Summary
Background
The tumor suppressor gene p53 is a key regulator of cell division and/or apoptosis. Survivin is a multifunctional member of the inhibitor of apoptosis family. Survivin and p53 represent diametrically opposed signals that influence the apoptotic pathway.
Material/Methods
To determine the role of p53 and survivin in basal cell carcinoma (BCC), we evaluated the expression pattern of both proteins with regard to the percentage of positively immunostained tumor cells, the intensity of staining, and subcellular localization among 31 subjects with BCC.
Results
Overexpression of p53 protein was found in 28 of 31 cases (90.3%), whereas survivin accumulation was seen in 27 (87.1%). For p53, moderate and/or strong immunoreactivity was seen in 20 of 28 cases (71.4%), and 26 of 28 cases (92.9%) showed more than 25% reactive tumor cells. Nuclear p53 staining was detected in 23 of 28 cases (82.1%), whereas combined nuclear and cytoplasmic localization was found in only 5 of 28 cases (17.9%). Survivin revealed mild intensity of immuno-reaction in 22 of 27 cases (71%), and 25 of 27 cases (92.6%) showed less than 25% labeled tumor cells. Combined nuclear and cytoplasmic survivin localization was present in 26 of 27 cases (96.3%). Statistically significant differences were detected in the assessed expression parameters between those proteins.
Conclusions
Our results suggest that overexpression of wild type p53 protein may suppress the expression of survivin and its antiapoptotic activity in BCC cells.
Keywords: p53, survivin, immunohistochemistry, basal, cell, carcinoma
Background
Basal cell carcinoma (BCC) is the most common form of human skin cancer. Its etiopathogenesis is still not fully understood. Generally, most common genetic alterations in human cutaneous cancers are found at the level of the p53 tumor suppressor gene [1,2]. The p53 tumor suppressor protein plays a critical role in both the induction of apoptosis and carcinogenesis [3].
Under normal conditions, the inactive form of p53 protein is usually present at low levels in the cytoplasm of cells. Cytoplasmic localization in normal cells is controlled by murine double minute (MDM2) oncogene [4]. Murine double minute is able to shuttle continually between the cytoplasm and nucleus because it contains both a nuclear export signal and a nuclear localization signal. In response to DNA damage, for example, by ultraviolet or ionizing radiation, p53 starts accumulating in the nucleus [1,5]. Deoxyribonucleic acid damage can cause activation of p53 by phosphorylation and, consequently, the interaction of p53 and MDM2 is disrupted [6,7]. Tumor protein 53 becomes temporarily free of MDM2 and then accumulates in the nucleus. The intracellular content of p53 depends on the DNA damage response.
Higher content of p53 directs the cells toward apoptosis, whereas low and/or moderate p53 levels result in cell cycle arrest, providing time for DNA repair [8,9]. On one hand, the activated p53 can induce apoptosis or cell cycle arrest through up-regulation of several associated genes. On the other hand, there are genes with antiapoptotic activity that are repressed by p53 at the transcription or translation levels, and this negative regulation is important for the induction of apoptosis [2,3]. Survivin, a unique member of the inhibitor of apoptosis protein (IAP) family, is known to be repressed by the wild-type p53. This repression is one of the mechanisms inducing apoptosis by the activation of the mitochondrial pathway [2,10].
Proteins that belong to the IAP family play a key role in negative regulation of apoptosis. Eight human IAP family members have been identified: c-IAP1, c-IAP2, NAIP, ILP-2, XIAP, apollon, ML-IAP/livin, and survivin [11,12]. Multifunctional survivin possesses a number of distinct features not shared with other IAP members: (1) it is the shortest polypeptide consisting of 142 amino acid residues; (2) the expression of this protein is cell cycle-regulated and occurs in the G2/M phase; (3) it is undetectable in most normal differentiated adult tissues, but is frequently expressed in embryonic and fetal organs, as well as in developed human malignant tumors; (4) it can be found in different subcellular localizations; and (5) it functions in both inhibition of apoptosis and regulation of cell division in contrast to other IAP members regularly restricted to 1 of these 2 functions [13–15]. Survivin is currently undergoing intensive research as a potential tumor marker and prognostic factor [14,16–19,39].
The down-regulation of survivin by wild type p53 in cell cycle is generally well known [3,19], but data that describe the expression pattern and the interaction between survivin and p53 in BCC cells are scarce [20]. Recently, more research groups have been dealing with this topic, mainly in the framework of apoptosis regulation. A variety of non-malignant and malignant cells are being studied, including human melanocytic and keratolytic lesions. The majority of skin lesion studies have focused primarily on squamous cell carcinoma and malignant melanoma [19,21–23]. However, this is not the case with BCC. Review articles on the molecular etiology and pathogenesis of BCC explain the genetic aberrations in human skin cancers at the level of the p53 gene; however, data on survivin expression are limited [20]. Therefore, we evaluated the expression pattern of both proteins with respect to intensity, relative number of positively stained cells, and cellular localization as detected by immunohistochemical methods and the relationship between their expression modes in cutaneous basal cell carcinoma.
Material and Methods
The sample included 31 subjects with cutaneous BCC. The hematoxylin and eosin-stained slides from each subject were independently reviewed by 2 pathologists to ascertain the diagnosis based on morphologic and immunohistochemical parameters and were correlated with clinical data. The standard diagnostic histomorphologic and immunohistochemical criteria were applied as previously described [24–26]. Immunohistochemical staining was performed using monoclonal mouse anti-p53 antibody (Dako, Carpinteria, CA) and monoclonal mouse antisurvivin antibody (Dako, Carpinteria, CA). Negative controls were obtained by simply omitting the primary antibodies.
In each case, the following features were assessed: (1) the intensity of staining, (2) the relative number of positively stained cells and (3), the subcellular localization of p53 and survivin antigens. Microsoft Excel software package was used to perform statistical analyses. Survivin and p53 expression and the correlation between them were analyzed by χ2 test. A P value less than.05 was considered to indicate statistical significance.
Results
Both p53 and survivin were found in BCC cells either in the nucleus (N), the cytoplasm (C), or both the nucleus and cytoplasm (NC). The expression of antigens among the 31 subjects was scored semiquantitatively as follows:
Intensity of p53 staining: (a) absent or barely detectable; 3/31 cases, 9.7%; (b) weak; 8/31 cases, 25.8%; (c) moderate; 18/31 cases, 58.1%; and (d) strong; 2/31 cases, 6.4%.
Intensity of survivin staining: (a) absent or barely detectable; 4/31 cases, 12.9%; (b) week: 22/31 cases, 71%; (c) moderate: 5/31 cases, 16.1%; and (d) strong: 0/31 cases, 0%.
Number of p53 positively stained cells: (a) more than 25% per field of view: 26/28 cases, 92.9%; (b) less than 25% per field of view: 2/28 cases, 7.1%.
Number of survivin positively stained cells: (a) more than 25% per field of view: 2/27 cases, 7.4%; (b) less than 25% per field of view: 25/27 cases (92.6%).
Subcellular localization of p53 staining: (a) N localization only: 23/28 cases, 82.1%; (b) C localization only: 0/28 cases, 0%; and (c) NC localization: 5/28 cases, 17.9%.
Subcellular localization of survivin staining: (a) N localization only: 1/27 cases, 3.7%; (b) C localization only: 0/27 cases, 0%; (c) NC localization: 26/27 cases, 96.3%.
The results of all expression profiles are listed in Table 1. Tables 2 and 3 summarize the number of p53 and survivin positively stained cells and their cellular localization, respectively. The relation between the expression intensity of p53 and survivin is shown in Table 4. Increased p53 expression was present also in adjacent cells of normal epidermis. The χ2 analysis confirmed a significant correlation between the intensity of survivin and p53 immunoreactivity (P<.05), a significant difference in the percentage of survivin- and p53-labeled cells (P<.001), and a significant difference in N and NC localization of survivin and p53 (P<.001).
Table 1.
Panel of 31 basal cell carcinoma cases.
| p53 | Survivin | Diagnosis -subtype | Age | Sex | |||||
|---|---|---|---|---|---|---|---|---|---|
| I | P (%) | s. l. | I | P (%) | s. l. | ||||
| 1. | ++ | >25 | N | ++ | <25 | NC | Micronodular | 54 | M |
| 2. | + | >25 | N | + | <25 | NC | Cystic pigmented | 79 | M |
| 3. | ++ | >25 | N | + | <25 | NC | Solid | 50 | F |
| 4. | +++ | >25 | NC | + | <25 | NC | Multicentric | 53 | M |
| 5. | ++ | >25 | N | + | >25 | NC | Multicentric | 61 | M |
| 6. | ++ | >25 | N | ++ | <25 | NC | Multicentric | 82 | F |
| 7. | +++ | >25 | N | + | <25 | NC | Cystic | 84 | M |
| 8. | ++ | >25 | NC | ++ | <25 | NC | Mixed form | 79 | F |
| 9. | ++ | >25 | N | + | <25 | NC | Cystic | 77 | F |
| 10. | ++ | >25 | N | + | <25 | NC | Multicentric | 56 | F |
| 11. | ++ | >25 | N | + | <25 | NC | Solid | 50 | M |
| 12. | ++ | >25 | N | + | <25 | NC | Cystic | 88 | M |
| 13. | ++ | >25 | N | + | >25 | NC | Solid | 57 | M |
| 14. | ++ | >25 | N | ++ | <25 | NC | Cystic | 72 | M |
| 15. | + | >25 | N | + | <25 | NC | Mixed | 64 | M |
| 16. | ++ | >25 | N | + | <25 | NC | Multicentric | 31 | F |
| 17. | ++ | >25 | N | + | <25 | NC | Cystic | 78 | M |
| 18. | + | >25 | N | + | <25 | NC | Multicentric | 84 | M |
| 19. | + | >25 | N | 0 | – | – | Multicentric | 52 | M |
| 20. | ++ | >25 | N | 0 | – | – | Cystic | 53 | M |
| 21. | + | >25 | NC | + | <25 | NC | Solid | 78 | M |
| 22. | + | >25 | N | + | <25 | NC | Solid | 87 | F |
| 23. | ++ | >25 | N | + | <25 | NC | Multicentric | 39 | M |
| 24. | ++ | >25 | N | + | <25 | NC | Multicentric | 68 | M |
| 25. | ++ | >25 | NC | + | <25 | NC | Adenoid-cystic | 75 | F |
| 26. | ++ | >25 | N | ++ | <25 | NC | Solid | 79 | M |
| 27. | + | <25 | NC | + | <25 | NC | Multicentric | 71 | M |
| 28. | + | <25 | N | + | <25 | N | Mixed form | 78 | M |
| 29. | 0 | – | – | 0 | – | – | Cystic | 72 | F |
| 30. | 0 | – | – | 0 | – | – | Cystic | 70 | F |
| 31. | 0 | – | – | + | <25 | NC | Cystic | 59 | M |
I – intensity of immunoreactivity: 0 – no positivity, s. l. – subcellular localization of survivin and p53; + mild, ++ moderate, +++ strong; C – cytoplasm positivity; P – percentage of labeled cells; N – nuclear positivity; NC – nuclear as well as cytoplasmic positivity.
Table 2.
Number of p53 and survivin reactive cells.
| Percentage of stained cells | |||
|---|---|---|---|
| <25% | >25% | Total | |
| P53 | 2 (7.1%) | 26 (92.9%) | 28/31 |
| Survivin | 25 (92.6%) | 2 (7.4%) | 27/31 |
Table 3.
Cellular localization of p53 and survivin.
| Cellular localization | ||||
|---|---|---|---|---|
| n | C | NC | Total | |
| P53 | 23 (82.1%) | 0 (0%) | 5 (17.9%) | 28/31 |
| Survivin | 1 (3.7%) | 0 (0%) | 26 (96.3%) | 27/31 |
N – nuclear positivity’ C – cytoplasmic positivity; NC – nuclear as well as cytoplasmic positivity.
Table 4.
Intensity of survivin versus p53 expression.
| Survivin | 0 | + | ++/+++ | Total |
|---|---|---|---|---|
| p53 | ||||
| 0 | 2 (6.5%) | 1 (3.2%) | 0 (0.0%) | 3 (9.7%) |
| + | 1 (3.2%) | 7 (22.6%) | 0 (0.0%) | 8 (25.8%) |
| ++/+++ | 1 (3.2%) | 14 (45.2%) | 5 (16.1%) | 20 (64.5%) |
| Total | 4 (12.9%) | 22 (71.0%) | 5 (16.1%) | 31 (100.0%) |
0 – no positivity; + mild; ++ moderate; +++ strong.
Discussion
The results of the current study show significant differences in the intensity of immunoreactions and in the percentage of labeled cells between p53 and survival expression. There were also significant differences in subcellular compartmentalization of both proteins. Positive p53 immunoreactivity was observed in 28 of 31 cases (90.3%). In accord with previous papers [20,26–28], we noted a high frequency of p53 overexpression in BCC. Interestingly, basal layers of the epidermis, adjacent to the tumor lesion, expressed mild and/or moderate immunopositivity. All of our subjects showed cytoplasmic p53-positive reactions only. These findings may be related to early accumulation of p53 immunopositive clones as a consequence of sun exposure but without any pathological proliferation [29]. Calli-Demirkan and associates [30] found statistically significant differences between the immunoreactivity of p53 protein in normal epithelia adjacent to BCC and age of patients. Bäckvall and associates [31] pointed out that the foci of normal p53-positive keratinocytes are under regulation of more genetic events.
Generally, cells labeled by p53 display nuclear staining, but in some cases, cytoplasmic positivity has also been described [32]. The antiapoptotic protein survivin is also known to be localized both in cytoplasm and nucleus [33,34]. Similar to p53, nuclear export signal has been identified in survivin, as well [35]. It has been reported that survivin is highly expressed in many malignant tissues and is rarely detected in normal differentiated adult tissue [33]. In our study, survivin immunoreactivity was observed in 27 of 31 subjects (86.1%).
For p53, the number of positively stained cells in the majority of cases was more than 25% per field of view, whereas a greater number of subjects with positively stained survivin cells had less than 25% immunopositive cells (Table 2). Consistent with previous data, cellular localization of both proteins showed measurable differences. Results of our study showed nuclear p53 expression in 23 of 28 cases (82.1%) and combined nuclear and cytoplasmic staining in 5 of 28 cases (17.9%; Table 3).
A more-complicated situation seems to occur with compartmentalization of survivin. Conflicting data were reported with regard to survivin expression in various benign tissues and different types of tumors. Survivin exists in 5 different isoforms as a result of alternatively spliced transcripts: survivin, survivin-2B, survivin-Ex3, survivin-3B, and survivin-2a [36–38]. Survivin and survivin-2B are localized in the cytoplasm, particularly in mitochondria; survivin-Ex3 is primarily nuclear, survivin-2a is located in both the cytoplasm and nucleus, and the subcellular localization of survivin-3B remains to be determined [7,38,39].
These 5 survivin splicing variants and their unique subcellular compartmentalization create a delicate balance between induction and inhibition of the apoptotic process [36,38]. Discrepancies in the results regarding cytoplasmic versus nuclear survivin localization proceed from the fact that survivin basically exists in 2 subcellular compartments as a result of the properties of different polypeptides specified by alternatively spliced transcripts [40]. In general, malignant tumors are characterized mainly by nuclear localization [33,34,39]. In our study, nuclear or combined nuclear and cytoplasmic survivin immunoreactivity was observed in all of the 27 positive cases (Table 3). Thus, for the first time, we demonstrated a combined nuclear and cytoplasmic pattern of survivin expression in the majority of cutaneous BCC cases (26/27, 96.3%). As shown in our subjects, there were significant differences in the number of positively stained cells and cellular localization between p53 and survivin (P<.001).
There are several genes that play a role in the control of the G2/M phase of the cell cycle that are repressed following induction of wild type p53. Survivin is identified as a gene that is potently repressed at both the RNA and protein levels [3,30]. In the present study, we investigated this interplay at the protein level. Immunoreactivity of p53 was characterized by a predominance of moderate and strong intensity (20/31 cases, 64.5%) over mild intensity (8/31 cases. 25.8%). However, mild intensity of survivin staining dominated (22/31 cases; 71%). By comparison, moderate and strong p53 immunopositivity was accompanied by mild survivin immunopositivity in 16 cases (51.6%). Equal intensity of p53 and survivin was detected in 14 cases (45.2%). Only one case (3.2%) showed no p53 expression accompanied by mild intensity of survivin reaction (Table 4). Our immunohistochemical findings indicated that there are statistically significant differences between the expression intensity of both examined proteins (P<.05).
The basic function of p53 is associated with gene regulations that control apoptosis. Cell cycle arrest induced by p53 permits the repair of DNA damage or p53 can promote apoptosis by activation of the mitochondrial pathway. Wild type p53 leads to the release of mitochondrial cytochrome c, which is considered critical for the intrinsic apoptotic pathway [41–43]. Cytochrome c induces the formation of a large multimeric complex “apoptosome” within cytoplasm, which consists of cytochrome c, the adapter protein Apaf-1, adenosine triphosphate, and procaspase-9. The special 3-dimensional structure of this complex recruits and mediates the autoactivation of initiator caspase-9 with a subsequent caspase cascade [43]. Death receptor-mediated caspase activation may also induce p53-dependent apoptosis [44]. The precise mechanism for how survivin inhibits apoptosis is not fully understood. Some research groups have reported direct suppression of caspase-3 and caspase-7 [45]. Other investigators have suggested that survivin lacks structural linker sequence, which allows its binding to caspase-3 [46]. Marusawa and associates [47] found that survivin needs to use the cofactor, interacting with the hepatitis B virus coded X protein and binding to and inhibiting procaspase-9. It is known that survivin also may indirectly suppress apoptosis via intermediate proteins, for example, through binding and inactivating Smac/DIABLO [48,49].
Previous studies revealed that many genes involved in the G2/M phase of cell cycle, including a number of genes encoding microtubule components, are subject to negative regulation by wild type p53 [50,51]. Survivin is only expressed in the G2/M phase [52]. It is associated with mitotic spindle by interaction with tubulin during mitosis [50,53]. p53 interacts with the survivin promoter, which is demonstrated as the first promoter to confer p53-dependent repression. Survivin promoter is repressed by both direct (p53 binding) and indirect (induction of p21 protein) mechanisms. Each mechanism may depend on particular stresses, phases of the cell cycle, and cell types, as well [3]. Furthermore, p53 can interfere with bcl-2 proteins in mitochondria, resulting in cytochrome c release [54].
Conclusions
The results of our study suggest that overexpression of wild type p53 protein may suppress the expression of survivin and its antiapoptotic activity in BCC cells. In addition, de novo production or stabilization of p53 is fundamental to triggering apoptosis in BCC [55].
Figure 1.
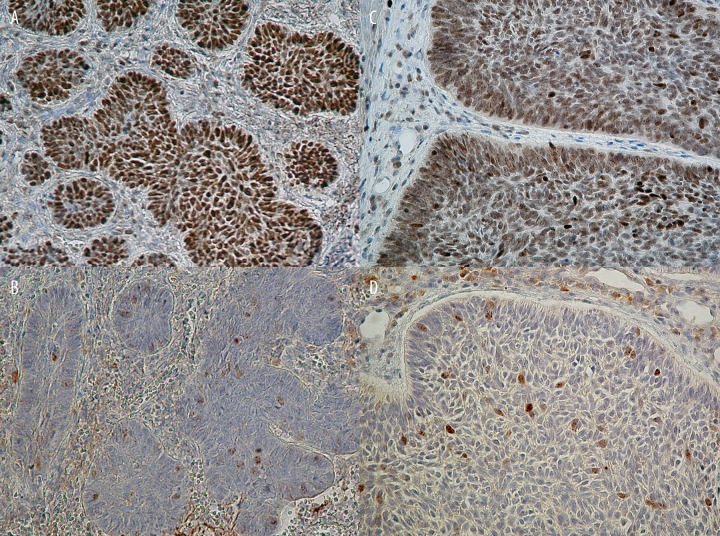
Different number of positively stained cells, subcellular localization, and intensity of immunoreactions of p53 and survivin antigens in BCC cells. (A) More than 25% of p53 positively stained tumor cells (case 1, original magnification: ×240). (B) Less than 25% of survivin positively stained cells, combined cytoplasmic and nuclear immunoreactivity (case 1, original magnification: ×240). (C) Heterogenous intensity of nuclear p53 immunoreactivity (case 8, original magnification: ×480). (D) Combined cytoplasmic and nuclear immunopositivity of survivin, nuclear staining exhibited punctuate pattern, and heterogenous intensity of immunoreaction (case 8, original magnification: ×480).
Acknowledgments
We gratefully acknowledge Professor Shigeo Mori for his careful review of this article and helpful comments. We thank Margareta Kondekova, Monika Letrichova, Agata Resetarova, Jana Visnovcova, and Slavka Drahosova for their skillful technical assistance.
Abbreviations
- NAIP
Neuronal Apoptosis Inhibitory Protein
- ILP-2
IAP-like protein-2
- XIAP
X-linked IAP
- ML-IAP
Melanoma-associated IAP
- c-IAP
cellular IAP
- Apaf-1
Apoptosis protease activating factor-1
- LSAB
labeled streptavidin biotin method
- MDM2
murine double minute 2 oncogene
- Smac/DIABLO
Second mitochondrial activator of caspases, a.k.a Diablo
Footnotes
Ethical consideration
The study was approved by local Ethics Committee of Jessenius Faculty of Medicine in Martin, registered in Office for Human Research Protection, US Department of Health and Human Services under No: IORG0004721.
Authorship
All authors have read and approved the final version of the manuscript, and all author listed as coworkers met the criteria for authorship.
Conflict of Interest
The authors declared no conflict of interest in relation to the article.
Source of support: This work was supported by project “CREATING A NEW DIAGNOSTIC ALGORITHM FOR SELECTED CANCER DISEASES” co-financed from EC sources and European Regional Development Fund and by foundation of Slovak Ministry of Education-AV No: 4/2026/08
References
- 1.Kastan MB, Onkyekwere O, Sidransky D, et al. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–11. [PubMed] [Google Scholar]
- 2.Mirza A, McGuirk M, Hockenberry TN, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–22. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman WH, Biade S, Zilfou JT, et al. Transcriptional Repression of the Anti-apoptotic survivin Gene by Wild Type p53. J Biol Chem. 2002;277:3247–57. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 4.Momand J, Wu HH, Dasgupta G. MDM2-master regulator of the p53 tumor supressor protein. Gene. 2000;242:15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- 5.Fritsche M, Haessler C, Brandner G. Induction of nuclear accumulation of the tumor-supressor protein p53 by DNA-damaging agents. Oncogene. 1993;8:307–18. [PubMed] [Google Scholar]
- 6.Siciliano JD, Canman CE, Taya Y, et al. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–81. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raj D, Brash DE, Grossman D. Keratinocyte Apoptosis in Epidermal Development and Disease. J Invest Dermatol. 2006;126:243–57. doi: 10.1038/sj.jid.5700008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ronen D, Schwartz D, Teitz Y, et al. Induction of HL-60 cells to undergo apoptosis is determined by high levels of wild type p53 protein whereas differentiation of the cells is mediated by lower p53 levels. Cell Growth Different. 1996;7:21–30. [PubMed] [Google Scholar]
- 9.Eberle J, Fecker LF, Forschner T, et al. Apoptosis pathways as promosing targets for skin cancer therapy. Br J Dermatol. 2007;156:18–24. doi: 10.1111/j.1365-2133.2007.07855.x. [DOI] [PubMed] [Google Scholar]
- 10.Schuler M, Bossy-Wetzel E, Goldstein JC, et al. p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J Biol Chem. 2000;275:7337–42. doi: 10.1074/jbc.275.10.7337. [DOI] [PubMed] [Google Scholar]
- 11.Deveraux QL, Reed JC. IAP family proteins-suppressors of apoptosis. Genes Dev. 1999;13:239–52. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 12.Amiri KI, Richmond A. Role of nuclear factor-k B in melanoma. Cancer Metastasis Rev. 2005;24:301–13. doi: 10.1007/s10555-005-1579-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F. Survivin study: what is the next wave? J Cell Physiol. 2003;197:8–29. doi: 10.1002/jcp.10327. [DOI] [PubMed] [Google Scholar]
- 14.Li F. Role of Survivin and its Splice Variants in Tumorigenesis. Br J Cancer. 2005;92:212–16. doi: 10.1038/sj.bjc.6602340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li F, Brattain MG. Role of the survivin gene in pathobiology. Am J Pathol. 2006;169:1–11. doi: 10.2353/ajpath.2006.060121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Y, Prieto VG, Zhang PS, et al. Nuclear expression of the antiapoptoptic protein survivin in malignant melanoma. Cancer. 2006;106:1123–29. doi: 10.1002/cncr.21727. [DOI] [PubMed] [Google Scholar]
- 17.Vetter CS, Miller-Blech K, Schrama D, et al. Cytoplasmic and nuclear expression of survivin in melanocytic skin lesions. Arch Dermatol Res. 2005;297:26–30. doi: 10.1007/s00403-005-0572-x. [DOI] [PubMed] [Google Scholar]
- 18.Moon WS, Tarnawski AS. Nuclear translocation of survivin in hepatocellular carcinoma: a key to cancer cell growth? Human Pathol. 2003;34:1119–26. doi: 10.1053/j.humpath.2003.07.016. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W, Hanks AN, Boucher K, et al. UVB-induced apoptosis drives clonal expansion during skin tumor development. Carcinogenesis. 2005;26:249–57. doi: 10.1093/carcin/bgh300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tilli CMLJ, Van Steensel MAM, Krekels GAM, et al. Molecular aetiology and pathogenesis of basal cell carcinoma. Br J Dermatol. 2005;152:1108–24. doi: 10.1111/j.1365-2133.2005.06587.x. [DOI] [PubMed] [Google Scholar]
- 21.Chiodino C, Cesarino AM, Ottani D, et al. Communication: expression of the novel inhibitor of apoptosis survivin in normal and neoplastic skin. J Invest Dermatol. 1999;113:415–18. doi: 10.1046/j.1523-1747.1999.00711.x. [DOI] [PubMed] [Google Scholar]
- 22.de Freitas Sousa J, Espreafico EM. Melanoma survival strategies: the intrinsic apoptotic pathway-upstream and downstream regulations. Braz J morphol Sci. 2006;23:43–56. [Google Scholar]
- 23.Raj D, Liu T, Samadashwily G, et al. Survivin repression by p53, Rb and E2F2 in normal human melanocytes. Carcinogenesis. 2008;29:194–201. doi: 10.1093/carcin/bgm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sexton M, Jones DB, Maloney ME. Histologic pattern analysis of basal cell carcinoma. J Am Acad Dermatol. 1990;23:1118–26. doi: 10.1016/0190-9622(90)70344-h. [DOI] [PubMed] [Google Scholar]
- 25.Rippey JJ. Why classify basal cell carcinoma? Histopathol. 1998;32:393–98. doi: 10.1046/j.1365-2559.1998.00431.x. [DOI] [PubMed] [Google Scholar]
- 26.Crowson AN. Basal cell carcinoma: biology, morphology and clinical implications. Modern Pathol. 2006;19:S127–47. doi: 10.1038/modpathol.3800512. [DOI] [PubMed] [Google Scholar]
- 27.Shea CR, McNutt NS, Volkenandt M, et al. Overexpression of p53 protein in basal cell carcinomas of human skin. Am J Pathol. 1992;141:25–29. [PMC free article] [PubMed] [Google Scholar]
- 28.Rajabi M-A, Rajabi P, Afshar-Moghaddam N. Determination of p53 expression in basal cell carcinoma tissues and adjacent nontumoral epidermis from sun-exposed areas of the head and neck. Arch Iranian Med. 2006;9:46–48. [PubMed] [Google Scholar]
- 29.Tabata H, Nagano T, Ray AJ, et al. Low frequency of genetic change in p53 immunopositive clones in human epidermis. J Invest Dermatol. 1999;113:972–76. doi: 10.1038/sj.jid.5600549.x. [DOI] [PubMed] [Google Scholar]
- 30.Calli Demirkan N, Colakoglu N, Duzcan E. Value of p53 protein in biological behavior of basal cell carcinoma and in normal epithelia adjacent to carcinomas. Pathol Oncol Res. 2000;6:272–74. doi: 10.1007/BF03187330. [DOI] [PubMed] [Google Scholar]
- 31.Bäckvall H, Stromberg S, Gustafsson A, et al. Mutation spectra of epidermal p53 clones adjacent to basal cell carcinoma and squamous cell carcinoma. Exp Dermatol. 2004;13:643–50. doi: 10.1111/j.0906-6705.2004.00211.x. [DOI] [PubMed] [Google Scholar]
- 32.Kontogeorgos G, Kapranos N, Thodou E, et al. Immunocytochemical accumulation of p53 in corticotroph adenomas: Relationship with heat shock proteins and apoptosis. Pituitary. 1999;1:207–12. doi: 10.1023/a:1009929704018. [DOI] [PubMed] [Google Scholar]
- 33.Duffy MJ, O’Donovan N, Brennan DJ, et al. Survivin: A promising tumor biomarker. Cancer Lett. 2007;249:49–60. doi: 10.1016/j.canlet.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 34.Adamkov M, Lauko L, Balentova S, et al. Expression pattern of antiapoptotic protein survivin in dysplastic nevi. Neoplasma. 2009;56:130–35. doi: 10.4149/neo_2009_02_130. [DOI] [PubMed] [Google Scholar]
- 35.Knauer SK, Kramer OH, Knosel T, et al. Nuclear export is essential for the tumor-promoting activity of survivin. FASEB J. 2007;21:207–16. doi: 10.1096/fj.06-5741com. [DOI] [PubMed] [Google Scholar]
- 36.Mahotka C, Wenzel M, Springer E, et al. Survivin-deltaEx3 and survivin-2B: 2 novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res. 1999;59:6097–102. [PubMed] [Google Scholar]
- 37.Johnson ME, Howerth EW. Survivin: A Bifunctional Inhibitor of Apoptosis Protein. Vet Pathol. 2004;41:599–607. doi: 10.1354/vp.41-6-599. [DOI] [PubMed] [Google Scholar]
- 38.Caldas H, Honsey LE, Altura RA. Survivin 2á: a novel survivin splice variant expressed in human malignancies. Mol Cancer. 2005;4:11. doi: 10.1186/1476-4598-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piras F, Murtas D, Minerba L, et al. Nuclear survivin is associated with disease recurrence and poor survival in patients with cutaneous malignant melanoma. Histopathol. 2007;50:835–42. doi: 10.1111/j.1365-2559.2007.02695.x. [DOI] [PubMed] [Google Scholar]
- 40.Fortugno P, Wall NR, Giodini A, et al. Survivin exists in immunohistochemically distinct subcellular pools and is involved in spindle micro-tubule function. J Cell Sci. 2002;115:575–85. doi: 10.1242/jcs.115.3.575. [DOI] [PubMed] [Google Scholar]
- 41.Daniel PT, Schulze-Osthoff K, Belka C, et al. Guardians of cell death: the BCL-2 family proteins. Essays Biochem. 2003;39:73–88. doi: 10.1042/bse0390073. [DOI] [PubMed] [Google Scholar]
- 42.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12:440–50. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Shi Y. Mechanical aspects of apoptosome assembly. Curr Opin Cell Biol. 2006;18:677–84. doi: 10.1016/j.ceb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Sheikh MS, Fornace AJ., Jr Death and decoy receptors and p53-mediated apoptosis. Leukemia. 2000;14:1509–13. doi: 10.1038/sj.leu.2401865. [DOI] [PubMed] [Google Scholar]
- 45.Shin S, Sung BJ, Cho YS, et al. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and −7. Biochemistry. 2001;40:1117–23. doi: 10.1021/bi001603q. [DOI] [PubMed] [Google Scholar]
- 46.Banks DP, Plescia J, Altieri DC. Survivin does not inhibit caspase-3 activity. Blood. 2000;96:4002. [PubMed] [Google Scholar]
- 47.Marusawa H, Matsuzawa S, Welsh K, et al. HBXIP functions as a cofactor of survivin in apoptosis supression. EMBO J. 2003;22:2729–40. doi: 10.1093/emboj/cdg263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du C, Fang M, Li Y, et al. Smac a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 49.Song Z, Yao X, Wu M. Direct interaction between survivin and Smac is essential for the anti-apoptotic activity of survivin during taxol-induced apoptosis. J Biol Chem. 2003;278:23130–40. doi: 10.1074/jbc.M300957200. [DOI] [PubMed] [Google Scholar]
- 50.Li F, Ambrosini G, Chu EY, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–83. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 51.Zhao R, Gish K, Murphy M, et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Rev. 2000;14:981–93. [PMC free article] [PubMed] [Google Scholar]
- 52.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–53. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 53.Olie RA, Simoes-Wust AP, Baumann B, et al. A novel antisense oligonucleotide targeting survivin expression induces apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer Res. 2000;60:2805–9. [PubMed] [Google Scholar]
- 54.Chipuk JE, Kuwana T, Bouchier-Hayes L, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–14. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 55.Jee SH, Shen SC, Tseng CR, et al. Curcumin induces a p53-dependent apoptosis in human basal cell carcinoma cells. J Invest Dermatol. 1998;111:656–61. doi: 10.1046/j.1523-1747.1998.00352.x. [DOI] [PubMed] [Google Scholar]