Abstract
Type 2 diabetes (T2D) is a disease of relative insulin deficiency resulting from both insulin resistance and beta cell failure. We have previously used heterogeneous stock (HS) rats to fine-map a locus for glucose tolerance. We show here that glucose intolerance in the founder strains of the HS colony is mediated by different mechanisms: insulin resistance in WKY and an insulin secretion defect in ACI, and we demonstrate a high degree of variability for measures of insulin resistance and insulin secretion in HS rats. As such, our goal was to use HS rats to fine-map several diabetes-related traits within a region on rat chromosome 1. We measured blood glucose and plasma insulin levels after a glucose tolerance test in 782 male HS rats. Using 97 SSLP markers, we genotyped a 68 Mb region on rat chromosome 1 previously implicated in glucose and insulin regulation. We used linkage disequilibrium mapping by mixed model regression with inferred descent to identify a region from 198.85 to 205.9 that contains one or more quantitative trait loci (QTL) for fasting insulin and a measure of insulin resistance, the quantitative insulin sensitivity check index. This region also encompasses loci identified for fasting glucose and Insulin_AUC (area under the curve). A separate <3 Mb QTL was identified for body weight. Using a novel penalized regression method we then estimated effects of alternative haplotype pairings under each locus. These studies highlight the utility of HS rats for fine-mapping genetic loci involved in the underlying causes of T2D.
Keywords: obesity, Type 2 diabetes, glucose tolerance, metabolism
type 2 diabetes (t2d) is a disease of relative insulin deficiency resulting from a combination of insulin resistance and decreased beta-cell function (50, 57). This complex disorder is caused by a combination of both environmental and genetic factors. Although diet and exercise are the major environmental factors contributing to T2D (25), the genetic factors contributing to this disease are only beginning to be understood. Over the past several years, over 40 genes have been identified for T2D in human genome-wide association studies (GWAS) (see Refs. 38, 62, 78). Even when the effects of these genes are combined, however, they explain only 10% of the heritable variance (78), indicating that many genetic factors remain unidentified and highlighting our limited understanding of this disorder.
We have previously used heterogeneous stock (HS) rats to fine-map a locus for glucose tolerance to 2.44 megabases (Mb) (64). The HS rat colony at the Medical College of Wisconsin (MCW) comprises outbred animals derived by the National Institutes of Health (NIH) in 1984 from a set of eight genetically and phenotypically diverse inbred founder strains: ACI/N, BN/N, BUF/N, F344/N, M520/N, MR/N, WKY/N, WN/N (23). HS populations are powerful tools for genetic studies because they provide a basis for high resolution mapping of quantitative trait loci (QTL) in a relatively short time period (64, 72). Although none of the inbred founder strains of the HS rat colony are overtly diabetic, we have demonstrated that at least two of the HS founder strains, WKY and ACI, exhibit glucose intolerance (64). Previous studies have found that the WKY rat also has higher plasma insulin levels than other strains both at fasting (75) and in response to a glucose challenge (35, 63). In addition, when made genetically obese by mutating the leptin receptor, the WKY rat (fatty WKY) shows decreased insulin sensitivity relative to Zucker rats (27). To our knowledge, insulin levels have not been measured in the other founder inbred strains, including the glucose-intolerant ACI, or in the HS rat colony.
To understand better the differences in glucose regulation between the founders that make up the HS colony, we initially measured multiple metabolic phenotypes in six of the inbred founder strains, including the quantitative insulin sensitivity check index (QUICKI) as a measure of insulin resistance, and the insulinogenic index (IGI) as a measure of beta cell function. We found that the WKY, M520, and BUF founder strains have decreased insulin sensitivity, whereas the ACI rat shows an insulin secretion defect. Because of the phenotypic variation between the inbred founder strains, we hypothesized that the HS rat colony would be useful for genetically fine-mapping these traits. Importantly, human GWAS have confirmed the utility of quantitative traits such as fasting glucose and fasting proinsulin in nondiabetic individuals for elucidating genetic mechanisms involved in glucose control, some of which have also been found to play a role in T2D (15, 66).
We were specifically interested in a 68 Mb region on rat chromosome 1 that has previously been identified for multiple metabolic traits (including glucose tolerance, body weight, and postprandial insulin) in several linkage studies in the rat (11, 13, 19–22, 34, 42, 53, 64, 79). Importantly, we previously mapped both postprandial insulin and body weight to this chromosome 1 region in a WKY × F344 F2 intercross (63), suggesting that these alleles would likely segregate in the HS rat colony. We demonstrate here successful fine-mapping of fasting glucose, insulin AUC (area under the curve), fasting insulin, QUICKI, and body weight. Although IGI (a marker of beta cell function) did not map to this region, the high degree of variability for this trait in the founder strains suggests that HS rats will be useful for mapping this trait genome-wide. These studies show that the HS rats provide a strong basis for fine-mapping of distinct genetic loci for insulin resistance and beta cell function and thereby can be used to accelerate the pace of discovery for novel genes involved in T2D.
METHODS
Animals
Founding inbred substrains.
The HS rat colony was initiated by NIH in 1984 from the following eight inbred founder strains: ACI/N, BN/SsN, BUF/N, F344/N, M520/N, MR/N, WKY/N, and WN/N (23). Most of these inbred founder strains are no longer available from outside vendors. We have cryo-resucitated the M520/N strain and are maintaining this strain at the MCW. Testing of the other founders was conducted in the following substrains (short names to be used throughout the rest of the manuscript are in parentheses): ACI/Eur or ACI/Seg (ACI), BN/NHsdMcwi or BN/SsnHsd (BN), BUF/NHsd (BUF), F344/NHsd (F344), and WKY/NHsd (WKY). These substrains were chosen based on similarities in origin to the original founders as determined by data obtained from the Rat Genome Database: http://rgd.mcw.edu/strains/. ACI/Eur and BN/NHsdMcwi were obtained from colonies maintained at MCW. ACI/Seg, BN/SsnHsd, BUF/NHsd, F344/NHsd, and WKY/NHsd were ordered from Harlan Sprague-Dawley (Indianapolis, IN). We saw no phenotypic differences between ACI/Eur and ACI/Seg substrains, nor between BN/NHsdMcwi and BN/SsnHsd substrains, and we combined these substrains as ACI and BN, respectively. All experiments were conducted only in males. We used 10–21 male rats per strain in the experiments outlined below. Two of the strains (MR/N and WN/N) can no longer be obtained from outside vendors, so these strains were not tested. All protocols were approved by the Institutional Animal Care and Use Committee at MCW.
Heterogeneous stock colony.
Origin of the heterogeneous stock colony has been described previously (64). Experiments were conducted in 782 NMcwi:HS male progeny. HS rats descended from 160 male-female pairs with an average of 4.89 full sibs from each pair. Glucose tolerance has previously been mapped in 515 of these animals (64). In the current work we phenotyped the original 515 plus an additional 267 rats, as described below, for fine-mapping these additional traits.
Phenotyping Protocol
In both founder substrains and HS rats, we conducted an intraperitoneal glucose tolerance test (IPGTT) at 16 wk of age. As previously described (64), animals were fasted overnight for 16 ± 1 h. In the morning, a basal sample of blood was taken from the tail and subsequent samples were collected 15, 30, 60, and 90 min after a 1 g/kg body wt glucose injection. We used the Ascensia Elite system for reading blood glucose values (Bayer, Elkhart, IN). We also collected blood at each time point for subsequent analysis of plasma insulin levels. Plasma insulin levels were determined with an ultrasensitive ELISA kit from Alpco Diagnostics (Salem, NH). Using a trapezoidal analysis of insulin, we calculated area under the curve for insulin during the glucose tolerance test (Insulin_AUC), a general measure of overall insulin secretion.
Measures of insulin sensitivity and beta cell function.
To determine insulin sensitivity, we calculated two related measures: the QUICKI and the homeostatic model assessment of insulin resistance (HOMA_IR). Both measures have been used in humans to assess insulin sensitivity (36, 44) and have been validated in the rat (7). In rat, QUICKI shows a stronger correlation with the glucose clamp method of determining insulin sensitivity than HOMA_IR or the fasting glucose-to-insulin ratio (7). We calculated QUICKI as 1/[log I0 + log G0], where I0 is fasting insulin and G0 is fasting glucose, and HOMA_IR as cI0G0, where c is a species-specific constant. Since c is unknown for rat, and because its value would anyway not alter comparisons within species, we set c = 1, such that QUICKI = 1/log(HOMA_IR).
To assess the sensitivity of the beta cell to glucose, we calculated the IGI, a measure validated in humans that combines glucose and insulin values from an oral glucose tolerance test (55, 69). IGI has recently been used as a way to understand function of genetic loci identified for fasting glucose, fasting proinsulin, and T2D (28). It is calculated as the change in plasma insulin during the IPGTT divided by the change in glucose during the same period. Specifically, if It and Gt are the levels of plasma insulin and glucose respectively after t min, then IGI_t = (It − I0)/(Gt − G0). We considered IGI_15 and IGI_30.
Genotyping
Tail samples from the original eight inbred founders were obtained from the NIH. We extracted DNA from tail tissue by an ethanol extraction procedure. Genotyping was conducted in founders and the 782 HS rats with 97 SSLP markers within a 68.26 Mb on rat chromosome 1 (from 198.85–267.11 Mb). The number of alleles per marker ranged from 2 to 6, with an average ± SE of 3.67 ± 0.10. Coordinates of markers are based on rat genome assembly version 3.4. Markers were spaced an average of 711 Kb apart. PCR was conducted as previously described (47) and analyzed on the ABI 3730.
Statistical Analysis
Analysis of phenotypes in the inbred and HS rats.
To determine whether there were statistical differences between inbred founder substrains for glucose and insulin levels during the IPGTT we fitted the following linear mixed model. Let yti be the concentration of glucose (or insulin) measured at time point t, where t corresponds to 0, 15, 30, 60, or 90 min, then we model
| (1) |
where μt is the intercept (i.e., the baseline level) for time t, strains is the increase over all time points associated with founder substrain s, s[i] denotes the strain to which rat i belongs, and rati and residualti are normally distributed random intercepts for rat and rat-specific time point, respectively. Significance of an effect of founder substrain was evaluated by an F-test (56). Significance of substrain effects for all other measures (fasting glucose, fasting insulin, Insulin_AUC, QUICKI, HOMA_IR, IGI_15, IGI_30, and body weight) was judged by ANOVA. Post hoc comparisons between pairs of strains were performed with Tukey honestly significant difference (HSD) test. Tests were performed in R (58), with mixed effects modeling using the add-on package nlme (56).
Mapping QTL.
We fine-mapped QTL in the 68 Mb region by linkage disequilibrium mapping, testing for the significance of association between a given phenotype and inferred haplotype descent at each locus. Our approach closely follows that used previously (64) with the following exceptions: here we use only the “full” genetic model (i.e., we test for additive and dominant genetic effects simultaneously rather than separately); our mapping is applied independently to multiple phenotypic measures, each of which is first subject to a normalizing transformation; and, depending on what is relevant to each phenotype, different covariates are used. In brief, haplotype descent along each HS rat genome was inferred by the haplotype reconstruction method HAPPY (49), which applies a hidden Markov model simultaneously to the genotypes of the eight founder strains and the 782 HS rats. At each interval m = 1, . . . , 96 between adjacent pairs of markers, HAPPY produces for each rat i a vector gi(m) containing the probabilities of descent from each of the 8(8 + 1)/2 = 36 unique haplotype pairs (diplotypes) (71). This is used in the following mixed model regression to predict rat i's phenotype yi:
| (2) |
where f(yi) is a transformed version of the phenotype, covariatesi = ∑c∈C βcTxi(c) models the effect of a set of covariates C with values xi(c) for c∈C, QTLi(m) = βTgi(m) models the putative effect of the QTL, sibshipk[i] is the (random) effect of the sibship k to which i belongs, and residuali accounts for individual variation (further parameters defined as in Ref. 64). The use of a simple random effect for sibship is an approximation to a polygenic term, that is, to a random effect modeling expected covariance between differentially related families as advocated in Ref. 37 and more recently in, e.g., Ref. 10. Although the use of this approximation can lead to a loss in power, since it assumes sibships are about evenly related and thereby overfits the effects of sibships that are more closely related, in our HS rats it is highly computationally convenient and only moderately suboptimal, because the circular breeding system that maintains this population leads to a highly regular pedigree. We defined the set of covariates C to include the following pretreatment indicators: location (2 buildings), injector (2 injectors), collector (person who collected the blood, 2 collectors), and the number of glucose injections the rat received (1 or 2; a small percentage of animals received a second glucose injection when the initial injection failed to lead to a glucose response). The model above assumes that both sibshipk and residuali are normally distributed. To make this assumption reasonable, for each phenotype we used the Box-Cox procedure to find a transformation f that would ensure residual normality when fitting the null model (5), that is, Eq. 1 without the QTL term. Specifically, for reported phenotypes we used the following transformations (y = original value): fasting glucose (), Insulin_AUC (inverse normal transformation), fasting insulin (y1/3), QUICKI (−y−2), IGI_15 ([y + 0.1]−2), body weight (y). Models were fitted by restricted estimate maximum likelihood, the significance of association at each interval m was assessed by comparing the fit of Eq. 1 with that of the null model. Region-wide significance thresholds were estimated by parametric bootstrap from the fitted null model (see Refs. 64, 71).
Confidence intervals (CI) for detected QTL were estimated using the 1.5-logarithm to the base 10 of the odds (lod) drop method (16, 41) (hereafter, lod-drop), an approximation to the 95% CI for QTL mapping. This method is commonly used in QTL mapping of F2 populations (6) but has also been applied to more recombinant populations such as advanced intercross lines (54, 61) and the Collaborative Cross (1). Because we test association at discrete genomic intervals defined by the marker positions, rather than approximating a curve of association between markers, we report the lod-drop conservatively, defining it by the markers that enclose the genomic interval that lies just outside the 1.5 lod-drop. For cases in which the lod score drops <1.5 and then comes back up (fasting insulin and QUICKI), we accept the larger connected interval as the most likely CI (6). To provide a complementary estimate of how sensitive our localization of the top QTL peak is to the sampling of the rats, we also used the general statistical method of nonparametric bootstrapping (24), in which the test statistic (in this case, the peak location) is re-estimated under alternative datasets based on the original, each alternative data created by resampling the individuals with replacement (this having originally been applied to QTL mapping in Ref. 77). QTL scans were performed using BAGPIPE (71), making use of the R packages lme4 (2), evd (65), and MASS (76).
Estimating heritability and QTL effect size.
To estimate the heritability (h2) of each trait we fit Eq. 1 excluding the QTL term and then combine the point estimates of the sibship variance (τ̂sibship2) and residual variance (σ̂2) as ĥ2 = 2τ̂sibship2/(τ̂sibship2 + σ̂2) × 100%. This follows the approximation described in, for example, Ref. 73, but excludes effects of environmental covariates. Since sibship is confounded with, for example, maternal effects and, to a limited extent, common environment, we expect our estimates to have a slight upward bias. We report the effect size of a QTL based on the percentage of phenotypic variance it explains when fitted in Eq. 1, using the method of Ref. 73 to describe this for fixed effects embedded in a mixed model.
Estimating diplotype substitution effects at QTL.
The HS rats are descended from eight founders, and so at any given locus the colony may be segregating as many as d = 36 distinct haplotype pairs (i.e., 36 diplotypes), each diplotype potentially exerting a distinct effect on the phenotype. For some QTL the genetic architecture will be simple, and the effects well summarized by a “dosage” effect for each of the eight haplotypes, these combining an additive fashion (67, 71). In other cases, however, the QTL architecture may be more complex and include substantial elements of dominance and recessivity. In our previous mapping of a glucose tolerance QTL we found strong dominance effects that were poorly explained by an additive model (64). To characterize the allelic series for our QTL it is therefore necessary to look at effects for all 36 diplotypes. This is technically challenging and has not to our knowledge been achieved in any similar population (e.g., the Diversity Outbred cross in Ref. 67 or the mouse HS stock in Ref. 72). We do this here using a novel approximation.
Describing descent at locus m for all n = 782 individuals, let D be the n × d matrix such that the ith row Di* = gi(m)T describes individual i's descent for all diplotypes, whereas the jth column D*j = dj(m) describes descent of the jth diplotype for all individuals. Describe Eq. 1 more accurately in matrix form (e.g., as in Ref. 45) as
| (3) |
where y is the 782-length vector of phenotypic responses, μ1 represents the baseline intercept, XB represents the covariate effects, Z allocates the sibship effects from the 160-length vector u, modeled as u∼N(0, Iτsibship2), ε is the 782-length vector of residual errors, modeled as ε∼N(0, Iσ2), G concisely represents diplotypes at the locus m, where specifically G = D−(1)() is the n × (h − 1) matrix formed from columns 2, . . . ; h of D (dropping the first column d1 because this will be absorbed into the intercept μ1), and θ is the corresponding vector of d − 1 estimable locus effects (described for simplicity of exposition in Eq. 2 as the d-vector β).
In practice, uncertainty about descent often makes G ill-conditioned, and this leads to wildly unstable estimates of θ and, therefore, of diplotype effects. This is not a problem for QTL mapping because it is straightforward to reduce the dimension of G during computation without compromising the fit to the data (see Appendix A in Ref. 71). Ill-conditioning of G is a problem, however, when it comes to reporting estimates of the 36 diplotype substitution effects, since doing so would require G to remain intact. One approach to obtaining stable estimates of allele effects is to constrain the values θ can take by penalization or formal hierarchical modeling. Until now, this has been achieved only for simple additive effect models that exclude covariate or random effects: specifically in inbred plants lines or the mouse Collaborative Cross using multiple imputation (17, 39), or in the mouse Diversity Outbred population (akin to a rodent HS) using ridge regression (67).
To estimate diplotype effects while simultaneously accounting for covariates and random sibship effects, we introduce a versatile new approximation. Adapting the ridge penalty proposed recently by Refs. 29 and 30, we substitute G with Gξ, where Gξ = Dξ−(1)(), with Dξ equal to the “ridge surrogate” of D defined as
| (4) |
where U, Λ, and V are defined the singular value decomposition D = UΛVT, and ξ is chosen by random 2/3-subsample validation in the mixed model (i.e., Eq. 2 with the Gξ substitution). Estimating diplotype substitution effects then proceeds in two further steps: extracting d-vector of estimates β̂D from parameters estimated by the mixed model, and then rescaling β̂D to reflect categorical substitutions of diplotype rather than quantitative shifts in diplotype proportions. Specifically, we first calculate β̂D = [μ̂, (θ̂ + μ̂)T]T, which describes the observed effects of changing D. Then, noting that when predicting contrasts in phenotype between hypothetical individuals, QTL descent will be categorical (i.e., treated as known, and representable by a matrix Δ of zeros and ones) rather than probabilistic (i.e., treated as uncertain, as from HMM inference, and represented by a matrix D of fractional numbers), we obtain a diplotype substitution estimate as β̂Δ = β̂D × sD/sΔ. Here, effects are rescaled approximately according to the standard deviation of columns in the observed D, with sD = d−1 ∑j=1d sd[dj(m)], relative to those in Δ, the latter being assumed multinomial with uniform diplotype probability pj = d−1 such that sΔ = d−1 ∑j=1d = (d − 1)1/2d−1.
RESULTS
Analysis of Phenotypes in the Inbred and HS Rats
Confirmation of glucose intolerance in WKY and ACI rats.
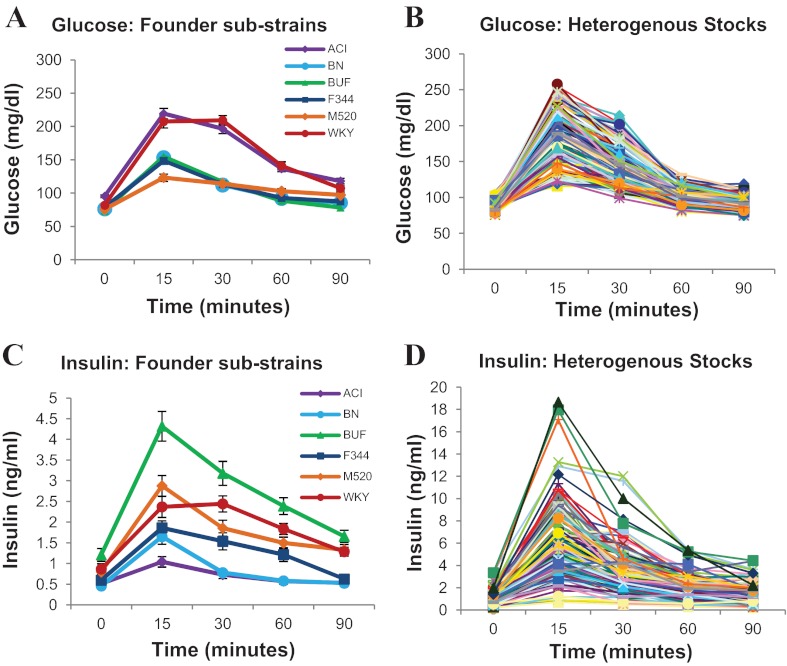
Founder strains vary considerably in their averages for fasting glucose (F5,92 = 21.14, P = 5.15e-14; see Figs. 1A and 2A) and glucose in response to a glucose challenge (glucose response; F5,87 = 54.19, P = 2.84e-25; see Fig. 1A). As shown previously (64), both ACI and WKY rats show higher levels of glucose response relative to the other rat strains ( P < 0.001, for all comparisons by Tukey-HSD), indicating glucose intolerance in these strains. The ACI rat also shows increased fasting glucose relative to all other strains (P < 0.001, for all comparisons by Tukey-HSD). Glucose levels in the M520/N strain, which was not included in our previous study, do not differ from the other normal strains (BN, BUF, F344). However, the 15-min glucose peak is slightly lower, and the 90 min time-point is slightly higher in M520/N relative to BN, BUF, and F344, such that the M520/N glucose response appears relatively flat.
Fig. 1.
Glucose and insulin in response to a glucose challenge in inbred founders (A, C) and heterogenous stock (HS) rats (B, D). Founder curves represent means ± SE of 10–21 animals within each inbred strain. See text for statistical differences between inbred strains. Each line in the HS graph represents an individual animal (200 representative HS rats are shown in graph). Because of the extremely high values for insulin in the HS rats (D), the y-axis differs between C and D, enabling differences between inbred strains (C) to be visualized.
Fig. 2.
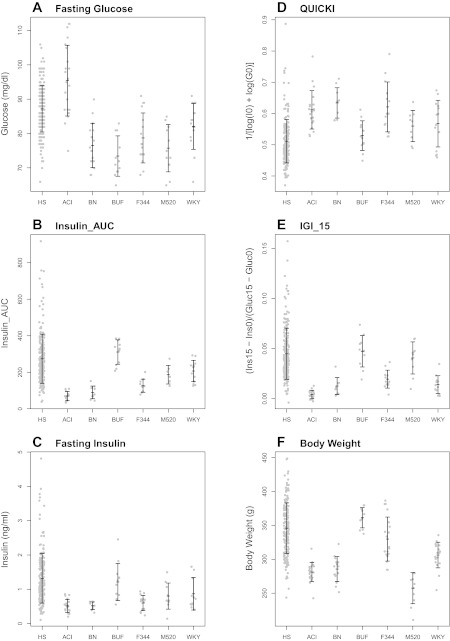
Fasting glucose (A), Insulin_Area Under the Curve (AUC) (B), fasting plasma insulin (C), quantitative insulin sensitivity check index (QUICKI) (D), insulinogenic index (IGI_15) (E), and body weight (F) in inbred founders and HS rats. Mean ± SD are shown. Gray dots represent individual animals for 200 representative HS rats or n = 10–21 founder strains. See text for statistical differences between founder strains.
WKY and ACI strains show a different insulin response to the glucose challenge.
Insulin levels, both at fasting (F5,79 = 9.27, P = 5.67e-07; see Figs. 1C and 2C) and in response to a glucose challenge (insulin response; F5,68 = 44.61, P = 3.63e-20; see Fig. 1C), differ significantly between the HS founder strains. There is a particularly striking difference between the glucose-intolerant strains, with the WKY rat showing high fasting insulin (P = 0.038 and 0.023 relative to ACI and BN, respectively; Tukey-HSD) and insulin response (P < 0.001 relative to ACI, BN, and F344; Tukey-HSD), and the ACI strain showing low insulin response (P < 0.05 relative to all strains except BN, Tukey-HSD). The BUF rat, a strain with normal glucose regulation, has significantly higher fasting insulin levels (P < 0.001 relative to ACI, BN, and F344; Tukey-HSD) and insulin response (P < 0.001 relative to all other strains, Tukey-HSD). The above differences are also clearly seen when measuring Insulin_AUC (F5,65 = 45.01, P = 7.9e-20; see Fig. 2B).
Decreased insulin sensitivity in BUF, M520, and WKY strains.
QUICKI (F5,77 = 7.19, P = 1.45e-05; see Fig. 2D) and HOMA_IR (F5,79 = 6.28, P = 5.87e-05; data not shown) vary significantly between the founder strains. When both measures are used, BUF shows the least insulin sensitivity (P ≤ 0.01 relative to ACI, BN, F344; Tukey-HSD) and BN shows the greatest (P < 0.01 relative to BUF, M520, WKY; Tukey-HSD). M520 and WKY rats have QUICKI and HOMA_IR levels that are similar to those of the BUF rat (P < 0.01 relative to BN, Tukey-HSD). HOMA_IR is commonly used to assess insulin sensitivity in humans (e.g., Ref. 44). However, because QUICKI has been shown to have a higher correlation with the glucose clamp method of insulin sensitivity than HOMA_IR (7), and because HOMA_IR and QUICKI exhibit similar statistical differences between the founder inbreds, we show the plot only for QUICKI here. Furthermore, as HOMA_IR and QUICKI are transformations of each other, and the genetic plots for both measures are essentially identical, we refer only to QUICKI throughout the rest of the text.
Decreased beta cell function in the ACI strain.
There are significant differences between founder strains for both IGI_30 (F5,67 = 10.47, P = 1.88e-07; data not shown) and IGI_15 (F5,74 = 34.24, P < 0.001; see Fig. 2E). ACI has the lowest IGI scores (P < 0.05 relative to all strains except WKY, Tukey-HSD), indicating that its beta cells are not responding appropriately to the glucose challenge. BUF and M520 have the highest IGI scores (P < 0.001 relative to all other strains, Tukey-HSD), demonstrating a strong response of beta cells to glucose, likely owing to the decreased insulin sensitivity in these strains. IGI scores in the WKY rat, by contrast, are relatively low, indicating that this strain does not compensate for its decreased insulin sensitivity (P < 0.001 relative to BUF and M520, Tukey-HSD). IGI_15 shows less within-strain variation than IGI_30, and so we hereafter use IGI_15 as our primary measure of IGI.
Increased body weight in the BUF strain.
Body weight varied significantly between founder strains (F5,85 = 37.01, P = 5.79e-20; see Fig. 2F), with BUF rats weighing the most (P < 0.001 relative to all other strains, Tukey-HSD) and the M520 strain weighing the least (P < 0.05 relative all strains except BN, Tukey-HSD).
HS rats show great variation for all traits measured.
In the HS rats, values for both fasting glucose and glucose response fall mostly between the average values for the inbred founder substrains, as previously shown (64); see Figs. 1, A and B, and 2A. In contrast, the range of fasting insulin and insulin response are much greater in the HS than that seen between the founders (see Figs. 1, C and D, and 2, B and C). For example, insulin levels 15 min after the glucose challenge range from 0.5 ng/ml to as high as 18 ng/ml in the HS, three times greater than in the BUF strain. This variation is also seen for Insulin_AUC (see Fig. 2B). The mean values for fasting insulin and Insulin_AUC in the HS are similar to those in the BUF, indicating that many HS rats are hyperinsulinemic.
For QUICKI, there is a broad range of insulin sensitivity in HS rats (see Fig. 2D). As with Insulin_AUC and fasting insulin, the mean for QUICKI in the HS animals is close to that of the insulin-resistant BUF strain (0.50 in HS vs. 0.53 in BUF), with 74.6% of the HS having a QUICKI score less than the BUF mean, indicating that many of the HS animals have some degree of insulin resistance. Interestingly, of those rats with a QUICKI score equal to or lower than the BUF strain, 25.1% also have glucose intolerance (i.e., area under the curve for glucose ≥ 1,200; data not shown). For IGI_15, there is also a high degree of variability in the HS rats (see Fig. 2E). Again, the mean for IGI_15 is similar to the BUF strain, indicating that beta cells in most HS rats are responding well to the glucose challenge, despite some degree of insulin resistance. In contrast, we determined that 4% of the HS rats show IGI_15 values equal to or smaller than the ACI rat, indicating a likely secretion defect in these animals. Lastly, there is a high degree of variability for body weight in the HS rats, with values generally falling in the range of values seen between the inbred strains (see Fig. 2F).
Correlations between traits reveal at least four independent diabetes-related phenotypes.
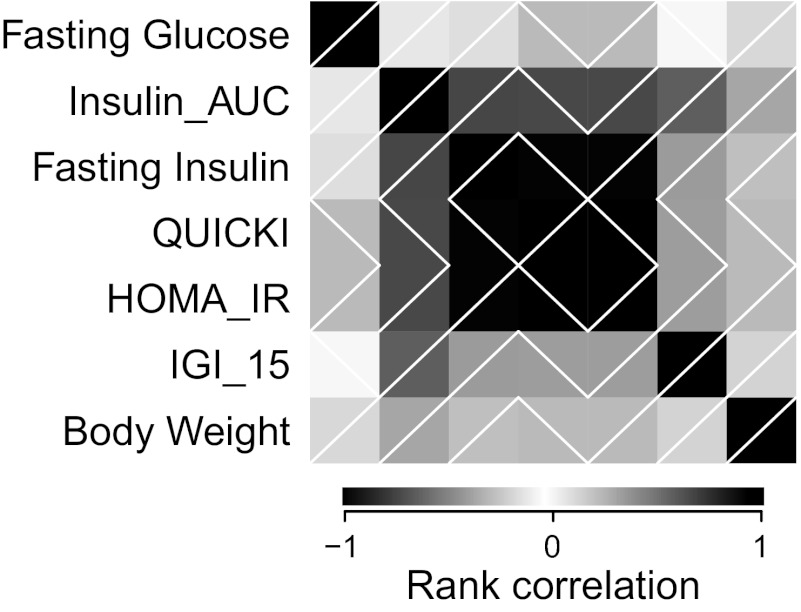
We estimated the Spearman rank correlation (rho) between traits in all 782 HS rats (see Fig. 3). Insulin_AUC was highly correlated with all other insulin traits including fasting insulin (rho = 0.729), QUICKI (rho = −0.719), HOMA_IR (rho = 0.719), and IGI_15 (rho = 0.632). As expected, QUICKI and HOMA_IR are in perfect correlation with each other (rho = −1) and are highly correlated with fasting insulin (rho = −0.987 and 0.987, respectively), but not with IGI_15 (rho = −0.377 and 0.377, respectively). Fasting glucose and body weight, on the other hand, show very little correlation with any of the insulin traits. Principal components analysis (PCA) of the transformed phenotypes shows that 97% of the variance in the seven traits listed can be explained by the first four principal components (PCs) (87% by the first 3 PCs, 99.978% by the first 5 PCs). This demonstrates that that despite some correlation, owing to mathematics or physiology, the set of seven phenotypes embodied at least four independent sources of information related to T2D susceptibility.
Fig. 3.
Correlations among phenotypes. Each block represents the Spearman rank correlation between 2 phenotypes, with the top-left to bottom-right diagonal of black blocks depicting correlation of +1 (i.e., a phenotype with itself). Shading indicates strength of correlation; white diagonal lines indicate positive vs. negative correlation.
Mapping QTL
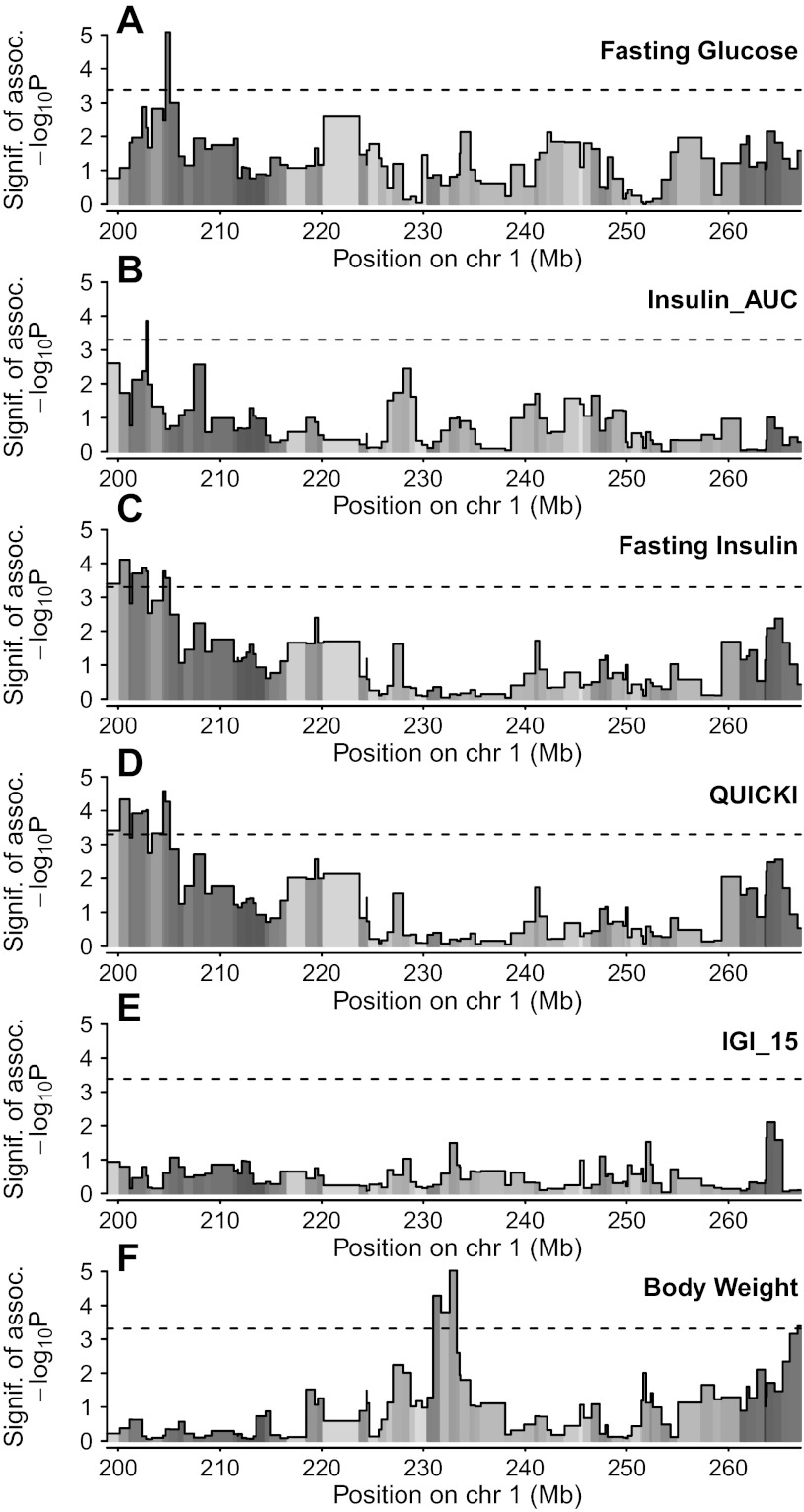
Using a combination of haplotype reconstruction and mixed model linear regression, which exploits information about the HS ancestry while taking into account sibling relationships, we identified a single sharp peak for fasting glucose (peak marker interval between D1Rat 168 and D1Rat70, 204.62–205.04 Mb; logP = 5.09; see Fig. 4A) and Insulin_AUC (peak marker interval between D1Rat181 and D1Arb24, 202.76–202.91 Mb; logP = 3.86; see Fig. 4B). We also identified a broader QTL for both fasting insulin (peak marker interval between D1Rat291 and D1Rat218, 200.17–201.13, logP = 4.11; see Fig. 4C) and QUICKI (peak marker interval between D1RatMgh10 and D1Rat168, 204.38–204.62, logP = 4.58; see Fig. 4D). Lastly, we identified a single peak for body weight (peak marker interval between D1Rat302 and D1Got225, 232.58–233.29 Mb, logP = 5.03; see Fig. 4F). All QTLs are listed in Table 1. All traits were mapped with a genetic model that incorporated both additive and dominant effects of QTL haplotypes. The region-wise 5% significance threshold for the full model was different for each phenotype (see Table 1), since this is affected by the heritability of the trait, but was on average logP = 3.33 (range: 3.3–3.39), where logP is defined as −log10(P value). IGI_15 did not map to this region on rat chromosome 1 (see Fig. 4E).
Fig. 4.
Mapping of fasting glucose (A), Insulin_AUC (B), fasting plasma insulin (C), QUICKI (D), IGI (E), and body weight (F) within a 68 Mb region on rat chromosome 1. The x-axis shows position in Mb, and the y-axis gives the −log10P value of association. Dashed lines represent region-wide 5% significance threshold for the full model, which are (in order, to 2 decimal places): 3.38, 3.30, 3.30, 3.30, 3.39, and 3.31. Shading under association line indicates selective information content, or how well genotype information informs haplotype pair descent at each marker interval, with black indicating that descent is known with certainty for all mice, and white indicating complete uncertainty.
Table 1.
Summary of QTL peaks, effect sizes, and CI
| Trait | Peak Marker Interval, Mb | LogP (region-wide threshold) | Effect Size, % of Total Phenotypic Variance Explained | 1.5-LOD Drop Interval in Mb (size) | Rat Genome Database Name (ID number) |
|---|---|---|---|---|---|
| Fasting glucose | D1Rat168-D1Rat70 (204.62–205.04) | 5.09 (3.38) | 6.33 | 204.38–205.90 (1.52 Mb) | Gluco64 (6480773) |
| Insulin_AUC | D1Rat181-D1Arb24 (202.76–202.91) | 3.86 (3.30) | 7.34 | 202.35–203.30 (0.95 Mb) | Insul17 (6480777) |
| Fasting insulin | D1Rat291-D1Rat218 (200.17–201.13) | 4.11 (3.30) | 6.24 | 198.85–205.90 (7.05 Mb) | Insul18 (6480780) |
| QUICKI | D1mgh10-D1Rat168 (204.38–204.62) | 4.58 (3.38) | 6.78 | 198.85–205.90 (7.05 Mb) | Insul19 (6480783) |
| Body weight | D1Rat302-D1Got225 (232.58–233.29) | 5.03 (3.31) | 6.17 | 231.73–233.55 (1.82 Mb) | Bw97 (6480786) |
QTL, quantitative trait locus; CI, confidence interval; LOD, logarithm of the odds. AUC, area under the curve; QUICKI, quantitative insulin sensitivity check index; LogP, negative logarithm to the base 10 of the P value.
Our method of genetic association is based on linkage disequilibrium mapping and allows us to model the effects of QTL that may be located in between genotyped markers. As such, it relies on inferred ancestry that may be better informed at some locations than others, which could potentially affect QTL location by making association in some regions intrinsically hard to detect. To explore this phenomenon, and the way it relates to QTL localization, we depict for each marker interval the average selective information content (see Fig. 4, shading under the association line), as defined in Ref. 60. This information is maximal (black) when the identity of the ancestral haplotypes can be inferred with certainty for all rats and minimal (white) when genotype information provides no information about haplotypes at this position. As expected, some regions have higher information content than others. Nonetheless, information content does not appear to drive estimated location of the QTL. The regions from 200–217 Mb and again from 261–267 Mb generally have relatively high information content, whereas the region from 217–261 Mb is scattered with marker intervals having lower information content, indicating that we may have less power to resolve QTL lying within these marker intervals.
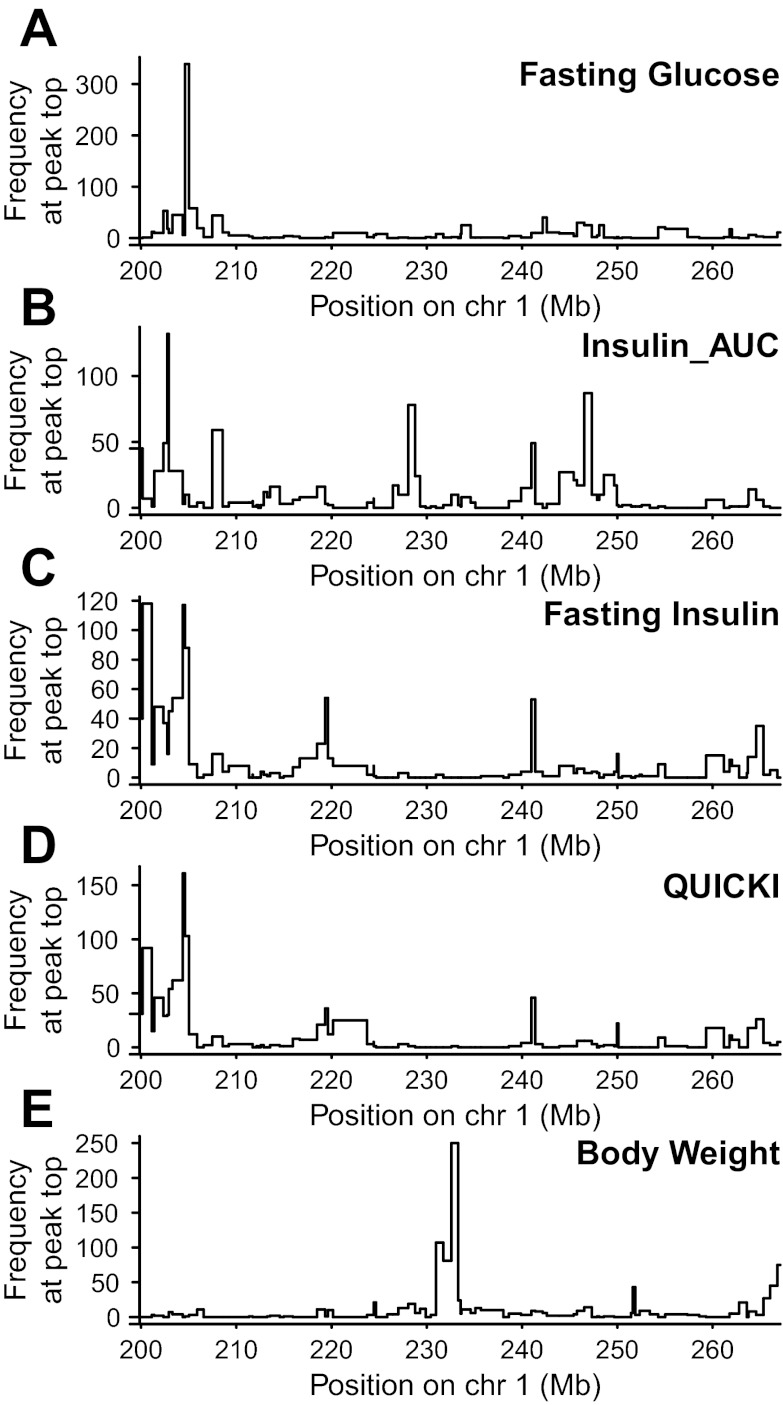
The 1.5 lod-drop intervals for all phenotypes ranged from 0.95 to 7.05 Mb and are reported in Table 1. These lod-drop intervals are estimates of the most likely region of the QTL and it is important to note that the causative gene or SNP may lie just outside this identified area. To estimate how sensitive the locations of the peaks in our analysis are to sampling of the individuals, we performed positional bootstrapping (after Ref. 77), in which the entire region is rescanned using a bootstrap sample of the rats, and the location of the topmost peak is recorded. Results from the bootstrapping complement those from the lod-drop method, providing additional information about the robustness of the mapping procedure that can help confirm the location of the lod-drop interval and determine whether more than one QTL may lie within (or outside of) the identified QTL. We ran 1,000 positional bootstraps across the entire 68 Mb region for each trait (see Fig. 5).
Fig. 5.
Positional bootstrapping across the entire 68 Mb region for fasting glucose (A), Insulin_AUC (B), fasting plasma insulin (C), QUICKI (D), and body weight (E). Histogram shows the number (y-axis) that each marker interval (x-axis, locations in Mb) harbored the strongest association (see text for details) when the analysis was applied to each of 1,000 bootstrapped sets of rats.
For fasting glucose, the 1.5 lod-drop CI is only 1.52 Mb (see Table 1), and this general region is confirmed in the region-wide bootstrap scan, with >300 resamples falling within the peak marker interval (see Fig. 5A) Although some of the resamples fall outside of the 1.52 Mb CI, this occurs at very low frequencies, indicating little support for QTL lying outside this region. The 1.52 Mb region overlaps Gluco56, the QTL we previously identified for Glucose_AUC (64).
Despite the sharp QTL peak for Insulin_AUC in the region-wide scan, and a 1.5 lod-drop interval of only 0.95 Mb, the topmost peaks from its bootstrap samples are more scattered, with only 132 resamples falling within the peak marker interval (see Fig. 5B), suggesting that additional loci may exist outside of this QTL. This is not surprising, as the region we are interrogating is complex (see Ref. 64 and references therein) and likely includes additional QTLs that we are currently underpowered to detect (see information content in Fig. 4).
The 1.5 lod-drop for fasting insulin and QUICKI is 7.05 Mb and overlaps the QTL identified for fasting glucose and Insulin_AUC (see Table 1). The bootstrap resamples for fasting insulin and QUICKI demonstrate two separate peaks within this 7.05 Mb interval (one at D1Rat291 with 118 and 92 resamples, respectively, and another at D1Mgh10 with 117 and 161 resamples, respectively; see Fig. 5, C and D). These results are consistent with a scenario in which more than one QTL underlies this locus. Similar to Insulin_AUC, we also see multiple peaks across the 68 Mb region, indicating additional loci, which we are currently underpowered to detect, may exist outside the identified region.
The 1.5 lod-drop interval for body weight is only 1.82 Mb. Positional bootstrapping supports this general region with 250 bootstraps falling right at the peak marker interval. An additional 107 resamples fall at neighboring marker interval between D1Mit34 and D1Got228 (230.96–231.73 Mb), suggesting the true CI for this QTL may lie in a slightly larger 2.59 Mb region from 230.96–233.55 Mb (see Fig. 5E). Although some bootstrap resamples fall outside of this 2.59 Mb region, they occur at low frequency indicating little support for the QTL lying outside of this region.
Heritability of Traits
We estimated the narrow-sense heritability of each trait in the HS based on how much phenotype values clustered by sibship (heritability in parentheses; see methods for calculation): Fasting glucose (59.12%), Insulin_AUC (44.31%), fasting insulin (53.88%), QUICKI (56.67%), HOMA (57.93%), IGI (35.55%), body weight (80.04%). Effect sizes for all QTL were ∼6–7% and are reported in Table 1.
Effects of Allele Substitution at Detected QTL
To characterize the way that substitutions of alternative haplotype pairs (i.e., diplotypes) at each QTL could be affecting each phenotype, we estimated the diplotype substitution effect for each of the 36 unique diplotypes based on our rat model. Figure 6 plots these effects for seven QTL detected in five phenotypes. For each QTL, the estimated effects of the 36 possible diplotype substitutions are plotted as a grayscale heat map, with a key on the right-hand side showing the numerical range of the substitution effects, and for comparison the range of the transformed phenotype. These values are estimated with a degree of error (see Fig. 6 legend for standard errors) and subtle contrasts will rarely represent statistically significant differences. As such Fig. 6 provides a less than definite guide to QTL architecture. Nonetheless, we note several features that stand out.
Fig. 6.
Estimated effects of diplotype substitutions at QTL. For each phenotype-QTL combination, shaded matrix indicates the estimated effect on the transformed phenotype of a given founder haplotype combination. Side bars show the scale of the phenotype (e.g., body weight varies from 202 to 464 g) and the scale of the effects, such that, e.g., for an otherwise average body weight, diplotype substitutions could push the phenotype anywhere from 296 to 374 g. The transformed value of the phenotypes, as described in methods, is used. These estimates act as a rough guide to the relative effects of substitution, with the order of closely ranked contrasts being highly sensitive to sampling error. In particular, the average standard error of estimates reported here was 0.166 (fasting glucose: D1Rat168-D1Rat70), 0.284 (Insulin_AUC: D1Rat181-D1Arb24), 0.0701 (fasting insulin: D1Mgh10-D1Rat168), 0.397 (QUICKI: D1Mgh10-D1Rat168), 0.140 (fasting insulin: D1Rat291-D1Rat218), 0.751 (QUICKI: D1Rat291-D1Rat218), and 27.4 (body weight: D1Rat302-D1Got225).
At marker interval D1Rat168-D1Rat70, the diplotype effects plot demonstrates a complex picture in which multiple haplotype combinations lead to either an increase or decrease in fasting glucose levels. The direction of the effect at this marker interval does not tend to match the phenotype seen in the inbred founders with the ACI haplotype leading mainly to decreased fasting glucose and several of the BN diplotypes resulting in increased fasting glucose.
At marker interval D1Rat181-D1Arb24, the WKY/WKY diplotype leads to the greatest increase in Insulin_AUC, whereas ACI/WKY leads to the lowest Insulin_AUC, following phenotypes seen in the founder strains and indicating ACI dominance. Despite the fact the BUF strain has the highest Insulin_AUC, the BUF allele does not appear to be playing a major role at this marker interval.
Similar to fasting glucose, diplotype effect plots for fasting insulin and QUICKI are complex. Importantly, effect plots at marker interval D1Mgh10-D1Rat168 differ from those at D1Rat291-D1Rat218, consistent with a scenario in which these marker intervals underlie separate QTL. For example, at D1Mgh10-D1Rat168, the ACI/WKY diplotype results in decreased fasting insulin levels and increased insulin sensitivity, while at D1Rat291-D1Rat218, ACI/WKY results in increased fasting insulin and decreased insulin sensitivity. Contrasting effects between these two marker intervals are also seen for several other haplotype combinations including: ACI/F344, BN/BN, BN/BUF, BN/WKY and M520/WKY.
At marker interval D1Rat302-D1Got225, the ACI/F344 diplotype leads to increased body weight, whereas the ACI/M520 diplotype results in decreased body weight, similar to increased body weight in the F344 founder strain and decreased body weight in the M520 strain.
DISCUSSION
T2D results from a combination of insulin resistance and decreased secretory capacity of pancreatic beta cells. Although >40 genes have recently been identified for T2D in human GWAS (38, 62, 78), in sum their effects explain only 10% of the genetic variance (78), indicating many genes have not yet been found. We demonstrate here that the HS rat colony harbors allelic variation that affects both insulin resistance and beta cell function. It is of particular interest that three of the HS founder strains (BUF, M520, and WKY) exhibit insulin resistance and one founder strain (ACI) exhibits an insulin secretion defect. We show that both insulin resistance and insulin secretion vary considerably among the founders and among individuals in the HS colony, suggesting that QTL for these traits segregate in the HS. We have fine-mapped both glucose and insulin traits within a region on rat chromosome 1 that has previously been identified for glucose tolerance and other metabolic traits (see Ref. 64 and references therein). Our results support the utility of HS rats for fine-mapping complex traits and suggest that the HS rat colony will be useful for fine-mapping loci genome-wide for both insulin resistance and beta cell function.
We demonstrate that the underlying cause of glucose intolerance in two of the HS founders is different: ACI exhibits an insulin secretion defect, whereas WKY shows decreased insulin sensitivity. In response to a glucose challenge, the ACI strain shows only a slight increase in plasma insulin levels, with a very low IGI, indicating decreased beta cell function. In addition to being glucose intolerant, the ACI is the only strain to exhibit increased fasting glucose levels, a known predictor of T2D (51, 52). In contrast to the ACI, the WKY strain shows elevated fasting insulin and an increased insulin response to a glucose challenge, as previously demonstrated (35, 63). We show that the WKY has relatively low QUICKI levels, implying decreased insulin sensitivity, and that their IGI levels are normal to low, indicating that their beta cell response is insufficient to make up for this. The combination of decreased insulin sensitivity coupled with poor beta cell response to glucose likely explains the glucose intolerance in this strain. In support of the WKY rat as a prediabetic model, previous studies have found that obesity, induced by mutating the leptin receptor, leads to overt diabetes with decreased insulin secretion over time in this strain (27).
Both the BUF and M520 strains also exhibit hyperinsulinemia. In contrast to the WKY, however, these strains exhibit a high IGI, indicating that the pancreas is responding appropriately to the insulin resistance with increased insulin secretion, thus protecting these strains from developing glucose intolerance. The BUF strain also has the highest body weight, typical of obesity-induced insulin resistance in the human population (33), whereas the M520 strain has the lowest body weight, representative of insulin resistance in the absence of obesity (46).
As expected, the variation between the founder strains for insulin resistance (as measured by QUICKI) and beta cell function (as measured by IGI) is recapitulated or exaggerated in the HS colony. Although variation for glucose tolerance in the HS falls between that of the founders, it is striking that insulin levels in response to a glucose challenge are extremely variable, with some values being five times higher than the insulin-resistant BUF. These data are not surprising in light of the fact that glucose homeostasis must be tightly regulated, whereas insulin varies as a means of keeping glucose under control. Our data demonstrate that a large proportion of the HS rats are insulin resistant. Of these insulin-resistant animals, only 25% also exhibit glucose intolerance. This is similar to what is seen in the human population in which not everyone with obesity or insulin resistance goes on to develop T2D (46, 59). There is also a smaller subset (4%) of HS animals that show decreased beta cell function, similar to the ACI founder strain. The large variation for both QUICKI and IGI suggests that these traits will be amenable to fine-mapping in the HS rat.
To investigate the utility of HS rats for fine-mapping these traits, we focused on a region on rat chromosome 1 that has previously been mapped for glucose tolerance and insulin using multiple F2 intercross and congenic strains (11, 13, 19–22, 34, 42, 53, 63, 64, 79). Although IGI did not map to this region, scores for body weight, fasting glucose, fasting insulin, Insulin_AUC, and QUICKI did. The most strongly associated region for the fasting glucose locus is only 1.52 Mb, located from 204.38–205.9 Mb, and containing 23 genes, none of which stand out as likely candidates. This single region is supported by the bootstrap resamples and falls within the larger QTL identified for fasting insulin and QUICKI. This locus also overlaps Gluco56, the region we previously identified for Glucose_AUC (64). Diplotype effect plots at this locus suggest that the BN haplotype tends to result in increased fasting glucose, whereas the ACI haplotype results in decreased fasting glucose. Of note, the ACI/WKY diplotype results in decreased fasting glucose at this locus, as well as decreased fasting insulin, decreased insulin sensitivity, and decreased Insulin_AUC at neighboring loci.
In contrast to fasting glucose, the peaks from bootstrap samples for Insulin_AUC, fasting insulin and QUICKI are scattered, with multiple peaks for each trait, consistent with a scenario in which multiple QTL within this region underlie these traits, most of which we are currently underpowered to detect. Despite multiple bootstraps falling outside of the identified QTL for Insulin_AUC, the bootstrap resamples support a single QTL at the peak marker interval. This region is only 0.95 Mb, from 202.35–203.30 Mb, and contains 30 genes. Our results suggest that the WKY/WKY diplotype leads to increased Insulin_AUC, whereas the ACI/WKY diplotype results in decreased Insulin_AUC at this locus. Within the larger 7.05 Mb CI for fasting insulin and QUICKI, we identified two separate bootstrap peaks: one at marker interval D1Rat291-D1Rat218 (200.17–201.13 Mb) and another at D1Mgh10-D1Rat168 (204.38–204.62 Mb), suggesting these are two separate loci. The diplotype substitution effect plots differ considerably at these two bootstrap peak marker intervals, also consistent with these two loci being independent. As an example, the ACI/WKY diplotype results in decreased fasting insulin and decreased insulin sensitivity at D1Mgh10-D1Rat168, with opposite effects at D1Rat291-D1Rat218.
There are >200 candidate genes within the 7.05 Mb region for fasting insulin and QUICKI. Although it may be premature at this time to speculate about likely causative genes within this region, Igf2 (insulin-like growth factor 2) and Kcnq1 (potassium voltage-gated channel, KQT-like subfamily, member 1) stand out as interesting potential candidates. Igf2, at 202.9 Mb, lies within the QTL for fasting insulin and QUICKI and is within the 0.95 Mb interval for Insulin_AUC. Igf2 is an imprinted gene involved in development and growth that plays a crucial role in development of the pancreas and beta cell mass (see Ref. 40). Igf1 has been associated with both fasting insulin and HOMA_IR in human GWAS (15, 26), and animal studies support a role for Igf2 in regulating beta cell mass and insulin levels at fasting and in response to glucose (8, 14). Another interesting candidate, Kcnq1, lies just outside the confidence intervals for Insulin_AUC and fasting glucose at 203.38–203.80 Mb. In human GWAS this gene has been associated with T2D (70, 81), fasting glucose (9, 68), and fasting insulin and HOMA_IR in response to a fast-acting insulin secretagogue (82). A role for Kcnq1 in both glucose and insulin phenotypes is also supported by a mouse knockout model (4), indicating this gene may be a candidate for regulating not only fasting insulin and QUICKI, but also potentially fasting glucose and Insulin_AUC in the HS rat. Because of the large number of genes and/or lack of clear biological candidates, however, additional strategies will be needed to identify the causative players within the identified QTL. One potential strategy will be to impute nongenotyped markers based on the strain distribution pattern of the founder strains, as previously described (32, 80). Identification of expression QTLs in this region can also be used to further prioritize candidate genes (i.e., Ref. 1).
To date, few of the genes identified in human GWAS play a role in insulin resistance, whereas many of the genes have been shown to influence insulin secretion (see Refs. 3, 18). In a review investigating the molecular mechanisms underlying 37 variants associated with T2D, 16 were found to play a role in insulin secretion or beta cell function, while only three were involved in insulin resistance (74). Heritability of insulin resistance is only slightly lower than for measures of beta cell function, which may suggest that the paucity of genes identified to date for insulin resistance results from beta cell function and insulin resistance having very different genetic architectures. For example, insulin resistance may be regulated by a larger number of genes with modest effect, more genes with rare variants, or genes with increased interactions with the environment relative to genes that regulate beta cell function (see Refs. 3, 28). In addition to differing genetic architectures, the lack of identified insulin-resistant genes may be because early human GWAS excluded obese patients, thus decreasing the likelihood that genes related to insulin resistance as a result of adiposity would be found (3). In support of this explanation, a recent study uncovered novel genes for insulin resistance by accounting for adiposity levels (43). Our study indicates that the HS rat colony may be a promising resource in which to identify at least some of these missing insulin-resistance genes, particularly those that act independently of body weight.
In addition to the region that encompasses QTL for fasting glucose, fasting insulin, Insulin_AUC, and QUICKI, we have also identified a separate QTL for body weight. Although the lod-drop confidence interval for this trait is only 1.82 Mb, the bootstrap resamples suggest the more likely region of this QTL is slightly larger, from 230.96 to 233.55 Mb or 2.59 Mb. At this locus, the F344/ACI diplotype leads to increased body weight and the ACI/M520 allele leads to decreased body weight. Within the more conservative 2.59 Mb region there are 32 genes. Jak2 (Janus kinase 2) is a tyrosine kinase that mediates leptin and insulin actions and stands out as a potential candidate gene as overeating and obesity results when the Jak2 pathways do not function properly (see Ref. 48). Further studies are needed to determine the gene in this region that is playing a causative role.
Mapping QTL in heterogeneous stocks presents both opportunities and challenges for the researcher. Greatly improved mapping resolution, over, say, F2 crosses, is bought at the cost of greater need for care in both the design and analysis of the experiment. These complications include the fact that the number of segregating alleles in a region is often greater than the number of marker alleles, that uneven relationships introduce effects of familial genetics that complicate QTL mapping and, depending on experimental design and subsequent modeling, may lead to more subtle confounding of genetics with environment, and that the comparatively richer genetic variation that helps detect QTL also makes subsequent characterization of their allelic series less clear cut. Although comprehensive methodology for analysis of HS remains a work in progress, we integrate a number of established and new techniques in the current study. Inference of ancestral haplotypes, and their use in linkage disequilibrium mapping, is well established for mapping in many multifounder populations, including HS rats (31, 64), HS mice (32, 49, 72), the mouse Collaborative Cross (12), and HS-like mouse Diversity Outbred (DO) population (67), as well as other populations including plants (39). Our modeling of family structure guards against much confounding but is less sophisticated than the fully polygenic models used in, e.g., Ref. 10, or those that use model selection over the whole genome (e.g., Ref. 71 and references therein). Nonetheless, in attempting to dissect allele effects at multifounder loci that exert dominance effects, we present a small methodological advance that will be immediately applicable to other HS and HS-like populations: a way to obtain stabilized estimates of diplotype effects through a ridge surrogate conditioning of the HAPPY diplotype probability matrix that, in contrast to, say, traditional ridge regression, is easily incorporated into existing regression modeling software.
In summary, we have demonstrated that the HS rat colony is useful for fine-mapping glucose and insulin traits, including a measure for insulin resistance, QUICKI. We have shown that the underlying mechanisms for glucose intolerance differ greatly between two founder strains of the HS rat colony, with the WKY strain exhibiting decreased insulin sensitivity and the ACI strain exhibiting decreased insulin secretion. Two additional founder strains, BUF and M520, were shown to exhibit decreased insulin sensitivity with normal glucose tolerance. Moreover, the HS rat colony itself exhibits great variation for all metabolic traits measured. Finally, we have fine-mapped QTL associated with fasting insulin and QUICKI to 7.05 Mb, and this region encompasses much smaller QTL for fasting glucose and Insulin_AUC. Body weight mapped to a separate region with a CI of <3 Mb. These studies demonstrate the promise of the HS rat colony as a platform for fine-mapping of genetic loci affecting insulin resistance and insulin secretion, and for providing information about genetic architecture of T2D that will complement that gained from human GWAS.
GRANTS
This work was funded by NIH Grants K01 DK-076977 and R01 DK-088975 and by the Individualized Medicine Institute at the MCW. W. Valdar and Y. Xie acknowledge partial support from the University of North Carolina Lineberger Comprehensive Cancer Center.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: L.C.S.W. conception and design of research; L.C.S.W. and K.L.H. performed experiments; L.C.S.W., K.L.H., D.O., Y.X., and W.V. analyzed data; L.C.S.W., D.O., and W.V. interpreted results of experiments; L.C.S.W., S.-W.T., and W.V. prepared figures; L.C.S.W. and W.V. drafted manuscript; L.C.S.W. and W.V. edited and revised manuscript; L.C.S.W., K.L.H., D.O., Y.X., S.-W.T., and W.V. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank John Radke and Alex Stoddard for programming assistance.
REFERENCES
- 1. Aylor DL, Valdar W, Foulds-Mathes W, Buus RJ, Verdugo RA, Baric RS, Ferris MT, Frelinger JA, Heise M, Frieman MB, Gralinski LE, Bell TA, Didion JD, Hua K, Nehrenberg DL, Powell CL, Steigerwalt J, Xie Y, Kelada SN, Collins FS, Yang IV, Schwartz DA, Branstetter LA, Chesler EJ, Miller DR, Spence J, Liu EY, McMillan L, Sarkar A, Wang J, Wang W, Zhang Q, Broman KW, Korstanje R, Durrant C, Mott R, Iraqi FA, Pomp D, Threadgill D, de Villena FP, Churchill GA. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res 21: 1213–1222, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bates D, Maechler M, Bolker B. lme4: linear mixed-effects models using S4 classes. R package version 0.999375–40, 2011, http://CRAN.R-project.org/package=lme4 [Google Scholar]
- 3. Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Ann NY Acad Sci 1212: 59–77, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boini KM, Graf D, Hennige AM, Koka S, Kempe DS, Wang K, Ackermann TF, Foller M, Vallon V, Pfeifer K, Schleicher E, Ullrich S, Haring HU, Haussinger D, Lang F. Enhanced insulin sensitivity of gene-targeted mice lacking functional KCNQ1. Am J Physiol Regul Integr Comp Physiol 296: R1695–R1701, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Box GEP, Cox DR. An analysis of transformations (with discussion). J Roy Stat Soc B26: 211–252, 1964 [Google Scholar]
- 6. Broman KW, Sen S. A Guide to QTL Mapping with R/qtl. New York: Springer, 2009 [Google Scholar]
- 7. Cacho J, Sevillano J, de Castro J, Herrera E, Ramos MP. Validation of simple indexes to assess insulin sensitivity during pregnancy in Wistar and Sprague-Dawley rats. Am J Physiol Endocrinol Metab 295: E1269–E1276, 2008 [DOI] [PubMed] [Google Scholar]
- 8. Calderari S, Gangnerau MN, Thibault M, Meile MJ, Kassis N, Alvarez C, Portha B, Serradas P. Defective IGF2 and IGF1R protein production in embryonic pancreas precedes beta cell mass anomaly in the Goto-Kakizaki rat model of type 2 diabetes. Diabetologia 50: 1463–1471, 2007 [DOI] [PubMed] [Google Scholar]
- 9. Chang YC, Chiu YF, Liu PH, Shih KC, Lin MW, Sheu WH, Quertermous T, Curb JD, Hsiung CA, Lee WJ, Lee PC, Chen YT, Chuang LM. Replication of genome-wide association signals of type 2 diabetes in Han Chinese in a prospective cohort. Clin Endocrinol (Oxf) 76: 365–372, 2012 [DOI] [PubMed] [Google Scholar]
- 10. Cheng R, Lim JE, Samocha KE, Sokoloff G, Abney M, Skol AD, Palmer AA. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics 185: 1033–1044, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chung WK, Zheng M, Chua M, Kershaw E, Power-Kehoe L, Tsuji M, Wu-Peng XS, Williams J, Chua SC, Jr, Leibel RL. Genetic modifiers of Leprfa associated with variability in insulin production and susceptibility to NIDDM. Genomics 41: 332–344, 1997 [DOI] [PubMed] [Google Scholar]
- 12. Collaborative Cross Consortium The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190: 389–401, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collins SC, Wallis RH, Wilder SP, Wallace KJ, Argoud K, Kaisaki PJ, Bihoreau MT, Gauguier D. Mapping diabetes QTL in an intercross derived from a congenic strain of the Brown Norway and Goto-Kakizaki rats. Mamm Genome 17: 538–547, 2006 [DOI] [PubMed] [Google Scholar]
- 14. Devedjian JC, George M, Casellas A, Pujol A, Visa J, Pelegrin M, Gros L, Bosch F. Transgenic mice overexpressing insulin-like growth factor-II in beta cells develop type 2 diabetes. J Clin Invest 105: 731–740, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Magi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparso T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proenca C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O'Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Bottcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jørgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martínez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orrù M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvänen AC, Tanaka T, Thorand B, Tichet J, Tönjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Ríos M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators, Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 42: 105–116, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dupuis J, Siegmund D. Statistical methods for mapping quantitative trait loci from a dense set of markers. Genetics 151: 373–386, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Durrant C, Mott R. Bayesian quantitative trait locus mapping using inferred haplotypes. Genetics 184: 839–852, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Florez JC. Newly identified loci highlight beta cell dysfunction as a key cause of type 2 diabetes: Where are the insulin resistance genes? Diabetologia 51: 1100–1110, 2008 [DOI] [PubMed] [Google Scholar]
- 19. Galli J, Fakhrai-Rad H, Kamel A, Marcus C, Norgren S, Luthman H. Pathophysiological and genetic characterization of the major diabetes locus in GK rats. Diabetes 48: 2463–2470, 1999 [DOI] [PubMed] [Google Scholar]
- 20. Galli J, Li LS, Glaser A, Ostenson CG, Jiao H, Fakhrai-Rad H, Jacob HJ, Lander ES, Luthman H. Genetic analysis of non-insulin dependent diabetes mellitus in the GK rat. Nat Genet 12: 31–37, 1996 [DOI] [PubMed] [Google Scholar]
- 21. Gauguier D, Froguel P, Parent V, Bernard C, Bihoreau MT, Portha B, James MR, Penicaud L, Lathrop M, Ktorza A. Chromosomal mapping of genetic loci associated with non-insulin dependent diabetes in the GK rat. Nat Genet 12: 38–43, 1996 [DOI] [PubMed] [Google Scholar]
- 22. Granhall C, Park HB, Fakhrai-Rad H, Luthman H. High-resolution quantitative trait locus analysis reveals multiple diabetes susceptibility loci mapped to intervals<800 kb in the species-conserved Niddm1i of the GK rat. Genetics 174: 1565–1572, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hansen C, Spuhler K. Development of the National Institutes of Health genetically heterogeneous rat stock. Alcohol Clin Exp Res 8: 477–479, 1984 [DOI] [PubMed] [Google Scholar]
- 24. Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction (2nd edition). New York: Springer, 2009 [Google Scholar]
- 25. Hossain P, Kawar B, El Nahas M. Obesity and diabetes in the developing world–a growing challenge. N Engl J Med 356: 213–215, 2007 [DOI] [PubMed] [Google Scholar]
- 26. Hu C, Zhang R, Wang C, Wang J, Ma X, Hou X, Lu J, Yu W, Jiang F, Bao Y, Xiang K, Jia W. Variants from GIPR, TCF7L2, DGKB, MADD, CRY2, GLIS3, PROX1, SLC30A8 and IGF1 are associated with glucose metabolism in the Chinese. PLoS One 5: e15542, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ikeda H, Shino A, Matsuo T, Iwatsuka H, Suzuoki Z. A new genetically obese-hyperglycemic rat (Wistar fatty). Diabetes 30: 1045–1050, 1981 [DOI] [PubMed] [Google Scholar]
- 28. Ingelsson E, Langenberg C, Hivert MF, Prokopenko I, Lyssenko V, Dupuis J, Magi R, Sharp S, Jackson AU, Assimes TL, Shrader P, Knowles JW, Zethelius B, Abbasi FA, Bergman RN, Bergmann A, Berne C, Boehnke M, Bonnycastle LL, Bornstein SR, Buchanan TA, Bumpstead SJ, Bottcher Y, Chines P, Collins FS, Cooper CC, Dennison EM, Erdos MR, Ferrannini E, Fox CS, Graessler J, Hao K, Isomaa B, Jameson KA, Kovacs P, Kuusisto J, Laakso M, Ladenvall C, Mohlke KL, Morken MA, Narisu N, Nathan DM, Pascoe L, Payne F, Petrie JR, Sayer AA, Schwarz PE, Scott LJ, Stringham HM, Stumvoll M, Swift AJ, Syvanen AC, Tuomi T, Tuomilehto J, Tonjes A, Valle TT, Williams GH, Lind L, Barroso I, Quertermous T, Walker M, Wareham NJ, Meigs JB, McCarthy MI, Groop L, Watanabe RM, Florez JC. Detailed physiologic characterization reveals diverse mechanisms for novel genetic Loci regulating glucose and insulin metabolism in humans. Diabetes 59: 1266–1275, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jensen DR, Ramirez DE. Anomalies in the foundations of ridge regression. Int Stat Rev 76: 89–105, 2008 [Google Scholar]
- 30. Jensen DR, Ramirez DE. Surrogate models in ill-conditioned systems. J Stat Plan Inf 140: 2069–2077, 2010 [Google Scholar]
- 31. Johannesson M, Lopez-Aumatell R, Stridh P, Diez M, Tuncel J, Blazquez G, Martinez-Membrives E, Canete T, Vicens-Costa E, Graham D, Copley RR, Hernandez-Pliego P, Beyeen AD, Ockinger J, Fernandez-Santamaria C, Gulko PS, Brenner M, Tobena A, Guitart-Masip M, Gimenez-Llort L, Dominiczak A, Holmdahl R, Gauguier D, Olsson T, Mott R, Valdar W, Redei EE, Fernandez-Teruel A, Flint J. A resource for the simultaneous high-resolution mapping of multiple quantitative trait loci in rats: the NIH heterogeneous stock. Genome Res 19: 150–158, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnsen AK, Valdar W, Golden L, Ortiz-Lopez A, Hitzemann R, Flint J, Mathis D, Benoist C. Genome-wide and species-wide dissection of the genetics of arthritis severity in heterogeneous stock mice. Arthritis Rheum 63: 2630–2640, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444: 840–846, 2006 [DOI] [PubMed] [Google Scholar]
- 34. Kanemoto N, Hishigaki H, Miyakita A, Oga K, Okuno S, Tsuji A, Takagi T, Takahashi E, Nakamura Y, Watanabe TK. Genetic dissection of “OLETF”, a rat model for non-insulin-dependent diabetes mellitus. Mamm Genome 9: 419–425, 1998 [DOI] [PubMed] [Google Scholar]
- 35. Katayama S, Inaba M, Maruno Y, Morita T, Awata T, Oka Y. Glucose intolerance in spontaneously hypertensive and Wistar-Kyoto rats: enhanced gene expression and synthesis of skeletal muscle glucose transporter 4. Hypertens Res 20: 279–286, 1997 [DOI] [PubMed] [Google Scholar]
- 36. Katz A, Nambi SS, Mather K, Baron AD, Follmann DA, Sullivan G, Quon MJ. Quantitative insulin sensitivity check index: a simple, accurate method for assessing insulin sensitivity in humans. J Clin Endocrinol Metab 85: 2402–2410, 2000 [DOI] [PubMed] [Google Scholar]
- 37. Kennedy BW, Quinton M, van Arendonk JA. Estimation of effects of single genes on quantitative traits. J Anim Sci 70: 2000–2012, 1992 [DOI] [PubMed] [Google Scholar]
- 38. Kooner JS, Saleheen D, Sim X, Sehmi J, Zhang W, Frossard P, Been LF, Chia KS, Dimas AS, Hassanali N, Jafar T, Jowett JB, Li X, Radha V, Rees SD, Takeuchi F, Young R, Aung T, Basit A, Chidambaram M, Das D, Grundberg E, Hedman AK, Hydrie ZI, Islam M, Khor CC, Kowlessur S, Kristensen MM, Liju S, Lim WY, Matthews DR, Liu J, Morris AP, Nica AC, Pinidiyapathirage JM, Prokopenko I, Rasheed A, Samuel M, Shah N, Shera AS, Small KS, Suo C, Wickremasinghe AR, Wong TY, Yang M, Zhang F, Abecasis GR, Barnett AH, Caulfield M, Deloukas P, Frayling TM, Froguel P, Kato N, Katulanda P, Kelly MA, Liang J, Mohan V, Sanghera DK, Scott J, Seielstad M, Zimmet PZ, Elliott P, Teo YY, McCarthy MI, Danesh J, Tai ES, Chambers JC. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet 43: 984–989, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet 5: e1000551, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kulkarni RN. New insights into the roles of insulin/IGF-I in the development and maintenance of beta-cell mass. Rev Endocr Metab Disord 6: 199–210, 2005 [DOI] [PubMed] [Google Scholar]
- 41. Lander ES, Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin JM, Ortsater H, Fakhrai-Rad H, Galli J, Luthman H, Bergsten P. Phenotyping of individual pancreatic islets locates genetic defects in stimulus secretion coupling to Niddm1i within the major diabetes locus in GK rats. Diabetes 50: 2737–2743, 2001 [DOI] [PubMed] [Google Scholar]
- 43. Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, Rybin D, Liu CT, Bielak LF, Prokopenko I, Amin N, Barnes D, Cadby G, Hottenga JJ, Ingelsson E, Jackson AU, Johnson T, Kanoni S, Ladenvall C, Lagou V, Lahti J, Lecoeur C, Liu Y, Martinez-Larrad MT, Montasser ME, Navarro P, Perry JR, Rasmussen-Torvik LJ, Salo P, Sattar N, Shungin D, Strawbridge RJ, Tanaka T, van Duijn CM, An P, de Andrade M, Andrews JS, Aspelund T, Atalay M, Aulchenko Y, Balkau B, Bandinelli S, Beckmann JS, Beilby JP, Bellis C, Bergman RN, Blangero J, Boban M, Boehnke M, Boerwinkle E, Bonnycastle LL, Boomsma DI, Borecki IB, Bottcher Y, Bouchard C, Brunner E, Budimir D, Campbell H, Carlson O, Chines PS, Clarke R, Collins FS, Corbaton-Anchuelo A, Couper D, de Faire U, Dedoussis GV, Deloukas P, Dimitriou M, Egan JM, Eiriksdottir G, Erdos MR, Eriksson JG, Eury E, Ferrucci L, Ford I, Forouhi NG, Fox CS, Franzosi MG, Franks PW, Frayling TM, Froguel P, Galan P, de Geus E, Gigante B, Glazer NL, Goel A, Groop L, Gudnason V, Hallmans G, Hamsten A, Hansson O, Harris TB, Hayward C, Heath S, Hercberg S, Hicks AA, Hingorani A, Hofman A, Hui J, Hung J, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 44: 659–660, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28: 412–419, 1985 [DOI] [PubMed] [Google Scholar]
- 45. McCulloch C, Neuhaus J, Searle SR. Generalized, Linear, and Mixed Models. Hoboken: Wiley, 2008 [Google Scholar]
- 46. Meigs JB, Wilson PW, Fox CS, Vasan RS, Nathan DM, Sullivan LM, D'Agostino RB. Body mass index, metabolic syndrome, and risk of type 2 diabetes or cardiovascular disease. J Clin Endocrinol Metab 91: 2906–2912, 2006 [DOI] [PubMed] [Google Scholar]
- 47. Moreno C, Kennedy K, Andrae JW, Jacob HJ. Genome-wide scanning with SSLPs in the rat. Methods Mol Med 108: 131–138, 2005 [DOI] [PubMed] [Google Scholar]
- 48. Morris DL, Rui L. Recent advances in understanding leptin signaling and leptin resistance. Am J Physiol Endocrinol Metab 297: E1247–E1259, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci USA 97: 12649–12654, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Muoio DM, Newgard CB. Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol 9: 193–205, 2008 [DOI] [PubMed] [Google Scholar]
- 51. Nguyen QM, Srinivasan SR, Xu JH, Chen W, Berenson GS. Fasting plasma glucose levels within the normoglycemic range in childhood as a predictor of prediabetes and type 2 diabetes in adulthood: the Bogalusa Heart Study. Arch Pediatr Adolesc Med 164: 124–128, 2010 [DOI] [PubMed] [Google Scholar]
- 52. Nichols GA, Hillier TA, Brown JB. Normal fasting plasma glucose and risk of type 2 diabetes diagnosis. Am J Med 121: 519–524, 2008 [DOI] [PubMed] [Google Scholar]
- 53. Nobrega MA, Woods LC, Fleming S, Jacob HJ. Distinct genetic regulation of progression of diabetes and renal disease in the Goto-Kakizaki rat. Physiol Genomics 39: 38–46, 2009 [DOI] [PubMed] [Google Scholar]
- 54. Parker CC, Sokoloff G, Cheng R, Palmer AA. Genome-wide association for fear conditioning in an advanced intercross mouse line. Behav Genet 42: 437–448, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Phillips DI, Clark PM, Hales CN, Osmond C. Understanding oral glucose tolerance: comparison of glucose or insulin measurements during the oral glucose tolerance test with specific measurements of insulin resistance and insulin secretion. Diabet Med 11: 286–292, 1994 [DOI] [PubMed] [Google Scholar]
- 56. Pinheiro J, Bates D. Mixed Effects Models in S and S-Plus. New York: Springer, 2000 [Google Scholar]
- 57. Polonsky KS, Sturis J, Bell GI. Seminars in Medicine of the Beth Israel Hospital, Boston. Non-insulin-dependent diabetes mellitus - a genetically programmed failure of the beta cell to compensate for insulin resistance. N Engl J Med 334: 777–783, 1996 [DOI] [PubMed] [Google Scholar]
- 58. R Development Core Team R: A Language and Environment for Statistical Computing. Vienna, Austria, 2012 [Google Scholar]
- 59. Reaven GM. Insulin resistance: the link between obesity and cardiovascular disease. Med Clin North Am 95: 875–892, 2011 [DOI] [PubMed] [Google Scholar]
- 60. Ronnegard L, Valdar W. Detecting major genetic loci controlling phenotypic variability in experimental crosses. Genetics 188: 435–447, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Samocha KE, Lim JE, Cheng R, Sokoloff G, Palmer AA. Fine mapping of QTL for prepulse inhibition in LG/J and SM/J mice using F(2) and advanced intercross lines. Genes Brain Behav 9: 759–767, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Saxena R, Elbers CC, Guo Y, Peter I, Gaunt TR, Mega JL, Lanktree MB, Tare A, Castillo BA, Li YR, Johnson T, Bruinenberg M, Gilbert-Diamond D, Rajagopalan R, Voight BF, Balasubramanyam A, Barnard J, Bauer F, Baumert J, Bhangale T, Bohm BO, Braund PS, Burton PR, Chandrupatla HR, Clarke R, Cooper-DeHoff RM, Crook ED, Davey-Smith G, Day IN, de Boer A, de Groot MC, Drenos F, Ferguson J, Fox CS, Furlong CE, Gibson Q, Gieger C, Gilhuijs-Pederson LA, Glessner JT, Goel A, Gong Y, Grant SF, Grobbee DE, Hastie C, Humphries SE, Kim CE, Kivimaki M, Kleber M, Meisinger C, Kumari M, Langaee TY, Lawlor DA, Li M, Lobmeyer MT, Maitland-van der Zee AH, Meijs MF, Molony CM, Morrow DA, Murugesan G, Musani SK, Nelson CP, Newhouse SJ, O'Connell JR, Padmanabhan S, Palmen J, Patel SR, Pepine CJ, Pettinger M, Price TS, Rafelt S, Ranchalis J, Rasheed A, Rosenthal E, Ruczinski I, Shah S, Shen H, Silbernagel G, Smith EN, Spijkerman AW, Stanton A, Steffes MW, Thorand B, Trip M, van der Harst P, van der AD, van Iperen EP, van Setten J, van Vliet-Ostaptchouk JV, Verweij N, Wolffenbuttel BH, Young T, Zafarmand MH, Zmuda JM, Boehnke M, Altshuler D, McCarthy M, Kao WH, Pankow JS, Cappola TP, Sever P, Poulter N, Caulfield M, Dominiczak A, Shields DC, Bhatt DL, Zhang L, Curtis SP, Danesh J, Casas JP, van der Schouw YT, Onland-Moret NC, Doevendans PA, Dorn GW, 2nd, Farrall M, FitzGerald GA, Hamsten A, Hegele R, Hingorani AD, Hofker MH, Huggins GS, Illig T, Jarvik GP, Johnson JA, Klungel OH, Knowler WC, Koenig W, März W, Meigs JB, Melander O, Munroe PB, Mitchell BD, Bielinski SJ, Rader DJ, Reilly MP, Rich SS, Rotter JI, Saleheen D, Samani NJ, Schadt EE, Shuldiner AR, Silverstein R, Kottke-Marchant K, Talmud PJ, Watkins H, Asselbergs FW, de Bakker PI, McCaffery J, Wijmenga C, Sabatine MS, Wilson JG, Reiner A, Bowden DW, Hakonarson H, Siscovick DS, Keating BJ. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet 90: 410–425, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Solberg Woods LC, Ahmadiyeh N, Baum A, Shimomura K, Li Q, Steiner DF, Turek FW, Takahashi JS, Churchill GA, Redei EE. Identification of genetic loci involved in diabetes using a rat model of depression. Mamm Genome 20: 486–497, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Solberg Woods LC, Holl K, Tschannen M, Valdar W. Fine-mapping a locus for glucose tolerance using heterogeneous stock rats. Physiol Genomics 41: 102–108, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stephenson AG. evd: extreme value distributions. RNews 2: 31–32, 2002 [Google Scholar]
- 66. Strawbridge RJ, Dupuis J, Prokopenko I, Barker A, Ahlqvist E, Rybin D, Petrie JR, Travers ME, Bouatia-Naji N, Dimas AS, Nica A, Wheeler E, Chen H, Voight BF, Taneera J, Kanoni S, Peden JF, Turrini F, Gustafsson S, Zabena C, Almgren P, Barker DJ, Barnes D, Dennison EM, Eriksson JG, Eriksson P, Eury E, Folkersen L, Fox CS, Frayling TM, Goel A, Gu HF, Horikoshi M, Isomaa B, Jackson AU, Jameson KA, Kajantie E, Kerr-Conte J, Kuulasmaa T, Kuusisto J, Loos RJ, Luan J, Makrilakis K, Manning AK, Martinez-Larrad MT, Narisu N, Nastase Mannila M, Ohrvik J, Osmond C, Pascoe L, Payne F, Sayer AA, Sennblad B, Silveira A, Stancakova A, Stirrups K, Swift AJ, Syvanen AC, Tuomi T, van 't Hooft FM, Walker M, Weedon MN, Xie W, Zethelius B, Ongen H, Malarstig A, Hopewell JC, Saleheen D, Chambers J, Parish S, Danesh J, Kooner J, Ostenson CG, Lind L, Cooper CC, Serrano-Rios M, Ferrannini E, Forsen TJ, Clarke R, Franzosi MG, Seedorf U, Watkins H, Froguel P, Johnson P, Deloukas P, Collins FS, Laakso M, Dermitzakis ET, Boehnke M, McCarthy MI, Wareham NJ, Groop L, Pattou F, Gloyn AL, Dedoussis GV, Lyssenko V, Meigs JB, Barroso I, Watanabe RM, Ingelsson E, et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes 60: 2624–2634, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, Chesler EJ, Palmer AA, McMillan L, Churchill GA. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics 190: 437–447, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tan JT, Nurbaya S, Gardner D, Ye S, Tai ES, Ng DP. Genetic variation in KCNQ1 associates with fasting glucose and beta-cell function: a study of 3,734 subjects comprising three ethnicities living in Singapore. Diabetes 58: 1445–1449, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tura A, Kautzky-Willer A, Pacini G. Insulinogenic indices from insulin and C-peptide: comparison of beta-cell function from OGTT and IVGTT. Diabetes Res Clin Pract 72: 298–301, 2006 [DOI] [PubMed] [Google Scholar]
- 70. Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G, Ng DP, Holmkvist J, Borch-Johnsen K, Jorgensen T, Sandbaek A, Lauritzen T, Hansen T, Nurbaya S, Tsunoda T, Kubo M, Babazono T, Hirose H, Hayashi M, Iwamoto Y, Kashiwagi A, Kaku K, Kawamori R, Tai ES, Pedersen O, Kamatani N, Kadowaki T, Kikkawa R, Nakamura Y, Maeda S. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 40: 1098–1102, 2008 [DOI] [PubMed] [Google Scholar]
- 71. Valdar W, Holmes CC, Mott R, Flint J. Mapping in structured populations by resample model averaging. Genetics 182: 1263–1277, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet 38: 879–887, 2006 [DOI] [PubMed] [Google Scholar]
- 73. Valdar W, Solberg LC, Gauguier D, Cookson WO, Rawlins NJ, Mott R, Flint J. Genetic and environmental effects on complex traits in mice. Genetics 174: 959–984, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. van de Bunt M, Gloyn AL. From genetic association to molecular mechanism. Curr Diab Rep 10: 452–466, 2010 [DOI] [PubMed] [Google Scholar]
- 75. van Den Brandt J, Kovacs P, Kloting I. Metabolic variability among disease-resistant inbred rat strains and in comparison with wild rats (Rattus norvegicus). Clin Exp Pharmacol Physiol 27: 793–795, 2000 [DOI] [PubMed] [Google Scholar]
- 76. Venables WN, Ripley BD. Modern Applied Statistics. New York: Springer, 2002 [Google Scholar]
- 77. Visscher PM, Thompson R, Haley CS. Confidence intervals in QTL mapping by bootstrapping. Genetics 143: 1013–1020, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, McCulloch LJ, Ferreira T, Grallert H, Amin N, Wu G, Willer CJ, Raychaudhuri S, McCarroll SA, Langenberg C, Hofmann OM, Dupuis J, Qi L, Segre AV, van Hoek M, Navarro P, Ardlie K, Balkau B, Benediktsson R, Bennett AJ, Blagieva R, Boerwinkle E, Bonnycastle LL, Bengtsson Bostrom K, Bravenboer B, Bumpstead S, Burtt NP, Charpentier G, Chines PS, Cornelis M, Couper DJ, Crawford G, Doney AS, Elliott KS, Elliott AL, Erdos MR, Fox CS, Franklin CS, Ganser M, Gieger C, Grarup N, Green T, Griffin S, Groves CJ, Guiducci C, Hadjadj S, Hassanali N, Herder C, Isomaa B, Jackson AU, Johnson PR, Jorgensen T, Kao WH, Klopp N, Kong A, Kraft P, Kuusisto J, Lauritzen T, Li M, Lieverse A, Lindgren CM, Lyssenko V, Marre M, Meitinger T, Midthjell K, Morken MA, Narisu N, Nilsson P, Owen KR, Payne F, Perry JR, Petersen AK, Platou C, Proenca C, Prokopenko I, Rathmann W, Rayner NW, Robertson NR, Rocheleau G, Roden M, Sampson MJ, Saxena R, Shields BM, Shrader P, Sigurdsson G, Sparso T, Strassburger K, Stringham HM, Sun Q, Swift AJ, Thorand B, Tichet J, Tuomi T, van Dam RM, van Haeften TW, van Herpt T, van Vliet-Ostaptchouk JV, Walters GB, Weedon MN, Wijmenga C, Witteman J, Bergman RN, Cauchi S, Collins FS, Gloyn AL, Gyllensten U, Hansen T, Hide WA, Hitman GA, Hofman A, Hunter DJ, Hveem K, Laakso M, Mohlke KL, Morris AD, Palmer CN, Pramstaller PP, Rudan I, Sijbrands E, Stein LD, Tuomilehto J, Uitterlinden A, Walker M, Wareham NJ, Watanabe RM, Abecasis GR, Boehm BO, Campbell H, Daly MJ, Hattersley AT, Hu FB, Meigs JB, Pankow JS, Pedersen O, Wichmann HE, Barroso I, Florez JC, Frayling TM, Groop L, Sladek R, Thorsteinsdottir U, Wilson JF, Illig T, Froguel P, van Duijn CM, Stefansson K, Altshuler D, Boehnke M, McCarthy MI; MAGIC investigators; GIANT Consortium Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 42: 579–589, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wei S, Wei K, Moralejo DH, Ogino T, Koike G, Jacob HJ, Sugiura K, Sasaki Y, Yamada T, Matsumoto K. Mapping and characterization of quantitative trait loci for non-insulin-dependent diabetes mellitus with an improved genetic map in the Otsuka Long-Evans Tokushima fatty rat. Mamm Genome 10: 249–258, 1999 [DOI] [PubMed] [Google Scholar]
- 80. Yalcin B, Flint J, Mott R. Using progenitor strain information to identify quantitative trait nucleotides in outbred mice. Genetics 171: 673–681, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H, Hirota Y, Mori H, Jonsson A, Sato Y, Yamagata K, Hinokio Y, Wang HY, Tanahashi T, Nakamura N, Oka Y, Iwasaki N, Iwamoto Y, Yamada Y, Seino Y, Maegawa H, Kashiwagi A, Takeda J, Maeda E, Shin HD, Cho YM, Park KS, Lee HK, Ng MC, Ma RC, So WY, Chan JC, Lyssenko V, Tuomi T, Nilsson P, Groop L, Kamatani N, Sekine A, Nakamura Y, Yamamoto K, Yoshida T, Tokunaga K, Itakura M, Makino H, Nanjo K, Kadowaki T, Kasuga M. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 40: 1092–1097, 2008 [DOI] [PubMed] [Google Scholar]
- 82. Yu W, Hu C, Zhang R, Wang C, Qin W, Lu J, Jiang F, Tang S, Bao Y, Xiang K, Jia W. Effects of KCNQ1 polymorphisms on the therapeutic efficacy of oral antidiabetic drugs in Chinese patients with type 2 diabetes. Clin Pharmacol Ther 89: 437–442, 2011 [DOI] [PubMed] [Google Scholar]