes representing 1,363 genes) were uploaded to DAVID (http://david.abcc.ncifcrf.gov), and functional annotation clustering and functional annotation chart were obtained. Microarray data were submitted to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo) under the accession number GSE37011.
Histology.
Liver samples were fixed with 10% buffered formalin overnight and embedded in paraffin. Liver sections were stained with guinea pig anti-Plin2 antibody (Fitzgerald) as previously described (37). Collagen was visualized with Sirius red staining (16). Other liver sections were stained with hematoxylin and eosin and analyzed by an observer blinded to the experiment. The liver tissues were examined with a Nikon 80i microscope, and the number of nuclei was counted within four 160 μm × 160 μm grids, at ×100 magnification. Photographs were captured with a DS-Qi1MC camera and image analysis system.
Statistics.
The data are presented as means ± SE. Differences between two groups were assessed with unpaired Student's t-test. P < 0.05 is considered significant.
RESULTS
Effects of Plin2-ASO treatment on hepatic and serum lipids.
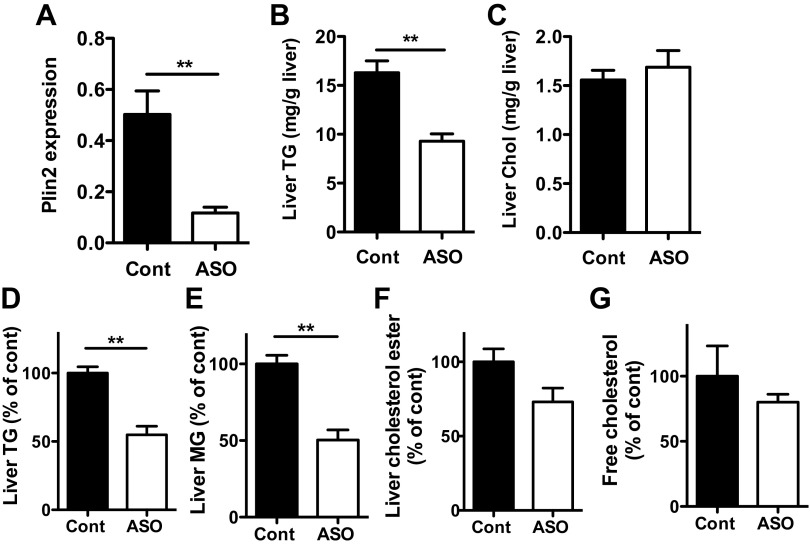
Plin2-ASO treatment of HF-fed mice reduced hepatic Plin2 expression and TG levels as previously reported (Fig. 1, A and B, P < 0.01; Ref. 37). Plin2-ASO treatment did not change body weight of the HF-fed mice, but the body fat content was significantly lower in Plin2-ASO compared with Cont (Table 1, P < 0.05). Plin2-ASO treatment of mice on HF decreased serum TG (Table 1, P < 0.005). In contrast, serum levels of glucose, NEFAs, and cholesterol and liver cholesterol content were not changed by Plin2-ASO treatment (Table 1, Fig. 1C) of HF-fed mice. TLC analysis of liver samples for HF-fed mice also showed significant reductions of TG and monoglycerides, but not free cholesterol or cholesterol ester, in Plin2-ASO-treated mice (Fig. 1, D–G).
Fig. 1.
Mice on high-fat diet (HF) were treated for 4 wk with Plin2-ASO or control oligo (Cont). A: Plin2 mRNA in the liver was determined by qPCR. Enzymatic colorimetric assay comparing liver triglycerides (TG) (B) and liver cholesterol (Chol) (C) levels. Liver lipids were also separated and quantified by TLC. D: TG, E: monoglycerides (MG), F: cholesterol ester, and G: free cholesterol. Data are means ± SE, n = 5 per group. **P < 0.01 vs. Cont.
Table 1.
Effects of Plin2-ASO treatment on metabolic parameters
| High-fat Diet |
Regular-chow Diet |
|||
|---|---|---|---|---|
| Cont | Plin2-ASO | Cont | Plin2-ASO | |
| Body weight, g | 31.3 ± 1.1 | 30.2 ± 0.3 | 26.5 ± 1.0 | 25.9 ± 0.8 |
| Body fat, % | 15.6 ± 2.1 | 10.7 ± 0.8* | 5.4 ± 0.9 | 3.7 ± 0.3 |
| Blood glucose, mg/dl | 203.0 ± 9.5 | 177.2 ± 15.9 | 159.2 ± 4.1 | 125.4 ± 4.8‡ |
| Serum triglycerides, mg/dl | 159.6 ± 10.8 | 89.4 ± 1.4† | 57.2 ± 3.2 | 55.6 ± 5.4 |
| Serum cholesterol, mg/dl | 132.3 ± 5.9 | 141.2 ± 6.3 | 83.7 ± 5.0 | 110 ± 4.6† |
| Serum NEFAs, mEq/ml | 0.80 ± 0.03 | 1.04 ± 0.15 | 0.64 ± 0.1 | 0.73 ± 0.10 |
Data are means ± SE; n = 5 per group. ASO, antisense oligonucleotide; NEFAs, nonesterified fatty acids. For respective diets,
P < 0.05 vs. Cont,
P < 0.005 vs. Cont,
P < 0.0001 vs. Cont.
Microarray analysis of hepatic gene expression in Plin2-ASO-treated HF mice.
The microarray analysis compared gene expression profile between the liver from Plin2-ASO and Cont mice on HF diet and identified 1,363 genes represented by 1,407 probes that were differentially regulated by Plin2-ASO treatment, when a fold change >1.5 and false discovery rate <1% were used as cut-off values (Supplementary Table S1).1 To elucidate the functional features of differentially regulated genes, the data were loaded on to DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov). Functional annotation clustering revealed that cell cycle/mitosis related categories including SP_PIR_KEYWORDS cell cycle and GOTERM_BP_FAT cell cycle were the most enriched group of genes, with enrichment score of 8.2. Kinetochore-related genes had an enrichment score of 4.5, microtubule/cytoskeleton-related genes 5.4, and extracellular matrix genes 4.2. When the functional annotation clustering was performed on the list of genes limited to those reduced in Plin2-ASO compared with Cont, genes related to steroid metabolism, including GOTERM_BP_FAT steroid metabolic process, were prominently reduced with an enrichment score of 4.1.
Plin2-ASO reduces expression of hepatic lipid metabolism genes in HF mice.
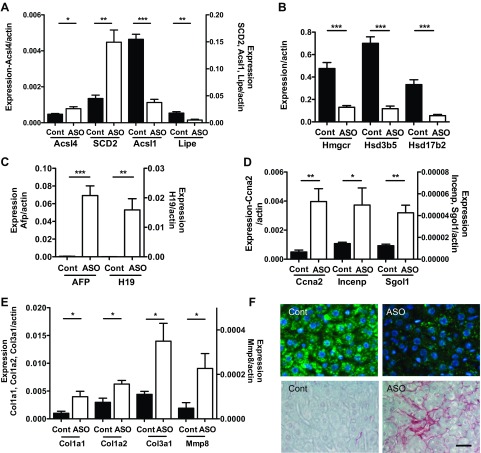
We previously reported that the reduction of hepatic TG by Plin2-ASO treatment of HF-fed mice was associated with decreased expression of genes related to TG synthesis, such as sterol regulatory element-binding protein 1 (SREBP1), fatty acid synthase (FAS), acetyl-CoA carboxylase (ACC), and diacylglycerol acyltransferase-2 (DGAT2) (37). Carnitine palmitoyltransferase 1A (CPT1a) was also reduced by Plin2-ASO treatment (37). Our unbiased screening by microarray revealed that other genes in lipid metabolism were differentially regulated by Plin2-ASO treatment (Supplementary Table S1A). We performed qPCR to validate representative genes in the cluster. The expression of stearoyl-coenzyme A desaturase 2 (Scd2) and acyl-CoA synthetase long-chain family member 4 (Acsl4) was significantly increased by Plin2-ASO treatment of HF-fed mice (Fig. 2A). In contrast, acyl-CoA synthetase long-chain family member 1 (Acsl1), an enzyme implicated in β-oxidation of fatty acids, and hormone-sensitive lipase (Lipe) were identified by microarray to be reduced after by Plin2-ASO treatment. qPCR confirmed significant reductions of Acsl1 (Fig. 2A, P < 0.005) and Lipe (Fig. 2A, P < 0.01) by Plin2-ASO.
Fig. 2.
Expression of genes in the livers of HF mice treated with Plin2-ASO (ASO) and Cont was measured with qPCR. A: genes associated with fatty acid metabolism, i.e., acyl-CoA synthetase long-chain family member 4 (Acsl4), stearoyl-Coenzyme A desaturase 2 (SCD2), acyl-CoA synthetase long-chain family member 1 (Acsl1), and hormone-sensitive lipase (Lipe). B: genes associated with cholesterol metabolism, i.e., 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr), hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 (Hsd3b5), and hydroxysteroid (17-beta) dehydrogenase 2 (Hsd17b2). C: α-fetoprotein (Afp) and H19 fetal liver (H19). D: genes associated with mitosis/kinetochore, i.e., cycline A2 (Ccna2), inner centromere protein antigens 135/155kDa (Incenp), and shugoshin-like 1 (Sgol1). E: genes associated with extracellular matrix (ECM), i.e., type 1 α1 collagen (Col1a1), type 1 α2 collagen (Col1a2), type 3 α1 collagen (Col3a1), and matrix metallopeptidase 8 (Mmp8). Data are means ± SE, n = 4–5 per group. *P < 0.05 vs. Cont, **P < 0.01 vs. Cont, ***P < 0.005 vs. Cont. F: histological study of livers from Cont- and Plin2-ASO-treated mice. Top: immunofluorescence showing Plin2 (green) staining in the cytoplasm and DAPI (blue) in the nuclei. Bottom: Sirius red staining of collagen in Cont vs. Plin2-ASO liver. Scale bar, 25 μm.
The effect of Plin2-ASO on hepatic gene expression in DIO mice was not limited to TG metabolism but was also seen in cholesterol and steroid metabolism. As mentioned before, genes related to steroid metabolism (GOTERM_BP_FAT steroid metabolic process) formed a prominent group of genes reduced in the Plin2-ASO-treated liver (Supplementary Table S1B). qPCR confirmed that 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr) was reduced to 0.27 (Fig. 2B, P < 0.005), and hydroxy-delta-5-steroid dehydrogenase 3 beta-hydroxy-delta-5-steroid dehydrogenase and steroid delta-isomerase 5 (Hsd3b5), an enzyme highly expressed in the liver of adult males and associated with hepatosteatosis, was reduced to 0.17 (Fig. 2B, P < 0.005; Refs. 1, 13). The expression of 17-beta hydroxysteroid dehydrogenase 2 (Hsd17b2), an enzyme involved in estrogen and testosterone metabolism, was also reduced to 0.17 in Plin2-ASO liver (Fig. 2B, P < 0.005, Ref. 24). Therefore, both microarray and qPCR analyses demonstrated global changes in expression of fatty acid metabolism genes in liver, in response to Plin2-ASO treatment.
Plin2-ASO increases expression of hepatic genes regulating cell division and extracellular matrix.
The microarray analysis showed that the majority of differentially regulated genes in the functional cluster of mitosis/kinetochore were upregulated by Plin2-ASO treatment of HF-fed mice (Supplementary Table S1, C and D). Moreover, genes associated with hepatocyte proliferation, such as α fetoprotein (Afp) and H19 fetal liver mRNA (H19) were among the top genes that are increased in the microarray analysis of Plin2-ASO liver (Supplementary Table S1E; Refs. 22, 32). Although the expression levels of Afp and H19 were low in Cont liver, reflecting a limited proliferation in adult liver, qPCR showed a 127-fold increase in Afp expression and 548-fold increase in H19 expression in the Plin2-ASO-treated liver (Fig. 2C, P < 0.001 for Afp and P < 0.01 for H19). qPCR also confirmed that three genes with functions in mitosis/kinetochore identified by microarray were increased in Plin2-ASO-treated liver (Supplementary Table S1, C and D; Fig. 2D). Plin2-ASO treatment increased cyclin A2 (Ccna2) 8.0-fold (P < 0.01), inner centromere protein antigens 135/155kDa (Incenp) 3.5-fold (P < 0.05), and shugoshin-like 1 (Sgol1) 3.4-fold (P < 0.01). However, we did not observe a significant change in the number of cells, determined by counting nuclei per area of paraffin-embedded liver sections (3,720 ± 291 in Cont vs. 3,365 ± 291 per 1 mm2 in Plin2-ASO).
Since extracellular matrix-related genes formed another cluster of genes significantly enriched in the Plin2-ASO-treated liver from HF-fed mice in the microarray analysis (Supplementary Table S1F), we validated their expression using qPCR. Type 1α1 collagen (Col1a1) was increased 4.8-fold (P < 0.05), Type 3α1 collagen (Col3a1) 4.0-fold (P < 0.05), and matrix metallopeptidase 8 (Mmp8) 4.8-fold (P < 0.05) by Plin2-ASO treatment (Fig. 2E). Histological analysis of the liver supported our qPCR results. Plin2 immunostaining was reduced, and collagen staining was more prominent in the liver from Plin2-ASO-treated mice on HF compared with Cont mice (Fig. 2F).
Effects of Plin2-ASO in regular chow-fed mice.
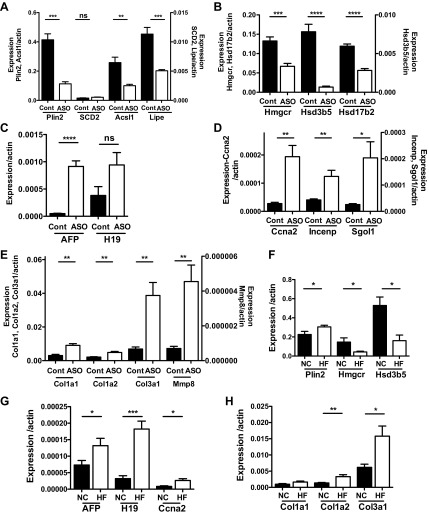
Plin2-ASO treatment of HF-fed mice resulted in the differential regulation of genes involved in lipid metabolism, growth, and extracellular matrix. It is possible that these changes were secondary to improvement of steatosis or due to independent effects of Plin2-ASO. Therefore, we determined whether Plin2-ASO treatment had similar effects on hepatic gene expression in chow-fed mice. Plin2-ASO treatment did not change body weight or fat content compared with Cont (Table 1). Serum TG levels were not altered by Plin2-ASO, but glucose was decreased and cholesterol was increased (Table 1). Plin2-ASO treatment decreased hepatic Plin2 expression to 27% of the level in Cont (Fig. 3A, P < 0.005). Although chow-fed mice do not develop steatosis, the liver TG content was reduced to 61% of Cont by Plin2-ASO (12.5 ± 1.6 mg/g in Cont vs. 7.7 ± 0.7 mg/g liver in Plin2-ASO, P < 0.01). rtPCR analyses showed that expression of a wide array of fatty acid and cholesterol metabolism genes including Acsl1, Lipe, Hmgcr, Hsd3b5, and Hsd17b2 were reduced by Plin2-ASO treatment of regular chow-fed mice (Fig. 3, A and B). As in HF-fed liver, the expression of genes associated with hepatocyte proliferation and cell cycling were also increased in Plin2-ASO-treated mice on chow diet (Fig. 3, C and D). Extracellular matrix-related genes, another group of genes identified by microarray analysis of the liver from Plin2-ASO-treated HF-fed mice, were upregulated in the liver after Plin2-ASO treatment of chow-fed mice (Fig. 3E). Overall these effects of Plin2-ASO on hepatic gene expression pattern were similar between HF- and chow-fed mice, indicating that fatty liver is not a major determinant of the regulation of hepatic genes by Plin2-ASO.
Fig. 3.
A–E: effects of Plin2-ASO on hepatic gene expression in regular chow-fed mice. A: Plin2 and genes associated with fatty acid metabolism, i.e., SCD2, Acsl1, and Lipe. B: genes associated with cholesterol metabolism, i.e., Hmgcr, Hsd3b5, Hsd17b2. C: Afp and H19. D: genes associated with mitosis/kinetochore, i.e., Ccna2, Incenp, and Sgol1. E: genes associated with extracellular matrix, i.e., Col1a1, Col1a2, Col3a1, and Mmp8. F–H: hepatic gene expression in livers from regular chow (NC) and HF livers without Plin2-ASO treatment. F: genes associated with cholesterol metabolism, i.e., Hmgcr, Hsd3b5. G: Afp, H19, and Ccna2. H: ECM genes, i.e., Col1a1, Col1a2, and Col3a1. Data are means ± SE, n = 4 per group. *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001, and n.s. (no significant difference) between the groups.
We further examined the effects of HF and chow diets on hepatic gene expression in the absence of Plin2-ASO treatment. A cohort of C57BL/6J mice was fed HF or normal chow diet (NC), without Plin2 or Cont-ASO treatment. The HF diet significantly increased body weight (47.2 ± 0.6 g) compared with NC (34.3 ± 1.6 g); n = 5, P < 0.005. Liver TG was also increased by the HF diet (14.5 ± 1.6 mg/g) compared with NC (10.2 ± 1.05 mg/g); n = 5, P < 0.05. As shown in Fig. 3F, the expression of Plin2 in the HF liver was increased by 1.35-fold, confirming our previous observation (37). This was associated with a reduction in the expression of genes involved in cholesterol metabolism in HF liver, i.e., Hmgcr to 0.29 and Hsd3b5 to 0.31 compared with NC (Fig. 3F). The expression of genes associated with hepatocyte proliferation and extracellular matrix was increased in HF liver compared with NC, i.e., Afp by 1.8-fold, H19 by 5.7-fold, Ccna2 by 3.1-fold, Col1a2 by 2.4-fold, and Col3a1 by 2.6 (Fig. 3B). Col1a1 expression also showed a trend of being increased in HF liver compared with NC liver (Fig. 3C). Thus, some of the differences between HF and NC livers were opposite to HF-Plin2-ASO vs. HF-Cont liver. These results confirm that Plin2-ASO regulates hepatic gene expression independently of its effect on TG content.
DISCUSSION
In the present study, we confirmed that Plin2-ASO treatment decreases hepatic and serum triglycerides in mice fed a HF diet (37). Plin2 (also known as adipose differentiation-related protein or adipophilin) is found on the surface of lipid droplets in most tissues. Plin2 expression is strongly induced by lipid loading in hepatocytes and fatty liver associated with obesity (15, 23, 31). On the other hand, ablation of plin2 (adfp) gene reduces fatty liver and enhances insulin sensitivity in DIO and Lepob mice (15, 37). We previously showed that Plin2-ASO treatment decreased the expression of genes involved in TG synthesis, including ACC, FAS, and DGAT2, and fatty acid oxidation, i.e., CPT1 (37). The current microarray analysis showed that genes involved in TG hydrolysis, i.e., lipe and ascl1 were also downregulated in Plin2-ASO-treated liver. Thus, the reduction in hepatic TG by Plin2-ASO treatment is associated with downregulation of genes involved in both TG synthesis and utilization. The reduction in the transcription factors involved in the regulation of lipid metabolism is one potential pathway by which Plin2-ASO affects a broad range of genes. Although not identified as differentially regulated according to the stringent criteria in our microarray analysis, we previously noted that Plin2-ASO treatment resulted in significant reduction in the expression of peroxisome proliferator-activated receptor alpha (PPARα) and SREBP1 (37). The reduction of TG content by Plin2-ASO may also affect PPARα activity through a decrease in TG hydrolysis. Studies have suggested that lipid metabolites generated by TG hydrolysis regulate peroxisome PPARα activity, thus altering the expression of oxidative genes such as CPT1 (14, 29).
In addition to TG and fatty acid metabolism, the microarray analysis showed that Plin2-ASO treatment significantly decreased the expression of genes involved in cholesterol and steroid metabolism. Hmgcr expression in Plin2-ASO-treated HF liver was reduced to 0.27 of the level in Cont HF liver. However, the levels of free cholesterol and cholesterol ester in liver and serum cholesterol levels were not altered significantly by the partial reduction in Hmgcr expression by Plin2-ASO. This result is consistent with a report that Hmgcr heterozygous mice have no apparent defects of serum or liver chemistry (26). We found that Hsd3b5 expression was decreased in Plin2-ASO HF liver, consistent with a report of a negative correlation of Hsd3b5 expression and steatosis in apo-E null mice (13). Further studies are needed to delineate the specific roles of Plin2 in TG and steroid metabolism in liver.
Afp and H19, two genes closely associated with hepatocyte proliferation, were among the genes with the highest induction by Plin2-ASO in the microarray analysis of HF mouse liver and confirmed by qPCR. Moreover, Plin2-ASO increased the expression of large numbers of genes with roles in cell division and extracellular matrix. A possible mechanism is the induction of these genes in hepatic stellate cell (HSC) by Plin2-ASO. Quiescent HSC serves as the major storage of vitamin A and possesses lipid droplets filled with retinyl esters (11). It has been known that lipid droplets in HSC are coated with Plin2 (35). Upon activation, HSCs lose their lipid droplet content and develop myofibroblast-like features, including the production of collagen and α-smooth muscle actin (3). Furthermore, HSC activation increases the production of chemokines and cytokines that can drive hepatocyte proliferation (11). Type I and type III collagens, whose expression were found to be increased in Plin2-ASO HF liver, are known to be the major collagens that are increased in fibrotic tissues and liver cirrhosis (25). Lee et al. (19) reported in their ex vivo model that the downregulation of Plin2 induced features of HSC activation in immortalized human HSC, LX-2, suggesting that Plin2 plays a critical role in the maintenance of quiescent status of HSC. Our finding of an increase in fibrotic genes in Plin2-ASO-treated liver is consistent with the results of Lee et al. (19). Since the liver contains hepatocytes, HSCs, and Kupffer cells, all of which express Plin2, cell type-specific ablation of plin2 gene is necessary to clarify the in vivo functions of Plin2 in lipid metabolism, inflammation, proliferation and fibrosis.
Plin2-ASO reverses several metabolic phenotypes of HF feeding in mice, including improvement in hepatic steatosis and insulin sensitivity. However, the changes in gene expression after Plin2-ASO treatment did not always parallel the differences in hepatic gene expression in HF vs. NC livers. Compared with HF Cont liver, Plin2-ASO treatment increased the expression of genes involved in steroid metabolism, hepatocyte proliferation, and extracellular matrix. In contrast, the expression of these genes was reduced in NC liver compared with HF liver. Moreover, Plin2-ASO treatment had similar effects on hepatic gene expression in both HF- and regular chow-fed mice, demonstrating that the changes in gene expression were not related to the ability of Plin2-ASO to reduce hepatic steatosis.
In conclusion, microarray analysis revealed differential effects of Plin2-ASO treatment on the expression of genes involved in regulation of hepatic lipid metabolism, growth, and extracellular matrix. Our microarray analysis results emphasize the power of unbiased screening, by showing Plin2-mediated changes in metabolic and structural pathways beyond our original observations of regulation hepatic TG content and insulin sensitivity (15, 37). The gene expression results indicate that Plin2-ASO decreases steatosis, while promoting proliferation and fibrosis. The latter changes are well known factors for cirrhosis, liver failure, and hepatocellular carcinoma. Therefore, a better understanding of how Plin2 and other lipid droplet proteins specifically regulate lipid hepatic lipid metabolism, growth, and fibrosis will provide novel insights into the pathogenesis and treatment of NAFLD and other liver diseases.
GRANTS
Microarray analysis was performed by the Penn Diabetes Research Center Functional Genomics Core, and body composition analysis was performed by the Mouse Metabolic Phenotyping Core, both supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant P30-DK-19525. Research support was provided by NIDDK Grants P01-DK-049210 (R. S. Ahima) and R01-DK-090490 (Y. Imai).
DISCLOSURES
M. J. Graham is an employee of Isis Pharmaceuticals, which provided Plin2 and Cont ASO for the experiments.
AUTHOR CONTRIBUTIONS
Author contributions: Y.I., S.B., G.M.V., E.C., X.Y., Raina Dhir, Ravindra Dhir, and M.J.G. performed experiments; Y.I., G.M.V., and R.S.A. analyzed data; Y.I. and R.S.A. interpreted results of experiments; Y.I. prepared figures; Y.I. drafted manuscript; Y.I., S.B., G.M.V., E.C., X.Y., Raina Dhir, Ravindra Dhir, M.J.G., and R.S.A. approved final version of manuscript; R.S.A. conception and design of research; R.S.A. edited and revised manuscript.
Supplementary Material
ACKNOWLEDGMENTS
Special thanks to Yui Machida for technical assistance.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1. Abbaszade IG, Clarke TR, Park CH, Payne AH. The mouse 3 beta-hydroxysteroid dehydrogenase multigene family includes two functionally distinct groups of proteins. Mol Endocrinol 9: 1214–1222, 1995. [DOI] [PubMed] [Google Scholar]
- 2. Angulo P. Nonalcoholic fatty liver disease. N Engl J Med 346: 1221–1231, 2002. [DOI] [PubMed] [Google Scholar]
- 3. Blaner WS, O'Byrne SM, Wongsiriroj N, Kluwe J, D'Ambrosio DM, Jiang H, Schwabe RF, Hillman EM, Piantedosi R, Libien J. Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage. Biochim Biophys Acta 1791: 467–473, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carman GM. Thematic minireview series on the lipid droplet, a dynamic organelle of biomedical and commercial importance. J Biol Chem 287: 2272, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carr RM, Patel RT, Rao V, Dhir R, Graham MJ, Crooke RM, Ahima RS. Reduction of TIP47 improves hepatic steatosis and glucose homeostasis in mice. Am J Physiol Regul Integr Comp Physiol 302: R996–R1003, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang BH, Li L, Paul A, Taniguchi S, Nannegari V, Heird WC, Chan L. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol Cell Biol 26: 1063–1076, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chang BH, Li L, Saha P, Chan L. Absence of adipose differentiation related protein upregulates hepatic VLDL secretion, relieves hepatosteatosis, and improves whole body insulin resistance in leptin-deficient mice. J Lipid Res 51: 2132–2142, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science 332: 1519–1523, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dowman JK, Armstrong MJ, Tomlinson JW, Newsome PN. Current therapeutic strategies in non-alcoholic fatty liver disease. Diabetes Obes Metab 13: 692–702, 2011. [DOI] [PubMed] [Google Scholar]
- 10. Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226: 497–509, 1957. [PubMed] [Google Scholar]
- 11. Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev 88: 125–172, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, Mashek DG. The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest 121: 2102–2110, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guillen N, Navarro MA, Arnal C, Noone E, Arbones-Mainar JM, Acin S, Surra JC, Muniesa P, Roche HM, Osada J. Microarray analysis of hepatic gene expression identifies new genes involved in steatotic liver. Physiol Genomics 37: 187–198, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haemmerle G, Moustafa T, Woelkart G, Buttner S, Schmidt A, van de Weijer T, Hesselink M, Jaeger D, Kienesberger PC, Zierler K, Schreiber R, Eichmann T, Kolb D, Kotzbeck P, Schweiger M, Kumari M, Eder S, Schoiswohl G, Wongsiriroj N, Pollak NM, Radner FP, Preiss-Landl K, Kolbe T, Rulicke T, Pieske B, Trauner M, Lass A, Zimmermann R, Hoefler G, Cinti S, Kershaw EE, Schrauwen P, Madeo F, Mayer B, Zechner R. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nat Med 17: 1076–1085, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Imai Y, Varela GM, Jackson MB, Graham MJ, Crooke RM, Ahima RS. Reduction of hepatosteatosis and lipid levels by an adipose differentiation-related protein antisense oligonucleotide. Gastroenterology 132: 1947–1954, 2007. [DOI] [PubMed] [Google Scholar]
- 16. Junqueira LC, Bignolas G, Brentani RR. Picrosirius staining plus polarization microscopy, a specific method for collagen detection in tissue sections. Histochem J 11: 447–455, 1979. [DOI] [PubMed] [Google Scholar]
- 17. Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res 51: 468–471, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kotronen A, Yki-Jarvinen H. Fatty liver: a novel component of the metabolic syndrome. Arterioscler Thromb Vasc Biol 28: 27–38, 2008. [DOI] [PubMed] [Google Scholar]
- 19. Lee TF, Mak KM, Rackovsky O, Lin YL, Kwong AJ, Loke JC, Friedman SL. Downregulation of hepatic stellate cell activation by retinol and palmitate mediated by adipose differentiation-related protein (ADRP). J Cell Physiol 223: 648–657, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Magnusson B, Asp L, Bostrom P, Ruiz M, Stillemark-Billton P, Linden D, Boren J, Olofsson SO. Adipocyte differentiation-related protein promotes fatty acid storage in cytosolic triglycerides and inhibits secretion of very low-density lipoproteins. Arterioscler Thromb Vasc Biol 26: 1566–1571, 2006. [DOI] [PubMed] [Google Scholar]
- 21. Mark N, de Alwis W, Day CP. Current and future therapeutic strategies in NAFLD. Curr Pharm Des 16: 1958–1962, 2010. [DOI] [PubMed] [Google Scholar]
- 22. Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One 2: e845, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Motomura W, Inoue M, Ohtake T, Takahashi N, Nagamine M, Tanno S, Kohgo Y, Okumura T. Up-regulation of ADRP in fatty liver in human and liver steatosis in mice fed with high fat diet. Biochem Biophys Res Commun 340: 1111–1118, 2006. [DOI] [PubMed] [Google Scholar]
- 24. Mustonen MV, Poutanen MH, Isomaa VV, Vihko PT, Vihko RK. Cloning of mouse 17beta-hydroxysteroid dehydrogenase type 2, and analysing expression of the mRNAs for types 1, 2, 3, 4 and 5 in mouse embryos and adult tissues. Biochem J 325: 199–205, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Neubauer K, Saile B, Ramadori G. Liver fibrosis and altered matrix synthesis. Can J Gastroenterol 15: 187–193, 2001. [DOI] [PubMed] [Google Scholar]
- 26. Ohashi K, Osuga J, Tozawa R, Kitamine T, Yagyu H, Sekiya M, Tomita S, Okazaki H, Tamura Y, Yahagi N, Iizuka Y, Harada K, Gotoda T, Shimano H, Yamada N, Ishibashi S. Early embryonic lethality caused by targeted disruption of the 3-hydroxy-3-methylglutaryl-CoA reductase gene. J Biol Chem 278: 42936–42941, 2003. [DOI] [PubMed] [Google Scholar]
- 27. Olofsson SO, Bostrom P, Andersson L, Rutberg M, Perman J, Boren J. Lipid droplets as dynamic organelles connecting storage and efflux of lipids. Biochim Biophys Acta 1791: 448–458, 2009. [DOI] [PubMed] [Google Scholar]
- 28. Ong JP, Younossi ZM. Epidemiology and natural history of NAFLD and NASH. Clin Liver Dis 11: 1–16, vii, 2007. [DOI] [PubMed] [Google Scholar]
- 29. Sapiro JM, Mashek MT, Greenberg AS, Mashek DG. Hepatic triacylglycerol hydrolysis regulates peroxisome proliferator-activated receptor alpha activity. J Lipid Res 50: 1621–1629, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sass DA, Chang P, Chopra KB. Nonalcoholic fatty liver disease: a clinical review. Dig Dis Sci 50: 171–180, 2005. [DOI] [PubMed] [Google Scholar]
- 31. Schadinger SE, Bucher NL, Schreiber BM, Farmer SR. PPARgamma2 regulates lipogenesis and lipid accumulation in steatotic hepatocytes. Am J Physiol Endocrinol Metab 288: E1195–E1205, 2005. [DOI] [PubMed] [Google Scholar]
- 32. Sell S. Alpha-fetoprotein, stem cells and cancer: how study of the production of alpha-fetoprotein during chemical hepatocarcinogenesis led to reaffirmation of the stem cell theory of cancer. Tumour Biol 29: 161–180, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith BW, Adams LA. Non-alcoholic fatty liver disease. Crit Rev Clin Lab Sci 48: 97–113, 2011. [DOI] [PubMed] [Google Scholar]
- 34. Smith BW, Adams LA. Nonalcoholic fatty liver disease and diabetes mellitus: pathogenesis and treatment. Nat Rev Endocrinol 7: 456–465, 2011. [DOI] [PubMed] [Google Scholar]
- 35. Straub BK, Stoeffel P, Heid H, Zimbelmann R, Schirmacher P. Differential pattern of lipid droplet-associated proteins and de novo perilipin expression in hepatocyte steatogenesis. Hepatology 47: 1936–1946, 2008. [DOI] [PubMed] [Google Scholar]
- 36. Takahashi N, Qi Y, Patel HR, Ahima RS. A novel aminosterol reverses diabetes and fatty liver disease in obese mice. J Hepatol 41: 391–398, 2004. [DOI] [PubMed] [Google Scholar]
- 37. Varela GM, Antwi DA, Dhir R, Yin X, Singhal NS, Graham MJ, Crooke RM, Ahima RS. Inhibition of ADRP prevents diet-induced insulin resistance. Am J Physiol Gastrointest Liver Physiol 295: G621–G628, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.