Abstract
In vertebrates, receptors of the rhodopsin G-protein coupled superfamily (GPCRs) play an important role in the regulation of feeding and energy homeostasis and are activated by peptide hormones produced in the brain-gut axis. These peptides regulate appetite and energy expenditure by promoting or inhibiting food intake. Sequence and function homologs of human GPCRs involved in feeding exist in the nematode roundworm, Caenorhabditis elegans (C. elegans), and the arthropod fruit fly, Drosophila melanogaster (D. melanogaster), suggesting that the mechanisms that regulate food intake emerged early and have been conserved during metazoan radiation. Nematodes and arthropods are the most diverse and successful animal phyla on Earth. They can survive in a vast diversity of environments and have acquired distinct life styles and feeding strategies. The aim of the present review is to investigate if this diversity has affected the evolution of invertebrate GPCRs. Homologs of the C. elegans and D. melanogaster rhodopsin receptors were characterized in the genome of other nematodes and arthropods and receptor evolution compared. With the exception of bombesin receptors (BBR) that are absent from nematodes, a similar gene complement was found. In arthropods, rhodopsin GPCR evolution is characterized by species-specific gene duplications and deletions and in nematodes by gene expansions in species with a free-living stage and gene deletions in representatives of obligate parasitic taxa. Based upon variation in GPCR gene number and potentially divergent functions within phyla we hypothesize that life style and feeding diversity practiced by nematodes and arthropods was one factor that contributed to rhodopsin GPCR gene evolution. Understanding how the regulation of food intake has evolved in invertebrates will contribute to the development of novel drugs to control nematodes and arthropods and the pests and diseases that use them as vectors.
Keywords: rhodopsin GPCR, feeding, conservation, evolution, invertebrates
Introduction
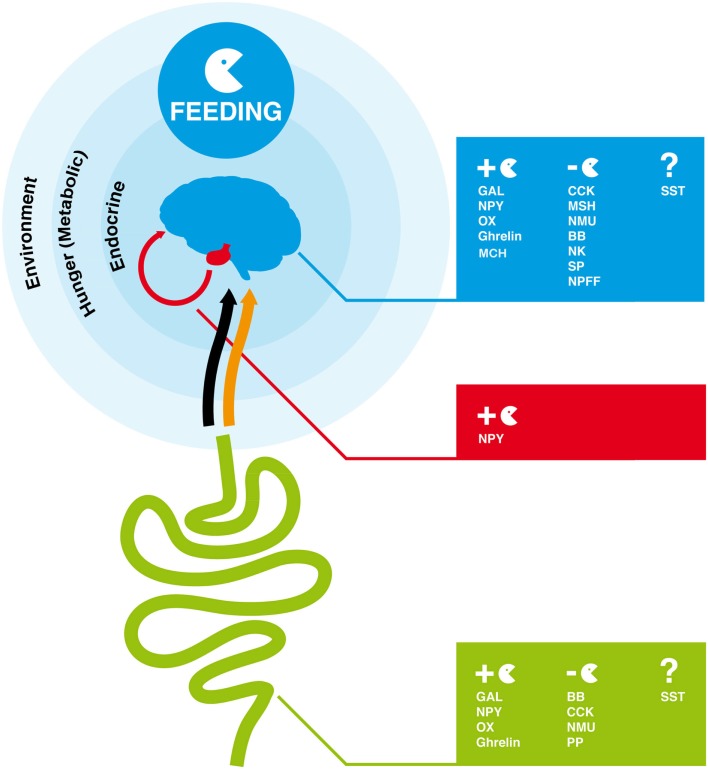
Feeding is the process by which food is obtained to provide energy. It must satisfy growth, survival, and reproductive requirements and has driven the evolution of specialized feeding behaviors and apparatus in metazoan. Regulation of feeding is a complex mechanism, which involves a combination of physical, chemical, and nutritional factors (Neary et al., 2004; Coll et al., 2007; Woods et al., 2008). Food-taking behavior is dependent on environmental signals (odors and taste), hunger signals (metabolic signals), and also endocrine satiety signals that via the blood stream or the vagal afferent terminals act on the hypothalamus, brain stem, or afferent autonomic nerves to modulate feeding response (Figure 1; Konturek et al., 2004; Stanley et al., 2005; Chaudhri et al., 2006; Woods et al., 2006, 2008). In mammals, psychological factors such as mood (emotions) and food reward have also been shown to affect eating behavior (Christensen, 1993; Berridge, 1996). In vertebrates, a group of small regulatory peptides that are produced by the brain-gut axis play a major role in the endocrine regulation of feeding and control of energy homeostasis (Figure 1; Coll et al., 2007; Chaudhri et al., 2008). These peptide hormones are divided into two groups, those that stimulate appetite (orexigenic peptides) and induce food intake and those that cause loss of appetite (anorexigenic peptides) and reduce food consumption and increase energy expenditure (Ahima and Osei, 2001; Wilding, 2002; Suzuki et al., 2010). The action of such peptides involves the activation of specific G-protein coupled receptors (GPCRs), which undergo conformational changes and promote the activation of intracellular signaling mechanisms that ultimately lead to a cellular response (Table 1; Marinissen and Gutkind, 2001; Xu et al., 2004; Fredriksson and Schioth, 2005).
Figure 1.
Overview of endocrine factors that regulate feeding behavior in the human brain-gut axis. In humans and other vertebrates, feeding is regulated by signals from the environment (odor and taste), hunger (metabolic signals), and endocrine signals produced by the gut and brain. The orange arrow represents the blood connection between gut and brain and the black arrow the nervous connection via the vagal afferent terminals through which peptides produced by the gut modulate the feeding response in the brain. GAL, NPY, OX, Ghrelin, and MCH are orexigenic peptides and promote appetite and feeding. CCK, MSH, NMU, BB, NK, SP, NPFF are anorexigenic. The role of SST peptides in feeding is unclear. The full peptide names are indicated in Table 1.
Table 1.
Rhodopsin GPCR family members and activating peptides that regulate food intake in mammals.
| Receptor | Subfamily | Members | Activating peptides | Effect on feed |
|---|---|---|---|---|
| α-Group | Melanocortin (MCR) | MC1R to 5R | Melanocortin peptides (MSH, ACTH, LPH) | Reduce |
| β-Group | Gastrin-cholecystokinin (CCKR) | CCK1R, 2R | Cholecystokinin (CCK), Gastrin | Reduce |
| Neurokinin (NKR) | NK1R to 3R | Substance P (SP), substance K (SK), neuromedin K (NK) | Reduce | |
| Neuropeptide FF (NPFFR) | NPFF1R, 2R | Neuropeptide FF (NPFF), neuropeptide AF (NPAF) | Reduce | |
| Orexin (OXR) | OX1R, 2R | Orexin-A and B (OXA, B) | Stimulate | |
| Neuropeptide Y (NPYR) | NPYRY1 to 6 | Neuropeptide Y (NPY), peptide YY (PYY), pancreatic polypeptide (PP) | Stimulate/reduce | |
| Bombesin (BBR) | BB1R to 3R | bombesin (BB), gastrin-releasing peptide (GRP), neuromedin C and B | Reduce | |
| Ghrelin/obestatin (GHSR/GPR39) | GHSR, GPR39 | Ghrelin (GHS), obestatin | Stimulate/reduce | |
| Neuromedin U (NMUR) | NMU1R, 2R | Neuromedin U (NMU) and S (NMS) | Reduce | |
| γ-Group | Somatostatin (SSTR) | SST1R to 5R | Somatostatin (SST) | Not clear |
| Galanin (GALR) | GAL1R to 3R | Galanin (GAL), galanin-like peptide (GALP) | Stimulate | |
| Melanin concentrating hormone (MCHR) | MCH1R, 2R | Melanin concentrating hormone (MHC) | Stimulate |
Receptor subfamily members, activating peptides and their effect on feed (stimulation or reduction) are indicated. For references please consult the text.
The involvement of GPCRs in the regulation of vertebrate feeding and appetite is well recognized (Shioda et al., 2008). Much less is known about their homologs and cognate activating peptides in non-vertebrates. However, comparative sequence approaches and functional studies suggest that the involvement of GPCRs in metazoa feeding behavior emerged early and has been maintained during the species radiation (Brody and Cravchik, 2000; Hewes and Taghert, 2001; Fredriksson and Schioth, 2005; Teng et al., 2008). GPCRs have emerged via gene or genome duplication events followed by selection of the gene duplicates. Understanding the origin of GPCRs represents a valuable tool for the characterization of basic physiological functions that have been maintained during evolution. The present review takes a comparative approach and targets rhodopsin GPCR subfamily members in the model species, C. elegans (a nematode) and D. melanogaster (a arthropod) that are sequence and function homologs of vertebrate GPCRs implicated in feeding regulation. To enrich the data and provide insight into how divergent life style and feeding strategies may have shaped receptor evolution in invertebrates the sequence of the target GPCRs were identified in other nematodes and arthropods with available genome data.
The Vertebrate GPCRs Superfamily and Their Role in Feeding
G-protein coupled receptors are one of the largest groups of receptors present in cells. Based upon their structure and sequence similarity five distinct superfamilies have been defined in human: glutamate (G), Rhodopsin (R), Adhesion (A), Frizzled (F), and Secretin (S) and are collectively known as GRAFS (Fredriksson and Schioth, 2005; Figure 2). GPCRs are characterized by a signature motif of seven conserved transmembrane spanning helix domains (TM) in vertebrates and non-vertebrates. Receptor activation is mediated by the extracellular N-terminal domain and also by TM and extracellular loops (receptor core domain) that interact with diverse types of molecules. The cellular response is provoked by the receptor C-terminal domain which activates a series of intracellular signaling cascades via the G-protein coupled pathway complex (Bockaert and Pin, 1999; Marinissen and Gutkind, 2001). Other molecular mechanisms such assembly of receptor heterodimers and allosteric receptor–receptor interactions in the cell membrane are also involved in GPCR regulation, activation and signaling (Prinster et al., 2005; Langmead and Christopoulos, 2006; Fuxe et al., 2012).
Figure 2.
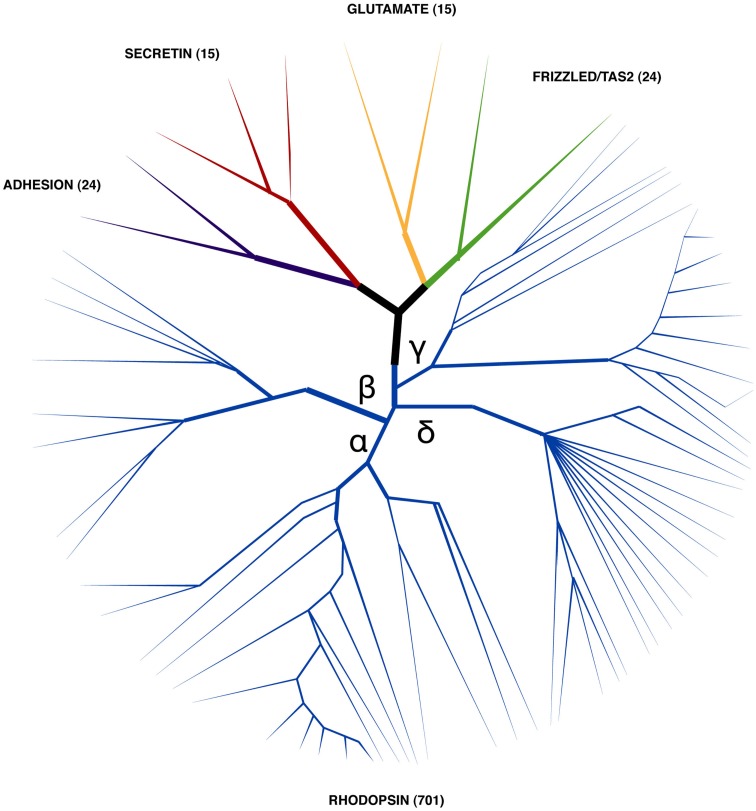
Stylized phylogenetic tree showing the relationship between human GRAFS. The number of human representatives identified within each superfamily is indicated within brackets (Fredriksson et al., 2003). Rhodopsin family members (which are represented by the blue branch) are the most numerous and their members are classified into four main sub-branches (α, β, γ, and δ). Human receptors, which are activated by peptides and have a role in feeding regulation, are members of the rhodopsin and secretin families.
G-protein coupled receptors are ubiquitous and involved in many different physiological functions. The glutamate receptors are involved in synaptic plasticity and participate in numerous functions in the central nervous system (CNS; Niswender and Conn, 2010). Rhodopsin receptors include receptors for hormones, neurotransmitters and photons and they are involved in taste, smell, and also regulate metabolism, reproduction, and neural function (Simoni et al., 1997; Murdoch and Finn, 2000; Gaillard et al., 2004; Waldhoer et al., 2004). Adhesion receptors participate in cell adhesion, signaling, and immune function (Bjarnadottir et al., 2007; Yona et al., 2008). Frizzled receptors are involved in the Wnt signaling pathway and in the control of cell proliferation and embryogenesis (van Amerongen and Nusse, 2009; Schulte, 2010). In contrast to other GPCRs, secretin family members are only activated by peptide hormones and they are implicated in brain-gut functions, calcium homeostasis, and in the stress response (McDermott and Kidd, 1987; Harmar, 2001; Bale and Vale, 2004; Moody et al., 2011). Due to their conserved structure and presence in many phyla, GPCRs are suggested to have a common evolutionary origin and to have arisen via gene/genome duplication early in the species radiation (Krishnan et al., 2012). With the exception of the glutamate family members, they are proposed to share a common ancestor with the cAMP receptors of primitive eukaryote species (Nordstrom et al., 2011).
In humans, more than 700 GPCR genes are predicted and a large proportion are orphans with unknown function (Figure 2). The rhodopsin family (a.k.a family A or class 1 GPCRs) comprise the most diverse receptor group and in humans they account for more than 80% of GPCRs and include members that are involved in regulation of feeding (Joost and Methner, 2002; Fredriksson et al., 2003). Rhodopsin family members possess a short N-terminal domain and are characterized by the presence of several conserved amino acid motifs such as N-S-x-x-N-P-x-x-Y within TM7 and the DRY (D(E)-R-Y(F)) motif between TM3 and intracellular loop (IL) 2 (Schioth and Fredriksson, 2005; Suwa et al., 2011). Based upon sequence similarity the human rhodopsin receptors are sub classified into four main groups (α, β, γ and δ; Figure 2; Fredriksson et al., 2003). The α-group contains clusters for the prostaglandin, amine, opsin, melatonin, melanocortin, endothelial, cannabinoid, and adenosine binding receptors. Members of the β-group include a subfamily of receptors for which known ligands are peptides such as orexin (OX), neuropeptide FF (NPFF), neurokinin (NK), gastrin-cholecystokinin (CCK), neuropeptide Y (NPY), endothelin-related (EDN), bombesin and related peptides (BB), neurotensin (NTS), ghrelin and obstatin, neuromedin (NMU), thyrotropin releasing hormone (TRH), arginine vasopressin (AVP), gonadotropin-releasing hormone (GNRH), and oxytocin (OXT). The γ group includes receptors for somatostatin (SST), opioids, galanin (GAL), melanin concentrating hormone (MCH), and chemokine peptides. The δ group contains the olfactory receptors (highly diverse > 400 members) as well as the glycoprotein, purine, and the MAS-related receptor clusters. In humans, twelve members of the rhodopsin family, which are activated by peptide hormones, play an important role in feed intake and stimulate or reduce food consumption (Table 1). The majority of these receptors are β group members and their role in the regulation of feed intake in mammals will now be briefly considered.
Receptors for melatonin (MT), gastrin-cholecystokinin (CCK), neurokinin (NK), neuropeptide FF (NPFF), bombesin and related peptides (BB), and neuromedin (NMU) have an inhibitory role in feed intake in vertebrates. Melanocortin receptors (MCR) are activated by melanocortin (ACTH, MSH, and lipotropin) peptides and administration of receptor agonists significantly reduces food consumption in rats (Irani and Haskell-Luevano, 2005). In addition mutant MC3R mice have increased fat mass (Coll et al., 2007) and ablation of the MC4R gene results in severe obesity (Coll et al., 2004; Millington, 2007). In rats, mutations of CCK1R are associated with obesity (Kopin et al., 1999) and peripheral administration of an NK1R antagonist leads to reduced weight gain after a high-fat diet (Karagiannides et al., 2011). Injection of NPFF provokes anorexia in mice and induces satiety (Murase et al., 1996; Bechtold and Luckman, 2006, 2007; Cline et al., 2009). BB peptides also mediate satiety (Hampton et al., 1998; Yamada et al., 2002; Gonzalez et al., 2008) and knockout BB2R mice have increased body weight (Ladenheim et al., 2002) and BB3R-deficient mice exhibit a mild obesity phenotype and increased food intake (Ohki-Hamazaki et al., 1997). Mice lacking the NMU gene are hyperphagic and have increased adiposity and obesity and amino acid variants in NMU are associated with human obesity (Brighton et al., 2004; Hainerova et al., 2006).
In contrast, orexin (OXs), neuropeptide Y (NPYs), galanin (GAL), and melanin concentrating hormone (MCH) receptors are activated by orexigenic peptides which stimulate feeding (Sakurai, 1999; Branchek et al., 2000; Chamorro et al., 2002; Lecklin et al., 2002; Lang et al., 2007; Wong et al., 2011). Administration of orexin-A and B stimulates food consumption in a dose-dependent manner (Sakurai et al., 1998; Matteri, 2001). NPY is one of the most potent orexigenic factors and NPY-induced feeding is markedly reduced in Y1-knockout mice and NPY Y1 receptor deficient mice lack appetite (Mercer et al., 2011; Pjetri et al., 2012). GAL1R-KO mice display increased food intake and body weight gain in response to an acute 3 day high-fat challenge (Zorrilla et al., 2007). MCH is a hypothalamic appetite-stimulating peptide that is high in obese mice (Kawauchi, 2006; Coll et al., 2007) and deletions in MCH1R confer resistance to diet-induced obesity (DIO) and MCH1R antagonists are effective in reducing body weight (Chung et al., 2011).
The role of SSTR and their activating peptides in vertebrates is unclear. In rats SSTR can stimulate or inhibit appetite although peptide injections in chickens have an orexigenic effect (Tachibana et al., 2009). In addition receptors for ghrelin-obestatin have opposing effects on feeding and ghrelin is associated with hunger scores and plasma ghrelin levels increase during fasting and decrease after food intake (Rocha-Sousa et al., 2010). Treatment of rats with obestatin suppresses food intake and decreases body weight gain (Zhang et al., 2005).
Other GPCR families activated by peptide hormones may also play a role in food intake and include members of the secretin receptor family: pituitary Adenylate-Cyclase Activating Peptide/Vasoactive Intestinal Peptide (PACR/VIPR; Morley et al., 1992; Chance et al., 1995); Glucagon and related peptide (GCGR/GLPR; McMahon and Wellman, 1997, 1998; Tang-Christensen et al., 2001; Woods et al., 2006); Calcitonin (CTR; Riediger et al., 2004) and Corticotrophin Releasing Factor (CRFR) receptors (Heinrichs and Richard, 1999; Bradbury et al., 2000; Richard et al., 2002). However, the secretin receptor family will not be considered in the present review.
The Invertebrate GPCRs Superfamily
Invertebrates are one of the most diverse animal groups and they represent more than 95% of the species on Earth. Protostomia comprise the majority of the species identified and are of both ecological and economic importance as they are involved in the nutrient cycle, plant fertilization, and include agricultural pests and vectors of human disease, such as malaria and sleeping sickness. The divergence of Protostomes from Deuterostomes occurred more than 700 million years ago (MYA) and their success is associated with adaptations to a variety of ecological niches and modifications in their feeding habits that allow them to live, survive and reproduce in many different environments. Invertebrates can be herbivores (eating plant tissue, nectar, and pollen), carnivores (feeding on other invertebrates as well as larger animals), parasites (living on plant and animals), and detritus feeders (eating dead animal and plants). Surprisingly few studies exist about the regulation of feed intake in invertebrates, despite its importance for their success and this is also a neglected target for alternative control strategies. The genome of several invertebrates has been sequenced and in the metazoan Ensembl genome database (www.ensemblgenomes.org) 48 invertebrate genomes are available. Comparative molecular studies represent an invaluable mechanism to better understand invertebrate biology and to characterize endocrine factors associated with feeding.
Homologs of the vertebrate GPCR repertoire have been described in many invertebrates and representatives of the five distinct human GRAFS families are proposed to have emerged before the split of nematodes from the chordate lineage (Table 2; Fredriksson and Schioth, 2005). The model organisms, the nematode roundworm C. elegans and the fruit fly D. melanogaster are the most studied Prostostomes. Their genomes have been completely sequenced and are fully annotated and a vast range of functional resources exists and numerous GPCRs have been characterized (Consortium, 1998; Adams et al., 2000; Keating et al., 2003). In the roundworm, GPCRs account for approximately 5% of the genome (there are more than 1000) and the chemoreceptor genes, which are involved in chemoreception of environmental stimuli are unique in nematodes and are also the most abundant and diverse (Schioth and Fredriksson, 2005; Robertson and Thomas, 2006; Nagarathnam et al., 2012). In the fruit fly, approximately 200 GPCRs (1% of the genome) are predicted and the gustatory/taste receptors (Montell, 2009) are specific to insects although a quarter share sequence homology with vertebrate neurohormone receptors (Keating et al., 2003; Fredriksson and Schioth, 2005; Hauser et al., 2006; Nagarathnam et al., 2012). Recently GPCRs were also characterized in the genome of two Platyhelminthes, the blood fluke Schistosoma mansoni and the planarian Schmidtea mediterranea and a similar gene repertoire to vertebrates has been characterized. A platyhelminth-specific rhodopsin subfamily (PROF1) and a planarian-specific Adhesion-like family (PARF1) have been identified suggesting lineage specific GPCRs evolved in invertebrates (Suwa et al., 2011; Zamanian et al., 2011).
Table 2.
Gene number and receptor subfamilies of the human rhodopsin GPCRs involved in feeding and the sequence homologs identified in C. elegans and D. melanogaster.
| Rhodopsin | Subfamily | Human | C. elegans | D. melanogaster |
|---|---|---|---|---|
| α-Group | Melanocortin | 5 | ni | ni |
| β-Group | Gastrin-cholecystokinin | 2 | 2 | 2 |
| Neurokinin | 3 | 6 | 5 | |
| Neuropeptide FF | 2 | |||
| Orexin | 2 | |||
| Neuropeptide Y | 6 | 12 | 4 | |
| Bombesin | 3 | ni | 2 | |
| Ghrelin/obestatin | 2 | 6 | 5 | |
| Neuromedin U | 2 | |||
| γ-Group | Somatostatin | 5 | 6 | 2 |
| Galanin | 3 | 3 | 2 | |
| Melanin concentrating hormone | 2 | ni | ni | |
| Total | 37 | 35 | 22 |
The total number of receptor genes in human, C. elegans and D. melanogaster is indicated. In C. elegans and D. melanogaster the homologs of the human Neurokinin/Neuropeptide FF/Orexin receptors and Ghrelin-Obestatin/Neuromedin U receptors were grouped due to their high sequence relatedness (Hewes and Taghert, 2001). ni, not identified.
Comparison of the neuroendocrine GPCR complement in the fruit fly and the honey bee Apis mellifera (A. mellifera) revealed that a similar gene complement is present (Hauser et al., 2006). In the malaria vector, the mosquito Anopheles gambiae (A. gambiae) genome, a total of 276 GPCRs are predicted and approximately 30 correspond to putative neuropeptide receptors (Hill et al., 2002). With the exception of C. elegans, very little is known about GPCRs in other nematodes despite availability of molecular data in public databases. The activating molecules for the roundworm and fruit fly GPCRs in common with other organisms are in general neurohormones (biogenic amines, protein hormones, and neuropeptides) and they play a central role in the control of behavior, reproduction, development, feeding, and many other physiological processes. This suggests that GPCR signaling has been conserved during evolution and that neuropeptide signaling plays a key role in both Proto and Deuterostomes (Grimmelikhuijzen and Hauser, 2012).
The present review provides a general overview of the evolution of the rhodopsin GPCR members that are implicated in feeding regulation. It will start by identifying and describing sequence homologs of human rhodopsin GPCRs in the model invertebrate organisms C. elegans and D. melanogaster followed by the characterization of their homologs in other nematodes and arthropods with distinctive feeding habits and life styles (Table 4). The C. elegans and D. melanogaster rhodopsin GPCR repertoire was obtained from published data and to enrich and confirm the dataset it was complemented with appropriate database searches using the human homologs (Table 3). A total of 35 rhodopsin GPCRs are present in C. elegans and 22 in D. melanogaster genomes (Table 2) and a conserved role in feeding regulation has been demonstrated.
Table 4.
Nematodes and arthropods used to analyze the rhodopsin GPCRs.
| Life style | Feeding type | Databases | |
|---|---|---|---|
| NEMATODES | |||
| Caenorhabditis elegans | Free-living | Bacteria | http://metazoa.ensembl.org |
| http://www.wormbase.org | |||
| Caenorhabditis briggsae | Free-living | Bacteria | http://metazoa.ensembl.org |
| Caenorhabditis japonica | Free-living | Bacteria, dead eggs and adult bugs | http://metazoa.ensembl.org |
| Pristionchus pacificus (necromenic nematode) | Parasitic | Bacteria, fungi and other nematodes | http://metazoa.ensembl.org |
| Haemonchus contortus (red stomach worm) | Parasitic | Bacteria, blood and tissue | http://www.sanger.ac.uk |
| Brugia malayi (filariasis worm) | Parasitic | Blood and lymphatic tissue | http://blast.ncbi.nlm.nih.gov |
| Trichinella spiralis (pork worm) | Parasitic | Mammalian cells and blood | http://metazoa.ensembl.org |
| Meloidogyne incognita (root-knot plant parasite) | Parasitic | Plant tissue | http://meloidogyne.toulouse.inra.fr |
| ARTHROPODS | |||
| Drosophila melanogaster (fruit fly) | Free-living | Yeast | http://metazoa.ensembl.org |
| http://www.flybase.org | |||
| Apis mellifera (honeybee) | Free-living | Nectar and pollen | http://metazoa.ensembl.org |
| Bombyx mori (silkworm) | Free-living | Plant leafs | http://metazoa.ensembl.org |
| Aedes aegypti (yellow fever mosquito) | Parasitic | Nectar and blood | http://metazoa.ensembl.org |
| Anopheles gambiae (malaria mosquito) | Parasitic | Nectar and blood | http://metazoa.ensembl.org |
| Ixodes scapularis (blacklegged tick) | Parasitic | Blood | http://metazoa.ensembl.org |
Information about life style, feeding type and the database interrogated is indicated.
Table 3.
The human C. elegans and D. melanogaster rhodopsin GPCRs used for comparative sequence analysis and their accession numbers.
| Human | C. elegans | D. melanogaster | |||
|---|---|---|---|---|---|
| Type | Accession number | Type | Accession number | Type | Accession number |
| BB1R | AAH95542.1 | ckr-1 | T23B3.4 | AlCR2 | CG13702 |
| BB2R | AAA88050.1 | ckr-2 | Y39A3B.5 | capaR | CG14575 |
| BB3R | AAT79496.1 | nmur-1 | C48C5.1 | CCHa1r | CG30106 |
| CCK1R | NP_000721.1 | nmur-2 | K10B4.4 | CCHa-2r | CG14593 |
| CCK2R | NP_795344.1 | nmur-3 | F02E8.2A | CCKL-R17D3 | CG32540 |
| GALR1 | NP_001471.2 | nmur-4 | C30F12.6 | CCKL-R17D1 | CG42301 |
| GALR2 | NP_003848.1 | npr-1 | C39E6.6 | DAR-1 | CG2872 |
| GALR3 | NP_003605.1 | npr-2 | T05A1.1A | DAR-2 | CG10001 |
| GHSR | AAI13548.1 | npr-3 | C10C6.2 | DTKR | CG7887 |
| GPR39 | AAC26082.1 | npr-4 | C16D6.2 | LKR | CG10626 |
| MC1R | AAD41355.1 | npr-5 | Y58G8A.4 | NepYr | CG5811 |
| MC2R | NP_000520.1 | npr-6 | F41E7.3 | NPFR1 | CG1147 |
| MCHR1 | NP_005288.3 | npr-7 | F35G8.1 | NKD | CG6515 |
| NK1R | AAR23925.1 | npr-8 | C56G3.1B | PK-1R | CG9918 |
| NK2R | AAH96842.1 | npr-9 | ZK455.3 | PK-2-R2 | CG8795 |
| NK3R | AAR23926.1 | npr-10 | C53C7.1A | PK-2-R1 | CG8784 |
| NMUR-1 | AAH51914.1 | npr-11 | C25G6.5 | SNPFR | CG7395 |
| NMUR-2 | EAW61653.1 | npr-12 | T22D1.12 | Star1-RA | CG7285 |
| NPFF1R | NP_071429.1 | npr-13 | ZC412.1 | CG10823 | CG10823 |
| NPFF2R | NP_004876.2 | npr-14 | W05B5.2 | CG30340 | CG30340 |
| NPY1R | AAA59947.1 | npr-15 | T27D1.3 | CG32547 | CG32547 |
| NPY2R | AAO92062.1 | npr-16 | F56B6.5 | CG34381 | CG34381 |
| NPY4R | NP_005963.3 | npr-17 | C06G4.5 | ||
| NPY5R | NP_006165.1 | npr-18 | C43C3.2 | ||
| OX1R | AAC39601 | npr-20 | T07D4.1 | ||
| OX2R | AAC39602.1 | npr-21 | T23C6.5 | ||
| SSTR1 | AAP84349.1 | npr-22 | Y59H11AL.1 | ||
| SSTR2 | AAO92064.1 | npr-24 | R106.2 | ||
| SSTR3 | AAP84354.1 | tkr-1 | C38C10.1 | ||
| SSTR4 | AAS55648.1 | tkr-3 | AC7.1 | ||
| SSTR5 | EAW85687.1 | C49A9.7 | C49A9.7 | ||
| C50F7.1 | C50F7.1 | ||||
| T02E9.1 | T02E9.1 | ||||
| Y116A8B.5 | Y116A8B.5 | ||||
| Y54E2A.1 | Y54E2A.1 | ||||
Feeding in Nematodes and Arthropods
Feeding in invertebrates in common with other animals involves a complex combination of physical, chemical, and nutritional factors (Chapman and De Boer, 1995). Taste and smell are important for feeding behavior and provide the CNS with information on quality and quantity of food and feeding behavior occurs mainly in response to both nutrient and nutritional storage status. Once feeding has been initiated and food ingested, the alimentary canal, and its associated glands triturate, lubricate, store, digest, and absorb the food material and excrete and expel unwanted remains (Audsley and Weaver, 2009).
The Nematoda is a highly diverse, complex, and specialized group of metazoans, about 30,000 species are currently known and many are renowned parasites (15%) and have specialized life cycles that depend on their host to survive and reproduce. Their success is associated with a protective, impermeable cuticle and by the diversity of the pharynx and feeding mechanisms (Coghlan, 2005). The shape and presence or absence of teeth, lancets, stylets, or other structures in the mouth reflects their distinct feeding methods. The majority of nematodes are free-living and inhabit soil and water and feed on microorganisms (bacteria, fungi, algae) and organic debris. The parasites feed on animal and plant tissues and some on vertebrate blood.
The Arthropoda represents the most diverse animal phyla and comprises over 80% of the species identified and the Insecta class is the most specious with approximately 920,000 species. Four main classes of feeding habits are recognized: plant feeders, predators (feed on aphids and mites), scavengers (feeding on dead and decaying organic matter), and parasites (of other insects and vertebrates), some of which are hematophagous. Within each of these classes, various types of feeding can be found such as biting and chewing on leaves or animal tissue and sucking from plant or animal cells or tissues. Despite this unique ability to use almost any organic substrate, most insect species restrict themselves to a particular category of food (Posnien et al., 2010) and feed primarily on a fluid diet (Prakash and Steele, 2010). The variety of feeding habits in arthropods is the result of anatomical and physiological adaptations to distinct food sources (Chapman and De Boer, 1995). The alimentary canal is composed of specialized regions that vary according to feeding habit and life stage.
The organisms selected for analysis of rhodospin GPCRs potentially involved in invertebrate feeding are members of different nematode and arthropod lineages. The specific life style and feeding habits of the invertebrates included in the analysis are indicated in Table 4.
Homologs of the Vertebrate Rhodopsin Family GPCRs Implicated in Feeding and Appetite Regulation in Non-Vertebrates
The following section describes the evolution and function of rhodopsin family members in nematodes and arthropods. It will start with an overview of those described in C. elegans and D. melanogaster involved in or candidates for feed intake regulation (Tables 2 and 5). Expression data when available from wormbase and flybase is included to provide insight into receptor function. It is followed by a section in which receptor evolution in invertebrates is discussed including homologs from non-model nematode and arthropod species.
Table 5.
An overview of the amino acid sequence similarity of the main subfamilies of C. elegans and D. melanogaster rhodopsin GPCRs and their human homologs.
| Rhodopsin | Subfamily | Characterized with function assigned |
Novel members with an unknown role in feeding |
||
|---|---|---|---|---|---|
| C. elegans (%) | D. melanogaster (%) | C. elegans (%) | D. melanogaster (%) | ||
| α-Group | Gastrin-cholecystokinin | 32–36 | 33–37 | ||
| β-Group | Neurokinin/neuropeptide FF/Orexin | 38–43 | 27–45 | 30–38 | 27–33 |
| Neuropeptide Y | 32–40 | 12–36 | 29–41 | 12–14 | |
| Bombesin | 38–40 | ||||
| Neuromedin U | 35–41 | 29–41 | 30–36 | 21–25 | |
| γ-Group | Somatostatin | 35–41 | 27–35 | ||
| Galanin | 36–40 | 37–43 | |||
Percentage of sequence similarity was calculated in the GeneDoc program (http://www.nr.bsc.org/gfx/genedoc/). The maximum and minimum sequence similarity of receptor subgroups between invertebrate and human homologs is indicated.
In general, no putative melatonin peptide receptors (MCR) or melanin concentrating hormone receptor (MCHR) homologs have been described or were identified in the present study in any of the selected nematodes or arthropods (Figure 3). In addition, in nematodes no homolog of the vertebrate and fruit fly bombesin receptors seem to exist (Table 2). Duplicates of the human receptor genes were identified in the genomes of nearly all target species and phylogenetic analysis suggests specific gene duplication/deletions occurred within the nematode and arthropod lineages (Figure 3).
Figure 3.
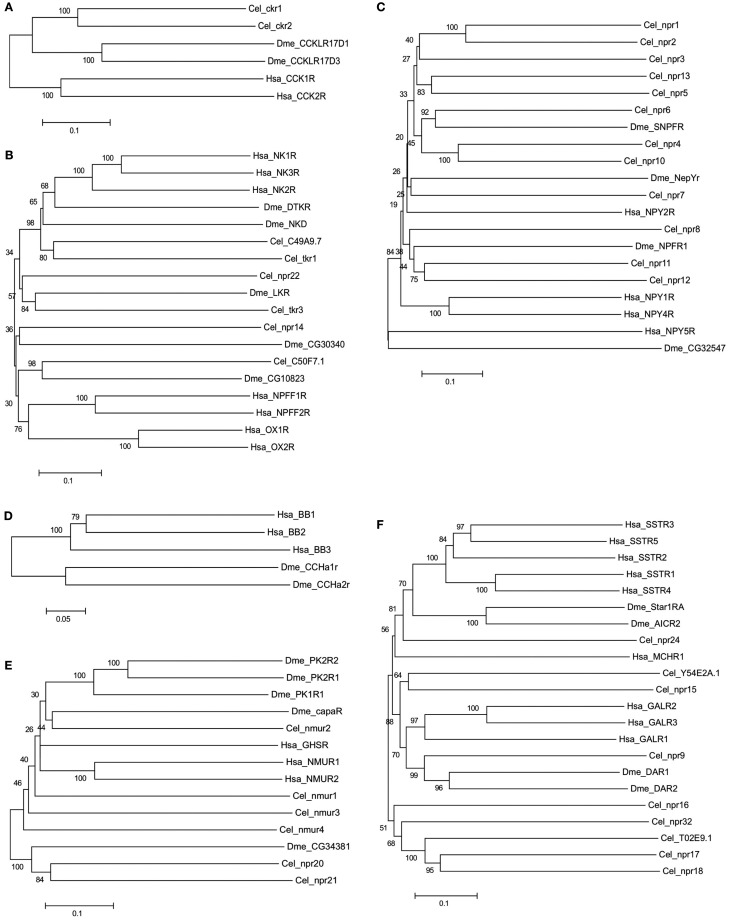
Phylogenetic relationship of the Human (Hsa) rhodopsin GPCRs involved in feeding with the nematode C. elegans (Cel) and arthropod D. melanogaster (Dme) sequence homologs. Trees were constructed using the neighbor joining method with 1000 bootstrap replicates (uniform rate among sites, pairwise deletion using the p-distance substitution model) built in the Mega5.1 program. Receptors were classified into six distinct subfamilies: (A) Gastrin-Cholecystokinin receptors; (B) Neurokinin/neuropeptide FF/orexin receptors, (C) Neuropeptide Y receptors, (D) Bombesin receptors, (E) Ghrelin/obstatin and Neuromedin U receptors, and (F) Somatostatin and galanin receptors. Accession numbers are described in Table 3.
The rhodopsin GPCRs in C. elegans and D. melanogaster genomes
Characterized and functionally assigned subfamily members
Gastrin-cholecystokinin receptor subfamily
In the genomes of C. elegans and D. melanogaster two putative Gastrin-CCK-like receptor homologs of the human members have been reported (Figure 3A; Keating et al., 2003; Janssen et al., 2008). In C. elegans, ckr-1, and ckr-2 have been described and functionally characterized. The ckr-1 is expressed in the nerve ring and functional RNAi knockdown studies reveal that loss of receptor activity provokes fat accumulation (McKay et al., 2007). However, if the receptors are ablated there is no apparent effect on feeding regulation but instead embryonic lethality and reduced brood size is observed (McKay et al., 2007). The neuropeptide nlp-12 is the ligand of nematode ckr-2 and the peptide receptor pair shares conserved biological activity with regards to fat storage with the human homolog (Janssen et al., 2008). A cognate peptide for nematode ckr-1 is yet to be identified.
In D. melanogaster the two existent CCK-like receptors were designated CCKL-R17D3 (DSKR1) and CCKL-R17D1 (Kubiak et al., 2002). They are mainly expressed in the CNS and are activated by Drosophila sulfakinin (DSK; Nichols et al., 1988), which is a structurally and functionally related peptide to the vertebrate CCK (Audsley and Weaver, 2009). Their role in feeding regulation has not yet been demonstrated in Drosophila but in other arthropods the homolog receptor stimulation by SK causes gut emptying and satiety (Nichols, 2007). Injections of SK peptides significantly reduce meal size in locusts (Schistocerca gregaria; Wei et al., 2000) and cockroach (Blattella germanica; Maestro et al., 2001), carbohydrate feeding in the blowfly (Phormia regina), and inhibit female horse flies from blood feeding (Downer et al., 2007).
Neurokinin/neuropeptide FF/orexin receptor subfamily
In C. elegans two putative neurokinin (a.k.a. tachykinins) receptors tkr-1 and tkr-3 have been described (Keating et al., 2003; Greenwood et al., 2005). In D. melanogaster three neurokinin-like receptors have been reported: the neurokinin receptor (NKD), the tachykinin receptor (DTKR; Li et al., 1991; Monnier et al., 1992; Rosay et al., 1995; Poels et al., 2009), and the leucokinin receptor (LKR; Radford et al., 2002). Phylogenetic analysis of the invertebrate receptors suggests that they arose from an ancestral Neurokinin/neuropeptide FF/orexin-like receptor gene by species-specific duplication events prior to the Proto-Deuterostome divergence (Figure 3B; Hewes and Taghert, 2001). Characterization of the C. elegans tkr-1 revealed expression is restricted to the socket cells (specialized nerve-accessory cells that act as an interface between the sensillum and hypodermis) and RNAi functional screens and the Nile Red fat assay revealed that this gene affects fat metabolism and fat droplet morphology and the pattern of fat deposition (Ashrafi et al., 2003). Knock down nematodes have a substantially lower fat content suggesting that this receptor is a key lipid storage regulator. Tkr-3 RNAi studies caused mild sluggishness and slowed locomotion in nematodes (Keating et al., 2003), which may be related to modifications in the nervous system. Tkr-3 is also present in the intestine but no role has yet been assigned in feeding and metabolism.
The D. melanogaster NKD and DTK receptors are expressed in the head of both larvae and adults and are activated by Drosophila tachykinin (DTK1–6) peptides, which are derived from the drosotachykinin (Dtk) gene (Birse et al., 2006; Poels et al., 2007) and also by substance P which is involved in the regulation of food intake and energy homeostasis in vertebrates (Birse et al., 2006; Poels et al., 2007). Knock down of DTKR in D. melanogaster modulated expression in both fed and starved flies of insulin-like peptides, which play a major role in the regulation of carbohydrates and lipid metabolism (Poels et al., 2009; Birse et al., 2011).
Neuropeptide Y receptor subfamily
In C. elegans four putative NPY-like receptors (npr-1, npr-2, npr-5, and npr-11) that share conserved sequence with the vertebrate NPYRs have been isolated and function characterized (de Bono and Bargmann, 1998; Keating et al., 2003; Kubiak et al., 2008; Cohen et al., 2009). Three NPY-like receptors have also been reported in D. melanogaster, these are the NepYr receptor and two neuropeptide F (NPF) receptors, the NPFR1 and the short NPFR (SNPFR; Figure 3C). The NPF peptide occurs as a long (NPF) and short (sNPF) isoform in arthropods (De Loof et al., 2001) and is the homolog of vertebrate neuropeptide Y (NPY; Li et al., 1992; de Jong-Brink et al., 2001).
In C. elegans, the nematode npr-1 was the first receptor found to influence social feeding behavior and is predominantly expressed in the nervous system (de Bono and Bargmann, 1998). This receptor is activated by flp-21 peptide (Rogers et al., 2003) and ablation of the peptide does not cause silencing of npr-1 functions, suggesting that it can be activated by other molecules. In fact, flp-18 peptide also activates npr-1 and this peptide is also the ligand of npr-5, which is involved, in chemosensory response, foraging behavior, and fat metabolism (Rogers et al., 2003). Nematode npr-5 is expressed in the head, neck, and body muscles and knock down and gene mutation studies revealed that in common with npr-2 it is associated with intestinal fat storage regulation (Keating et al., 2003; Cohen et al., 2009), dauer formation, and other food-dependent decisions (Cohen et al., 2009). The npr-11 has a role in reproduction and sensory dynamics of the olfactory system (Chalasani et al., 2010) but no role in feeding has yet been demonstrated (Chalasani et al., 2010).
The fruit fly NepYr and NPF receptors are expressed in the D. melanogaster CNS and NepYr is also present in the gut. NepYr is activated by dRYamide-1 and dRYamide-2, which has a C-terminal sequence similar to vertebrate NPY family peptides and in flies dRYamide suppresses feeding motivation (Ida et al., 2011). NPF and its receptors also modulate feeding behavior in D. melanogaster (Wu et al., 2003; Garczynski et al., 2005) and they promote feeding in larvae (Wu et al., 2003) and influence the effect of food deprivation in adult flies (Wu et al., 2003; Lingo et al., 2007). In other arthropods their functions have also been described and NPFR is involved in hindgut contraction in the bloodsucking bug (Rhodnius prolixus; Gonzalez and Orchard, 2009) and in ovarian maturation in locusts (Schoofs et al., 2001). In D. melanogaster sNPF is involved in the control of food intake and in the regulation of body size (Lee et al., 2004). Studies in mutant fruit flies over expressing sNPF peptide exhibit increased food intake and produce bigger and heavier flies, whereas sNPF loss-of-function mutants exhibit suppressed food intake (Lee et al., 2004). Gene expression studies with the red fire ant (Solenopsis invicta Buren) revealed SNPFR in brain is down-regulated during starvation (Chen and Pietrantonio, 2006) and expression of long NPF and its receptor in the malaria mosquito (A. gambiae) appear to be dependent on the insect nutritional status (Garczynski et al., 2005).
Bombesin receptor subfamily
Homologs of the vertebrate bombesin receptors have not been reported in nematodes and were not identified in the present study. Members of this family are only present in D. melanogaster and they correspond to the Allatostatin type B receptors (Stay, 2000). In D. melanogaster, two bombesin-like receptors have been isolated and function characterized: CCHamide-1r (CCHa1r; Johnson et al., 2003) and CCHamide-2r (CCHa-2r; Johnson et al., 2003; Hauser et al., 2008; Figure 3D).
In insects the function of the arthropod bombesin receptor is still poorly explored as a specific ligand has only recently been identified. CCHa-2r expression was detected in D. melanogaster brain and in the CNS and midgut of B. mori (Roller et al., 2008). Functional analysis reveals the receptors are activated by the peptides CCHamide-1 or CCHamide-2 that have been shown to suppress feeding activity in the cockroach, Blattella germanica (Audsley and Weaver, 2009).
Ghrelin-obestatin/neuromedin U receptor subfamily
In C. elegans four nmur-like receptors: nmur-1, nmur-2, nmur-3, and nmur-4 have been described. In D. melanogaster the capaR and three pyrokinin receptors PK-1R, PK-2-R1, and PK-2-R2 are the homologs of vertebrate NMURs (Iversen et al., 2002; Park et al., 2002; Figure 3E). The nematode nmur-1 is suggested to be involved in the sensory system and with processing information from specific food cues, which enables selection of different food types (Maier et al., 2010). C. elegans nmur-2 was also shown with its ligand peptide (derived from the nlp-44 precursor gene) to be involved in the regulation of food intake (Lindemans et al., 2009). To date no functional studies involving nmur-3 and nmur-4 have been reported although nmur-4 is expressed in the pharynx and intestine suggesting it may have a role in feeding.
The D. melanogaster capaR is mainly expressed in the Malpighian tubules and it is involved in the increase of fluid transport and diuresis and no direct role in feeding has yet been attributed (Terhzaz et al., 2012). CapaR is activated by two neuropeptides, capa-1 and -2 that are encoded by the capability gene and have antidiurectic actions in insects (Pollock et al., 2004; Coast and Garside, 2005; Paluzzi et al., 2010). The capability gene also encodes the pyrokinin-1 (PK1) peptide that is a specific activator of PK-1R. PK-2-R1 and PK-2-R2 are activated by pyrokinin-2 (PK2) and Hug-γ that are derived from the hugin (hug) prepropeptide (Cazzamali et al., 2005).
Phylogenetic analysis of the pyrokinin receptors suggests that they share common ancestry and that PK-2-R1 and R2 are the result of a recent duplication in the fly genome. The pyrokinin peptides are involved in rhythmic motor activity in arthropods (Saideman et al., 2007) and receptors are expressed in the abdomen (carcass) and nervous tissue and involvement in modulation of feeding behavior has been suggested. Overexpression of the hugin gene was found to suppress feeding in Drosophila, while blockage of the synaptic activity of hugin neurons caused the opposite effect (Meng et al., 2002; Melcher and Pankratz, 2005).
Somatostatin receptor subfamily
A homolog of human SSTR in the C. elegans genome was predicted in the 1990’s (Wilson et al., 1994). Characterization of the deduced protein revealed that the signature motif of the vertebrate SSTR was missing in TM7, suggesting that the receptor is probably activated by other ligands. Since no other homolog of vertebrate SSTR has been reported, the function of the putative SSTR-like receptors in nematodes remains to be explored. In arthropods, Allatostatin type-C receptors are the homologs of the vertebrate somastostatin receptors and in D. melanogaster, two receptors star1-RA and AlCR2 were described (Kreienkamp et al., 2002; Mayoral et al., 2010; Figure 3F).
The D. melanogaster star1-RA and AlCR2 receptors are detected in the CNS and they are activated by allatostatin-C peptides, which are potent modulators of hormone synthesis (Aguilar et al., 2003; Hergarden et al., 2012). These peptides inhibit or stimulate the corpora allata to synthesize juvenile hormone, which is an important regulator of development and reproduction in insects and may indirectly influence feeding behavior (Audsley and Weaver, 2009; Nassel and Winther, 2010).
Galanin receptor subfamily
In C. elegans and D. melanogaster a sequence and function homolog of vertebrate GALR has been described (Figure 3F). The C. elegans GALR-like receptor, npr-9 in common with the vertebrate homolog may be involved in food foraging and lipid storage (Bendena et al., 2008). The npr-9 is expressed in specific neurons around the posterior pharyngeal bulb and C. elegans receptor mutants are characterized by impaired food-related roaming behavior and accumulate intestinal fat as a result of fat ingestion and reduced energy expenditure (Lang et al., 2007; Bendena et al., 2008). Peptides involved in the activation of npr-9 have not been isolated, although nlp-5 and nlp-6, are candidate allatostatin-like peptides that in insects activate the GAL-like receptor (Nathoo et al., 2001).
In arthropods, the Allatostatin type-A receptors are homologs of the vertebrate GALRs (Birgul et al., 1999). Two receptors have been described in D. melanogaster, DAR-1 (a.k.a. AlstR) and DAR-2 (Birgul et al., 1999; Lenz et al., 2000; Figure 3F). AlstR is expressed in D. melanogaster head and CNS while DAR-2 is expressed in the gut suggesting they may have divergent functions. The receptors are activated by FGLamide neuropeptides (Pratt et al., 1991; Woodhead et al., 1994) that in arthropods inhibit food intake (Audsley and Weaver, 2009). Genetic epistasis assays in D. melanogaster indicate that FGLamide neuron activation inhibits or limits starvation-induced changes in feeding behavior (Hergarden et al., 2012).
Novel subfamily members with an unknown role in feeding regulation
Neurokinin/neuropeptide FF/orexin-like receptor subfamily
In C. elegans four additional NKRs members may exist: npr-14, npr-22 and the genes C49A9.7 and C50F7.1 (Keating et al., 2003). In D. melanogaster the SIFamide receptor and the gene CG10823 (Hewes and Taghert, 2001) also seem to be novel receptor members (Table 5, Figure 3B). In the phylogenetic tree, the C. elegans gene C49A9.7 clusters with tkr-1 suggesting they may be duplicates and the nematode npr-14 and C50F7.1 genes group with the fruit fly CG30340 and SIFamide receptor genes suggesting that they may have emerged from the same gene prior to the nematode-arthropod divergence. Functional studies of these receptors are scarce but those that exist indicate that the C. elegans MVRFamide neuropeptides but not tachykinin-like peptides activate the npr-22 receptor (Mertens et al., 2006). The function of D. melanogaster CG30340 gene, which is present in low abundance in the digestive and nervous system and of SIFamide receptors are unknown (Jorgensen et al., 2006).
Neuropeptide Y-like receptor subfamily
In C. elegans at least eight putative novel NPYR gene members are predicted: npr-3, npr-4, npr-6, npr-7, npr-8, npr-10, npr-12, and npr-13 and all remain to be validated and functionally characterized (Keating et al., 2003; Figure 3C). The receptors share between 30–40% amino acid sequence similarity with their human counterparts (Table 5) and are approximately 20% identical to the C. elegans homologs with a characterized function. The high sequence similarity and phylogenetic relationship between npr-5 and npr-13 (43%), npr-4 and npr-10 (50%) and npr-11 and npr-12 (44%) suggests that they may have arisen as a result of a recent duplication event in the nematode genome. These receptors are expressed in nervous tissue and intestine and their function is incompletely described and a specific role in feeding has not been demonstrated (Keating et al., 2003; Styer et al., 2008). In the D. melanogaster genome a putative novel insect NPY-like gene of unknown function (CG32547) may also exist (Hewes and Taghert, 2001) and seems to be expressed in the CNS (Figure 3C). The CG32547 gene shares less than 14% similarity with the human NPYR members (Table 5) and with the other insect family members, although this is probably due to its atypical size of 1008 amino acids, which makes family annotation ambiguous.
Ghrelin-obestatin/neuromedin U receptor subfamily
Two putative additional C. elegans nmur-like receptor genes the npr-20 and npr-21 were retrieved in the present study (Figure 3E). They share 30–36% amino acid sequence similarity with human homologs and are probably duplicates (Table 5). Expression of npr-21 in C. elegans occurs in nerves of the head, tail, and ventral nerve cord and also in the posterior intestine suggesting that it may have a role in brain-gut function associated with feeding regulation. Similarly in D. melanogaster a putative member of this family was also retrieved, the gene CG34381 (Table 5) and it clusters with nematode npr-20 and npr-21 suggesting that it may have shared common ancestry (Hewes and Taghert, 2001). Expression of the CG34381 gene occurred in the fruit fly head but so far no functional studies have been reported.
Somatostatin receptor subfamily
In the C. elegans genome at least eight putative SST-like receptor genes are predicted: npr-15, npr-16, npr-17, npr-18, npr-24, npr-32, and the Y54E2A.1 (Vashlishan et al., 2008) and T02E9.1 genes (Keating et al., 2003; Figure 3F). No additional putative SST-like receptors were identified or have been reported for D. melanogaster. Characterization of the nematode putative SST-like receptors revealed the C. elegans members share between 27–35% amino acid sequence similarity with the human SSTRs and that the npr-24 gene is the most closely related to the insect and human homologs suggesting that they may share a common ancestry (Table 5). Comparisons of the putative SSTR in C. elegans revealed they are highly divergent suggesting that after their emergence from an ancestral gene they underwent considerable change. Nematode npr-17 is most similar to npr-18 and to the T02E9.1 gene with which it shares 23% sequence identity and the three receptors tend to cluster with npr-16 and npr-32 suggesting they emerged in the nematode lineage.
The physiological role of the nematode SST-like receptors is poorly characterized but a role in metabolism and feeding behavior is probable. RNAi knockdown studies of npr-16, found to be expressed in head/tail neurons and the ventral nerve cord, increased fat deposition (Ashrafi et al., 2003). Ligand binding studies revealed that the peptide nlp-3 activates the receptor npr-17, which seems to be involved in food aversion and has a role in serotonergic modulation via ASH sensory neurons to modulate nematode behavior in response to an external stimuli (Harris et al., 2010). Deletion of the T02E9.1 gene resulted in an uncoordinated phenotype and nematodes moved slowly and with an increase in circular movement, although feeding was apparently unaffected (Keating et al., 2003). The function of npr-15, npr-18, npr-24, npr-32, and Y54E2A.1 remain to be explored.
Evolution of rhodopsin GPCR homologs in invertebrates
The evolution of the rhodopsin GPCRs in invertebrates was established (Figure 4) by identifying homologs in different nematode and arthropod lineages of the receptors present in C. elegans (Figure 5 and Table 6) and D. melanogaster (Figure 6 and Table 7). In general, the invertebrate GPCRs with a documented role in feeding or that are sequence homologs of mammalian seem to have evolved differently in nematodes and arthropods. A similar gene complement to that identified in C. elegans and D. melanogaster was identified in non-model nematodes and arthropods, respectively (Figure 4). Nematodes of the superfamily Rhabditoidea generally have more genes than other nematodes (Table 6). Gene duplicates in C. elegans and C. brigssae are more abundant than in arthropods (Lynch and Conery, 2000; Cutter et al., 2009) and a higher number of homologs of the human NPYRs and SSTRs occur in nematodes when compared to arthropods (Figures 5C and 6C). In arthropods, species-specific gene duplications exist rather than a conserved gene homolog complement suggesting that, despite their common ancestry, GPCRs have had distinct evolutionary trajectories in the different lineages (Table 7 and Figure 6).
Figure 4.
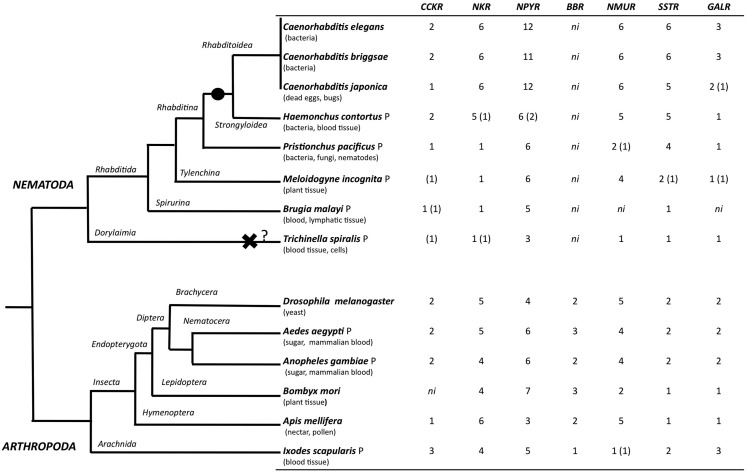
Distribution of rhodopsin subfamily members in nematodes and arthropods. The phylogenetic relationship of the species analyzed is represented on the right and their feeding habits are indicated. The black circle indicates a putative gene duplication event in the nematode radiation and the black cross potential gene deletion in the T. spiralis genome. Genes that were identified based upon sequence similarity but that were not considered for phylogenetic analysis are indicated within brackets “()”; ni- GPCR member not identified, and P represent parasitic nematode and arthropod. The evolutionary relationship within nematodes and arthropods was obtained from (Consortium, 2006; Sommer and Streit, 2011).
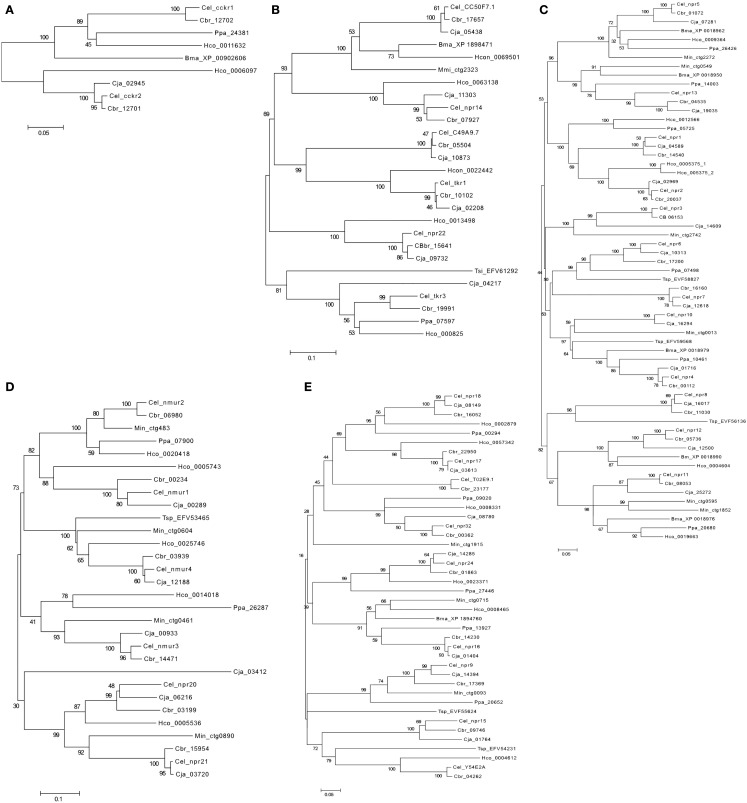
Figure 5.
Phylogenetic analysis of the nematode rhodopsin GPCRs. (A) Gastrin-cholecystokinin receptors; (B) Neurokinin/neuropeptide FF/orexin receptors, (C) Neuropeptide Y receptors, (D) Ghrelin-Obstatin/neuromedin U receptors, and (E) Somatostatin and galanin receptors. The C.elegans (Cel) receptors are annotated in bold. C. briggsae (Cbr), C. japonica (Cja), P. pacificus (Ppa) H. contortus (Hco) B. malayi (Bma), T. spiralis (Tsp), and M. incognita (Min). Accession numbers of the sequences used are indicated. Trees were constructed using the sequence alignment displayed in Figure S1 Supplementary Material using the methodology described in Figure 3.
Table 6.
Accession numbers of the C. elegans homologs in C. briggsae, C. japonica, P. pacificus, H. contortus, B. malayi, T. spiralis, and M. incognita.
| Receptor subfamily | C. elegans | C. briggsae | C. japonica | P. pacificus | H. contortus | B. malayi | T. spiralis | M. incognita |
|---|---|---|---|---|---|---|---|---|
| Gastrin-cholecystokinin | ckr-1 ckr-2 | CBG12702 CBG12701 | CJA02945 | PPA24381 | Supercontig0002945 | XP_001902606 | EFV58901* | MiV1ctg254* |
| Supercontig0006097 | XP_001895620* | |||||||
| Neurokinin/neuropeptide FF/orexin | tkr-1 | CBG10102 | CJA02208 | PPA07597 | Supercontig0022442 | XP_001898471 | EFV61292 | MiV1ctg2323 |
| tkr-3 | CBG19991 | CJA04217 | Supercontig000825 | EFV59206* | ||||
| npr-14 | CBG07927 | CJA11303 | Supercontig0069501 | |||||
| npr-22 | CBG15641 | CJA09732 | Supercontig0013498 | |||||
| C49A9.7 | CBG05504 | CJA10873 | Supercontig0000067* | |||||
| C50F7.1 | CBG17657 | CJA05438 | Supercontig0063138 | |||||
| Neuropeptide Y | npr-1 | CBG14540 | CJA04589 | PPA05725 | Supercontig0012566 | XP_001897991 | EFV58827 | MiV1ctg2742 |
| npr-2 | CBG20037 | CJA02969 | PPA10461 | SuperContig0005375 | XP_001895072 | EVF56136 | MiV1ctg13 | |
| npr-3 | CBG06153 | CJA14609 | PPA26426 | Supercontig0005375 | XP_001896282 | EVF59568 | MiV1ctg2272 | |
| npr-4 | CBG00112 | CJA01716 | PPA07498 | Supercontig0004842* | XP_001897675 | MiV1ctg595 | ||
| npr-5 | CBG01072 | CJA07281 | PPA20680 | Supercontig0009364 | XP_001899021 | MiV1ctg1852 | ||
| npr-6 | CBG17200 | CJA10313 | PPA14003 | Supercontig0019663 | MiV1ctg549 | |||
| npr.7 | CBG16160 | CJA12618 | Supercontig0004604 | |||||
| npr-8 | CBG11030 | CJA16017 | Supercontig0005938* | |||||
| npr-10 | CBG08053 | CJA16294 | ||||||
| npr-11 | CBG05736 | CJA25272 | ||||||
| npr-12 | CBG04535 | CJA12500 | ||||||
| npr-13 | CJA19035 | |||||||
| Ghrelin-obstatin/neuromedin U | nmur-1 | GBG00234 | CJA00289 | PPA17766* | Supercontig0005743 | ni | EFV53465 | MiV1ctg483 |
| nmur-2 | CBG06980 | CJA03412 | PPA07900 | Supercontig0020418 | MiV1ctg461 | |||
| nmur-3 | CBG14471 | CJA00933 | PPA26287 | Supercontig0014018 | MiV1ctg604 | |||
| nmur-4 | CBG03939 | CJA12188 | Supercontig0025746 | MiV1ctg890 | ||||
| npr-20 | CBG03199 | CJA06216 | Supercontig0005536 | |||||
| npr-21 | CBG15954 | CJA03720 | ||||||
| Somatostatin | npr-24 | CBG01863 | CJA14285 | PPA27446 | Supercontig0023371 | XP_001894760.1 | EFV55624 | MiV1ctg715 |
| npr-16 | CBG14230 | CJA01404 | PPA13927 | Supercontig0008465 | MiV1ctg1915 | |||
| npr-17 | CBG22950 | CJA03613 | PPA00294 | Supercontig0057342 | MiV1ctg1587* | |||
| npr-18 | CBG16052 | CJA08149 | PPA09020 | Supercontig0002879 | ||||
| npr-32 | CBG00362 | CJA08780 | Supercontig0008331 | |||||
| T02E9.1 | CBG23177 | |||||||
| Galanin | npr-9 | CBG17363 | CJA14394 | PPA20652 | Supercontig0004612 | ni | EFV54231 | MiV1ctg93 |
| npr-15 | CBG09746 | CJA01764 | MiV1ctg1567* | |||||
| Y54E2A.1 | CBG04262 | CJA18843* | ||||||
*Indicates sequences not used in the phylogenetic analysis due to poor sequence or non-identification of TM domains. ni, indicates gene not identified.
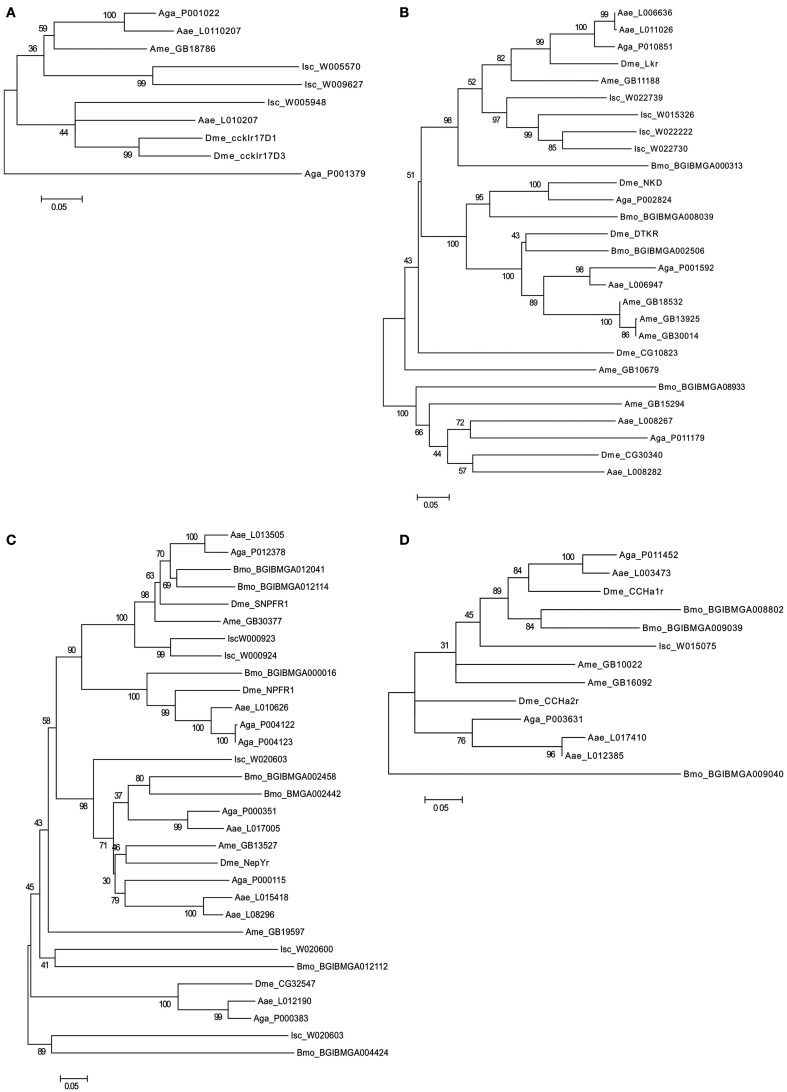
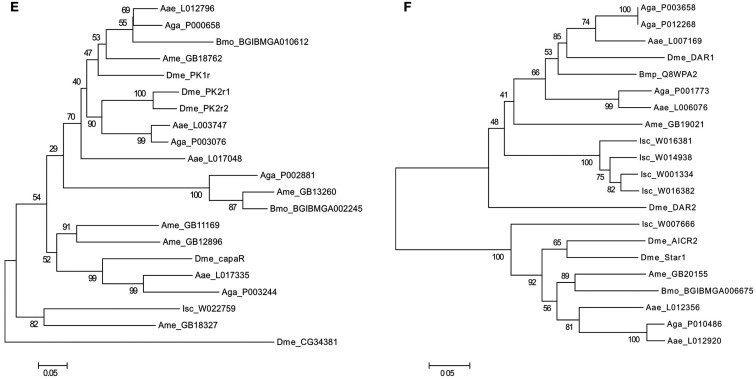
Figure 6.
Phylogenetic analysis of the arthropod rhodopsin GPCRs. (A) Gastrin-cholecystokinin receptors; (B) Neurokinin/neuropeptide FF/Orexin receptors, (C) Neuropeptide Y receptors, (D) Bombesin receptors, (E) Ghrelin-Obstatin/Neuromedin U receptors, and (F) Somatostatin and galanin receptors. The D. melanogaster (Dme) receptors are annotated in bold. A. gambiae (Aga), A. aegypti (Aae), A. mellifera (Ame), B. mori (Bmo), and I. scapularis (Isc). Accession numbers of the sequences used are indicated. Trees were constructed using the sequence alignment displayed in Figure S2 in Supplementary Material using a similar approach to that described in Figure 3.
Table 7.
Accession numbers of the D. melanogaster homologs in A. gambiae, A. aegypti, A. mellifera, B. mori and I. scapularis.
| Receptor subfamily | D. melanogaster | A. gambiae | A. aegypti | A. mellifera | B. mori | I. scapularis |
|---|---|---|---|---|---|---|
| Gastrin-cholecystokinin | CCKLR-17D3 | AGAP001022 | AAEL010207 | GB18786 | ni | ISCW005570 |
| CCKLR-17D1 | AGAP001379 | AAEL017238 | ISCW009627 | |||
| ISCW005948 | ||||||
| Neurokinin/neuropeptide FF/orexin | NKD | AGAP002824 | AAEL006947 | GB13925 | BGIBMGA008039 | ISCW022739 |
| DTKR | AGAP001592 | AAEL008267 | GB30014 | BGIBMGA002506 | ISCW015326 | |
| Lkr | AGAP011179 | AAEL008282 | GB18532 | BGIBMGA008933 | ISCW022222 | |
| CG30340 | AGAP010851 | AAEL011026 | GB15294 | BGIBMGA000313 | ISCW022730 | |
| CG10823 | AAEL006636 | GB11188 | ||||
| GB10679 | ||||||
| Neuropeptide Y | NepYr | AGAP000351 | AAEL017005 | GB13527 | BGIBMGA002442 | ISCW020603 |
| NPFR1 | AGAP000115 | AAEL008296 | GB19597 | BGIBMGA002458 | ISCW020600 | |
| SNPFR1 | AGAP000383 | AAEL015418 | GB30377 | BGIBMGA000016 | ISCW022779* | |
| CG32547 | AGAP004123 | AAEL012190 | BGIBMGA012112 | ISCW000923 | ||
| AGAP012378 | AAEL010626 | BGIBMGA012041 | ISCW000924 | |||
| AGAP004122 | AAEL007924 | BGIBMGA012114 | ||||
| AAEL013505 | ||||||
| Bombesin | CCHa1r | AGAP003631 | AAEL012385 | GB10022 | BGIBMGA008802 | ISCW015075 |
| CCHa-2r | AGAP011452 | AAEL017410 | GB16092 | BGIBMGA009039 | ||
| AAEL003473 | BGIBMGA009040 | |||||
| Ghrelin-obstatin/neuromedin U | capaR | AGAP003244 | AAEL017335 | GB11169 | BGIBMGA002245 | ISCW012018* |
| PK-1r | AGAP002881 | AAEL012796 | GB12896 | BGIBMGA010612 | ISCW022759 | |
| PK2r2 | AGAP000658 | AAEL003747 | GB18762 | |||
| PK2r1 | AGAP003076 | AAEL017048 | GB13260 | |||
| CG34381 | GB18327 | |||||
| Somatostatin | Star1-RA | AGAP010486 | AAEL012920 | GB20155 | BGIBMGA006675 | ISCW007666 |
| AlCR2 | AGAP012268 | AAEL012356 | ||||
| Galanin | DAR-1 | AGAP003658 | AAEL007169 | GB19021 | Q8WPA2 | ISCW001334 |
| DAR-2 | AGAP001773 | AAEL006076 | ISCW014938 | |||
| ISCW016381 | ||||||
| ISCW016382 |
*Indicates sequences not used in the phylogenetic analysis due to poor sequence or non-identification of TM domains. ni: indicates gene not identified.
A striking observation is the absence in nematodes of homologs of the arthropod bombesin receptors (BBR; Figure 4; Table 5). The reason for the loss of BBR in nematodes is unknown and their function and any link to feeding regulation remains to be established. In vertebrates, bombesin and its receptors are involved in smooth muscle contraction, exocrine, and endocrine secretion in the gut, pancreas, and pituitary and they also have a central role in food intake and energy homeostasis (Sano et al., 2004; Gonzalez et al., 2008). Three receptors have been isolated in humans and a similar number exist in arthropods and they share a common ancestry (Figure 6D).
A similar number of gastrin-CCK, NKR, NMUR, and GALR subfamily members were characterized in nematodes and arthropods (Figure 4). Two putative gastrin-CCK receptors were identified in invertebrates and in humans two gastrin-CCK receptors also exist suggesting that the evolution of the members of this family has been highly conserved. However, phylogenetic analysis suggests that the duplication, which delivered the two gene copies, was not common to all the species and occurred independently within each lineage. The two ckr that are present in nematodes resulted from a lineage specific duplication and homologs of the two C. elegans genes were identified in most nematode genomes analyzed (Figure 5A). In arthropods, a different situation exists and the two D. melanogaster genes are very similar and seem to have resulted from a species-specific duplication event (Figure 6A). Similarly in the blacklegged tick (I. scapularis) three putative gastrin-CCK receptors were also identified. In contrast, no putative homologs were identified in the plant feeding arthropod, the silkworm B. mori, even though they had a similar gene complement to other arthropods. It remains to be established if the absence of this receptor in B. mori is a consequence of its incomplete genome assembly (Xia et al., 2004) or represents an adaptation relative to feeding regulation.
Members of the NKR, NMUR, and GALR subfamilies have also evolved via lineage specific and species-specific duplication events. In nematodes, a similar number of NKR, NMUR, and GALR receptors exist in H. contortus and in the three representatives of the Caernohabitis genus analyzed (Figures 5B,D,E). In contrast, few genes of these families have been identified in other nematode taxa and a single NKR subfamily member was retrieved from P. pacificus, M. incognita, B. malayi, and T. spiralis. In arthropods, gene duplication of the D. melanogaster LKR receptor homologs was identified in the mosquito A. aegypti and also in I. scapularies in which four putative receptors exist (Figure 6B). In addition, in the honeybee (A. mellifera) three putative homologs of the fruit fly DTKR receptors were also identified. In contrast, no homologs of D. melanogaster NKD were detected in the honeybee and A. aegypti genomes. Within the NMUR family (Figure 6E), the D. melanogaster PK2Rs emerged as a consequence of a species-specific duplication event and two putative capaR were also identified in the honeybee, but only a single member was found in I. scapulars. In contrast, duplication of GALR occurred in the I. scapulars genome and four putative receptors were identified while other arthropods contained a single homolog of D. melanogaster DAR-1 and DAR-2 genes (Figure 6F).
The complete genome sequence of some of the species used in this study are not yet available, nonetheless gene representatives identified in the selected nematodes and arthropods provides a clear idea of the GPCR evolution in invertebrates. The majority of the C. elegans sequence homologs were identified in the target species and an increase in gene number seems to have occurred in Rhabditoidea and Strongyloidea (Abad et al., 2008; Dieterich et al., 2008; Mitreva et al., 2011). The exception was B. malayi in which representatives of NMUR and GALR were not identified possibly because of its incomplete genome assembly (Ghedin et al., 2007). The absence of the majority of the C. elegans receptor homologs in parasitic nematode genomes and the higher number of genes present in H. contortus and in other representatives of the Caernohabitis genus is curious. A general comparison of the gene content of T. spiralis with C. elegans revealed that the parasitic nematode genome contains fewer genes (15,808 compared to 20,060 and 19,507 in C. elegans and C. briggsae, respectively) and we hypothesize that gene absence is a consequence of the selective pressures provoked by the host on which they live and depend for survival (Mitreva et al., 2011; Sommer and Streit, 2011). The genome of P. pacificus is predicted to contain a higher gene number than C. elegans and suggests that a specific GPCR gene expansion occurred in the nematode lineage after their divergence (Dieterich et al., 2008; Sommer and Streit, 2011). Comparisons between T. spiralis and the other blood feeding parasitic nematode H. contortus revealed that the latter has a higher GPCR gene number than T. spiralis. One explanation may be related to their life cycles and while both nematodes need blood to survive T. spiralis is an obligate parasite, while H. contortus has a non-parasitic free-living stage. Intriguingly during the parasitic stage of H. contortus significant changes in the active transcriptome occurs when compared to the nematode free-living stage (Hoekstra et al., 2000) and it will be of interest to establish if this affects the diversity of rhodopsin GPCRs expressed.
In arthropods, GPCR gene evolution appears species dependent and specific gene duplications and deletions have occurred despite their common ancestry. The existence of specific gene duplicates in arthropods may indicate that a divergent regulatory system evolved in different species and the origin and maintenance of duplicates in the genome remain to be explored. Gene number in the two mosquito species analyzed are very similar and may reflect their identical life styles (Klowden, 1990). In the tick, which feeds exclusively on blood, a specific expansion of NKR and GALR gene families occurred. Further studies are required to determine the significance of the specific evolution of rhodopsin family GPCRs in arthropods and to consider how life style and feeding activity may have influenced receptor evolution.
Final Considerations
In general, the physiological processes involving GPCRs are conserved and sequence and function homologs of vertebrate rhodopsin GPCRs are present in invertebrates indicating they emerged early in evolution. In Nematoda and Arthropoda the rhodopsin GPCRs have evolved differently. Gene expansion is observed in nematodes with a free-living stage and specific gene deletions seem to have affected parasitic nematode genomes. In arthropods species-specific gene duplications occurred. We hypothesize that the evolving feeding regime and life style of invertebrates was one of the pressure forcing GPCR evolution and that this may explain some of the specific gene family expansions and deletions. Comparative studies of GPCRs gained or lost in the nematodes and arthropods and their relationship to feeding regulation may provide insights into how GPCRs contributed and shaped adaptation to new ecological niche. Studies of other nematodes and arthropods coupled with experiments to assign function and potential conserved role in feeding will be needed to test this hypothesis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Neuroendocrine_Science/10.3389/fendo.2012.00157/abstract
Sequence of the nematodes GPCR transmembrane (TM) domains from non-model nematodes within each receptor family were extracted by sequence homology using the roundworm C. elegans TM regions. To facilitate visualization the TM1, 3, 5, and 7 were annotated in gray.
Sequence of the Arthropod GPCR transmembrane (TM) domains used in for phylogenetic analysis. TM domains from non-model arthropods within each receptor family were extracted by sequence homology using the D. melanogaster TMs. To facilitate visualization the TM1, 3, 5 and 7 were annotated in gray.
Acknowledgments
This study was funded by the Portuguese Science Foundation PTDC/BIA-BCM/114395/2009 and CCMAR pluriannual grant. Vera G. Fonseca was supported by FCT grant SFRH/BPD/80447/2011.
References
- Abad P., Gouzy J., Aury J. M., Castagnone-Sereno P., Danchin E. G., Deleury E., et al. (2008). Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26, 909–915 10.1038/nbt.1482 [DOI] [PubMed] [Google Scholar]
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., Amanatides P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195 10.1126/science.287.5461.2185 [DOI] [PubMed] [Google Scholar]
- Aguilar R., Maestro J. L., Vilaplana L., Pascual N., Piulachs M. D., Belles X. (2003). Allatostatin gene expression in brain and midgut, and activity of synthetic allatostatins on feeding-related processes in the cockroach Blattella germanica. Regul. Pept. 115, 171–177 10.1016/S0167-0115(03)00165-4 [DOI] [PubMed] [Google Scholar]
- Ahima R. S., Osei S. Y. (2001). Molecular regulation of eating behavior: new insights and prospects for therapeutic strategies. Trends. Mol. Med. 7, 205–213 10.1016/S1471-4914(01)01989-X [DOI] [PubMed] [Google Scholar]
- Ashrafi K., Chang F. Y., Watts J. L., Fraser A. G., Kamath R. S., Ahringer J., et al. (2003). Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421, 268–272 10.1038/nature01279 [DOI] [PubMed] [Google Scholar]
- Audsley N., Weaver R. J. (2009). Neuropeptides associated with the regulation of feeding in insects. Gen. Comp. Endocrinol. 162, 93–104 10.1016/j.ygcen.2008.08.003 [DOI] [PubMed] [Google Scholar]
- Bale T. L., Vale W. W. (2004). CRF and CRF receptors: role in stress responsivity and other behaviors. Annu. Rev. Pharmacol. Toxicol. 44, 525–557 10.1146/annurev.pharmtox.44.101802.121410 [DOI] [PubMed] [Google Scholar]
- Bechtold D. A., Luckman S. M. (2006). Prolactin-releasing peptide mediates cholecystokinin-induced satiety in mice. Endocrinology 147, 4723–4729 10.1210/en.2006-0753 [DOI] [PubMed] [Google Scholar]
- Bechtold D. A., Luckman S. M. (2007). The role of RFamide peptides in feeding. J. Endocrinol. 192, 3–15 10.1677/JOE-06-0069 [DOI] [PubMed] [Google Scholar]
- Bendena W. G., Boudreau J. R., Papanicolaou T., Maltby M., Tobe S. S., Chin-Sang I. D. (2008). A Caenorhabditis elegans allatostatin/galanin-like receptor NPR-9 inhibits local search behavior in response to feeding cues. Proc. Natl. Acad. Sci. U.S.A. 105, 1339–1342 10.1073/pnas.0709492105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge K. C. (1996). Food reward: brain substrates of wanting and liking. Neurosci. Biobehav. Rev. 20, 1–25 10.1016/0149-7634(95)00033-B [DOI] [PubMed] [Google Scholar]
- Birgul N., Weise C., Kreienkamp H. J., Richter D. (1999). Reverse physiology in drosophila: identification of a novel allatostatin-like neuropeptide and its cognate receptor structurally related to the mammalian somatostatin/galanin/opioid receptor family. EMBO J. 18, 5892–5900 10.1093/emboj/18.21.5892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse R. T., Johnson E. C., Taghert P. H., Nassel D. R. (2006). Widely distributed Drosophila G-protein-coupled receptor (CG7887) is activated by endogenous tachykinin-related peptides. J. Neurobiol. 66, 33–46 10.1002/neu.20189 [DOI] [PubMed] [Google Scholar]
- Birse R. T., Soderberg J. A., Luo J., Winther A. M., Nassel D. R. (2011). Regulation of insulin-producing cells in the adult Drosophila brain via the tachykinin peptide receptor DTKR. J. Exp. Biol. 214, 4201–4208 10.1242/jeb.062091 [DOI] [PubMed] [Google Scholar]
- Bjarnadottir T. K., Fredriksson R., Schioth H. B. (2007). The adhesion GPCRs: a unique family of G protein-coupled receptors with important roles in both central and peripheral tissues. Cell. Mol. Life Sci. 64, 2104–2119 10.1007/s00018-007-7067-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockaert J., Pin J. P. (1999). Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 18, 1723–1729 10.1093/emboj/18.7.1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury M. J., McBurnie M. I., Denton D. A., Lee K. F., Vale W. W. (2000). Modulation of urocortin-induced hypophagia and weight loss by corticotropin-releasing factor receptor 1 deficiency in mice. Endocrinology 141, 2715–2724 10.1210/en.141.8.2715 [DOI] [PubMed] [Google Scholar]
- Branchek T. A., Smith K. E., Gerald C., Walker M. W. (2000). Galanin receptor subtypes. Trends Pharmacol. Sci. 21, 109–117 10.1016/S0165-6147(00)01446-2 [DOI] [PubMed] [Google Scholar]
- Brighton P. J., Szekeres P. G., Willars G. B. (2004). Neuromedin U and its receptors: structure, function, and physiological roles. Pharmacol. Rev. 56, 231–248 10.1124/pr.56.2.3 [DOI] [PubMed] [Google Scholar]
- Brody T., Cravchik A. (2000). Drosophila melanogaster G protein-coupled receptors. J. Cell Biol. 150, F83–F88 10.1083/jcb.150.2.F83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzamali G., Torp M., Hauser F., Williamson M., Grimmelikhuijzen C. J. (2005). The Drosophila gene CG9918 codes for a pyrokinin-1 receptor. Biochem. Biophys. Res. Commun. 335, 14–19 10.1016/j.bbrc.2005.07.038 [DOI] [PubMed] [Google Scholar]
- Chalasani S. H., Kato S., Albrecht D. R., Nakagawa T., Abbott L. F., Bargmann C. I. (2010). Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat. Neurosci. 13, 615–621 10.1038/nn.2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro S., Della-Zuana O., Fauchere J. L., Feletou M., Galizzi J. P., Levens N. (2002). Appetite suppression based on selective inhibition of NPY receptors. Int. J. Obes. Relat. Metab. Disord. 26, 281–298 10.1038/sj.ijo.0801948 [DOI] [PubMed] [Google Scholar]
- Chance W. T., Thompson H., Thomas I., Fischer J. E. (1995). Anorectic and neurochemical effects of pituitary adenylate cyclase activating polypeptide in rats. Peptides 16, 1511–1516 10.1016/0196-9781(95)02048-9 [DOI] [PubMed] [Google Scholar]
- Chapman R. F., De Boer G. (1995). Regulatory Mechanisms in Insect Feeding. New York: Chapman & Hall [Google Scholar]
- Chaudhri O., Small C., Bloom S. (2006). Gastrointestinal hormones regulating appetite. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1187–1209 10.1098/rstb.2006.1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhri O. B., Salem V., Murphy K. G., Bloom S. R. (2008). Gastrointestinal satiety signals. Annu. Rev. Physiol. 70, 239–255 10.1146/annurev.physiol.70.113006.100506 [DOI] [PubMed] [Google Scholar]
- Chen M. E., Pietrantonio P. V. (2006). The short neuropeptide F-like receptor from the red imported fire ant, Solenopsis invicta Buren (Hymenoptera: Formicidae). Arch. Insect Biochem. Physiol. 61, 195–208 10.1002/arch.20103 [DOI] [PubMed] [Google Scholar]
- Christensen L. (1993). Effects of eating behavior on mood: a review of the literature. Int. J. Eat. Disord. 14, 171–183 [DOI] [PubMed] [Google Scholar]
- Chung S., Parks G. S., Lee C., Civelli O. (2011). Recent updates on the melanin-concentrating hormone (MCH) and its receptor system: lessons from MCH1R antagonists. J. Mol. Neurosci. 43, 115–121 10.1007/s12031-010-9411-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M. A., Newmyer B. A., Smith M. L. (2009). The anorectic effect of neuropeptide AF is associated with satiety-related hypothalamic nuclei. J. Neuroendocrinol. 21, 595–601 10.1111/j.1365-2826.2009.01876.x [DOI] [PubMed] [Google Scholar]
- Coast G. M., Garside C. S. (2005). Neuropeptide control of fluid balance in insects. Ann. N. Y. Acad. Sci. 1040, 1–8 10.1196/annals.1327.001 [DOI] [PubMed] [Google Scholar]
- Coghlan A. (2005). “Nematode genome evolution,” in WormBook, ed. The C. elegans Research Community (WormBook) , Available at: http://www.wormbook.org 10.1895/wormbook.1.15.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M., Reale V., Olofsson B., Knights A., Evans P., De Bono M. (2009). Coordinated regulation of foraging and metabolism in C. elegans by RFamide neuropeptide signaling. Cell Metab. 9, 375–385 10.1016/j.cmet.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Coll A. P., Farooqi I. S., Challis B. G., Yeo G. S., O’Rahilly S. (2004). Proopiomelanocortin and energy balance: insights from human and murine genetics. J. Clin. Endocrinol. Metab. 89, 2557–2562 10.1210/jc.2004-0428 [DOI] [PubMed] [Google Scholar]
- Coll A. P., Farooqi I. S., O’Rahilly S. (2007). The hormonal control of food intake. Cell 129, 251–262 10.1016/j.cell.2007.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium T. C. E. S. (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018 10.1126/science.282.5396.2012 [DOI] [PubMed] [Google Scholar]
- Consortium T. H. G. S. (2006). Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931–949 10.1038/nature05260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter A. D., Dey A., Murray R. L. (2009). Evolution of the Caenorhabditis elegans genome. Mol. Biol. Evol. 26, 1199–1234 10.1093/molbev/msp048 [DOI] [PubMed] [Google Scholar]
- de Bono M., Bargmann C. I. (1998). Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94, 679–689 10.1016/S0092-8674(00)81609-8 [DOI] [PubMed] [Google Scholar]
- de Jong-Brink M., Ter Maat A., Tensen C. P. (2001). NPY in invertebrates: molecular answers to altered functions during evolution. Peptides 22, 309–315 10.1016/S0196-9781(01)00332-1 [DOI] [PubMed] [Google Scholar]
- De Loof A., Baggerman G., Breuer M., Claeys I., Cerstiaens A., Clynen E., et al. (2001). Gonadotropins in insects: an overview. Arch. Insect Biochem. Physiol. 47, 129–138 10.1002/arch.1043 [DOI] [PubMed] [Google Scholar]
- Dieterich C., Clifton S. W., Schuster L. N., Chinwalla A., Delehaunty K., Dinkelacker I., et al. (2008). The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 40, 1193–1198 10.1038/ng.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downer K. E., Haselton A. T., Nachman R. J., Stoffolano J. G., Jr. (2007). Insect satiety: sulfakinin localization and the effect of drosulfakinin on protein and carbohydrate ingestion in the blow fly, Phormia regina (Diptera: Calliphoridae). J. Insect Physiol. 53, 106–112 10.1016/j.jinsphys.2006.10.013 [DOI] [PubMed] [Google Scholar]
- Fredriksson R., Lagerstrom M. C., Lundin L. G., Schioth H. B. (2003). The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63, 1256–1272 10.1124/mol.63.6.1256 [DOI] [PubMed] [Google Scholar]
- Fredriksson R., Schioth H. B. (2005). The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol. Pharmacol. 67, 1414–1425 10.1124/mol.104.009001 [DOI] [PubMed] [Google Scholar]
- Fuxe K., Borroto-Escuela D. O., Marcellino D., Romero-Fernandez W., Frankowska M., Guidolin D., et al. (2012). GPCR heteromers and their allosteric receptor-receptor interactions. Curr. Med. Chem. 19, 356–363 10.2174/092986712803414259 [DOI] [PubMed] [Google Scholar]
- Gaillard I., Rouquier S., Giorgi D. (2004). Olfactory receptors. Cell. Mol. Life Sci. 61, 456–469 10.1007/s00018-003-3273-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garczynski S. F., Crim J. W., Brown M. R. (2005). Characterization of neuropeptide F and its receptor from the African malaria mosquito, Anopheles gambiae. Peptides 26, 99–107 10.1016/j.peptides.2004.07.014 [DOI] [PubMed] [Google Scholar]
- Ghedin E., Wang S., Spiro D., Caler E., Zhao Q., Crabtree J., et al. (2007). Draft genome of the filarial nematode parasite Brugia malayi. Science 317, 1756–1760 10.1126/science.1145406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N., Moody T. W., Igarashi H., Ito T., Jensen R. T. (2008). Bombesin-related peptides and their receptors: recent advances in their role in physiology and disease states. Curr. Opin. Endocrinol. Diabetes Obes. 15, 58–64 10.1097/MED.0b013e3282f3709b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Orchard I. (2009). Physiological activity of neuropeptide f on the hindgut of the blood-feeding hemipteran, Rhodnius prolixus. J. Insect Sci. 9, 1–14 10.1673/031.009.0301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood K., Williams T., Geary T. (2005). Nematode neuropeptide receptors and their development as anthelmintic screens. Parasitology 131(Suppl.), S169–S177 10.1017/S0031182005007559 [DOI] [PubMed] [Google Scholar]
- Grimmelikhuijzen C. J., Hauser F. (2012). Mini-review: the evolution of neuropeptide signaling. Regul. Pept. 177(Suppl.), S6–S9 10.1016/j.regpep.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Hainerova I., Torekov S. S., Ek J., Finkova M., Borch-Johnsen K., Jorgensen T., et al. (2006). Association between neuromedin U gene variants and overweight and obesity. J. Clin. Endocrinol. Metab. 91, 5057–5063 10.1210/jc.2006-1442 [DOI] [PubMed] [Google Scholar]
- Hampton L. L., Ladenheim E. E., Akeson M., Way J. M., Weber H. C., Sutliff V. E., et al. (1998). Loss of bombesin-induced feeding suppression in gastrin-releasing peptide receptor-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 95, 3188–3192 10.1073/pnas.95.6.3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar A. J. (2001). Family-B G-protein-coupled receptors. Genome Biol. 2, REVIEWS3013. 10.1186/gb-2001-2-12-reviews3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris G., Mills H., Wragg R., Hapiak V., Castelletto M., Korchnak A., et al. (2010). The monoaminergic modulation of sensory-mediated aversive responses in Caenorhabditis elegans requires glutamatergic/peptidergic cotransmission. J. Neurosci. 30, 7889–7899 10.1523/JNEUROSCI.5341-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser F., Cazzamali G., Williamson M., Blenau W., Grimmelikhuijzen C. J. (2006). A review of neurohormone GPCRs present in the fruitfly Drosophila melanogaster and the honey bee Apis mellifera. Prog. Neurobiol. 80, 1–19 10.1016/j.pneurobio.2006.07.005 [DOI] [PubMed] [Google Scholar]
- Hauser F., Cazzamali G., Williamson M., Park Y., Li B., Tanaka Y., et al. (2008). A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum. Front. Neuroendocrinol. 29, 142–165 10.1016/j.yfrne.2007.10.003 [DOI] [PubMed] [Google Scholar]
- Heinrichs S. C., Richard D. (1999). The role of corticotropin-releasing factor and urocortin in the modulation of ingestive behavior. Neuropeptides 33, 350–359 10.1054/npep.1999.0047 [DOI] [PubMed] [Google Scholar]
- Hergarden A. C., Tayler T. D., Anderson D. J. (2012). Allatostatin-A neurons inhibit feeding behavior in adult Drosophila. Proc. Natl. Acad. Sci. U.S.A. 109, 3967–3972 10.1073/pnas.1200778109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewes R. S., Taghert P. H. (2001). Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 11, 1126–1142 10.1101/gr.169901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. A., Fox A. N., Pitts R. J., Kent L. B., Tan P. L., Chrystal M. A., et al. (2002). G protein-coupled receptors in Anopheles gambiae. Science 298, 176–178 10.1126/science.298.5597.1317a [DOI] [PubMed] [Google Scholar]
- Hoekstra R., Visser A., Otsen M., Tibben J., Lenstra J. A., Roos M. H. (2000). EST sequencing of the parasitic nematode Haemonchus contortus suggests a shift in gene expression during transition to the parasitic stages. Mol. Biochem. Parasitol. 110, 53–68 10.1016/S0166-6851(00)00255-3 [DOI] [PubMed] [Google Scholar]
- Ida T., Takahashi T., Tominaga H., Sato T., Kume K., Ozaki M., et al. (2011). Identification of the novel bioactive peptides dRYamide-1 and dRYamide-2, ligands for a neuropeptide Y-like receptor in Drosophila. Biochem. Biophys. Res. Commun. 410, 872–877 10.1016/j.bbrc.2011.06.081 [DOI] [PubMed] [Google Scholar]
- Irani B. G., Haskell-Luevano C. (2005). Feeding effects of melanocortin ligands – a historical perspective. Peptides 26, 1788–1799 10.1016/j.peptides.2004.11.038 [DOI] [PubMed] [Google Scholar]
- Iversen A., Cazzamali G., Williamson M., Hauser F., Grimmelikhuijzen C. J. (2002). Molecular cloning and functional expression of a Drosophila receptor for the neuropeptides capa-1 and -2. Biochem. Biophys. Res. Commun. 299, 628–633 10.1016/S0006-291X(02)02709-2 [DOI] [PubMed] [Google Scholar]
- Janssen T., Meelkop E., Lindemans M., Verstraelen K., Husson S. J., Temmerman L., et al. (2008). Discovery of a cholecystokinin-gastrin-like signaling system in nematodes. Endocrinology 149, 2826–2839 10.1210/en.2007-1475 [DOI] [PubMed] [Google Scholar]
- Johnson E. C., Bohn L. M., Barak L. S., Birse R. T., Nassel D. R., Caron M. G., et al. (2003). Identification of Drosophila neuropeptide receptors by G protein-coupled receptors-beta-arrestin2 interactions. J. Biol. Chem. 278, 52172–52178 10.1074/jbc.M301846200 [DOI] [PubMed] [Google Scholar]
- Joost P., Methner A. (2002). Phylogenetic analysis of 277 human G-protein-coupled receptors as a tool for the prediction of orphan receptor ligands. Genome Biol. 3, RESEARCH0063. 10.1186/gb-2002-3-11-research0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen L. M., Hauser F., Cazzamali G., Williamson M., Grimmelikhuijzen C. J. (2006). Molecular identification of the first SIFamide receptor. Biochem. Biophys. Res. Commun. 340, 696–701 10.1016/j.bbrc.2005.12.062 [DOI] [PubMed] [Google Scholar]
- Karagiannides I., Stavrakis D., Bakirtzi K., Kokkotou E., Pirtskhalava T., Nayeb-Hashemi H., et al. (2011). Substance P (SP)-neurokinin-1 receptor (NK-1R) alters adipose tissue responses to high-fat diet and insulin action. Endocrinology 152, 2197–2205 10.1210/en.2011-1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi H. (2006). Functions of melanin-concentrating hormone in fish. J. Exp. Zoolog. A Comp. Exp. Biol. 305, 751–760 10.1002/jez.a.310 [DOI] [PubMed] [Google Scholar]
- Keating C. D., Kriek N., Daniels M., Ashcroft N. R., Hopper N. A., Siney E. J., et al. (2003). Whole-genome analysis of 60 G protein-coupled receptors in Caenorhabditis elegans by gene knockout with RNAi. Curr. Biol. 13, 1715–1720 10.1016/j.cub.2003.09.003 [DOI] [PubMed] [Google Scholar]
- Klowden M. J. (1990). The endogenous regulation of mosquito reproductive behavior. Experientia 46, 660–670 10.1007/BF01939928 [DOI] [PubMed] [Google Scholar]
- Konturek S. J., Konturek J. W., Pawlik T., Brzozowski T. (2004). Brain-gut axis and its role in the control of food intake. J. Physiol. Pharmacol. 55, 137–154 [PubMed] [Google Scholar]
- Kopin A. S., Mathes W. F., McBride E. W., Nguyen M., Al-Haider W., Schmitz F., et al. (1999). The cholecystokinin-A receptor mediates inhibition of food intake yet is not essential for the maintenance of body weight. J. Clin. Invest. 103, 383–391 10.1172/JCI4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreienkamp H. J., Larusson H. J., Witte I., Roeder T., Birgul N., Honck H. H., et al. (2002). Functional annotation of two orphan G-protein-coupled receptors, Drostar1 and -2, from Drosophila melanogaster and their ligands by reverse pharmacology. J. Biol. Chem. 277, 39937–39943 10.1074/jbc.M206931200 [DOI] [PubMed] [Google Scholar]
- Krishnan A., Almen M. S., Fredriksson R., Schioth H. B. (2012). The origin of GPCRs: identification of mammalian like Rhodopsin, Adhesion, Glutamate and Frizzled GPCRs in fungi. PLoS ONE 7:e29817. 10.1371/journal.pone.0029817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiak T. M., Larsen M. J., Bowman J. W., Geary T. G., Lowery D. E. (2008). FMRFamide-like peptides encoded on the flp-18 precursor gene activate two isoforms of the orphan Caenorhabditis elegans G-protein-coupled receptor Y58G8A.4 heterologously expressed in mammalian cells. Biopolymers 90, 339–348 10.1002/bip.20850 [DOI] [PubMed] [Google Scholar]
- Kubiak T. M., Larsen M. J., Burton K. J., Bannow C. A., Martin R. A., Zantello M. R., et al. (2002). Cloning and functional expression of the first Drosophila melanogaster sulfakinin receptor DSK-R1. Biochem. Biophys. Res. Commun. 291, 313–320 10.1006/bbrc.2002.6459 [DOI] [PubMed] [Google Scholar]
- Ladenheim E. E., Hampton L. L., Whitney A. C., White W. O., Battey J. F., Moran T. H. (2002). Disruptions in feeding and body weight control in gastrin-releasing peptide receptor deficient mice. J. Endocrinol. 174, 273–281 10.1677/joe.0.1740273 [DOI] [PubMed] [Google Scholar]
- Lang R., Gundlach A. L., Kofler B. (2007). The galanin peptide family: receptor pharmacology, pleiotropic biological actions, and implications in health and disease. Pharmacol. Ther. 115, 177–207 10.1016/j.pharmthera.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Langmead C. J., Christopoulos A. (2006). Allosteric agonists of 7TM receptors: expanding the pharmacological toolbox. Trends Pharmacol. Sci. 27, 475–481 10.1016/j.tips.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Lecklin A., Lundell I., Paananen L., Wikberg J. E., Mannisto P. T., Larhammar D. (2002). Receptor subtypes Y1 and Y5 mediate neuropeptide Y induced feeding in the guinea-pig. Br. J. Pharmacol. 135, 2029–2037 10.1038/sj.bjp.0704667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. S., You K. H., Choo J. K., Han Y. M., Yu K. (2004). Drosophila short neuropeptide F regulates food intake and body size. J. Biol. Chem. 279, 50781–50789 10.1074/jbc.M400857200 [DOI] [PubMed] [Google Scholar]
- Lenz C., Williamson M., Grimmelikhuijzen C. J. (2000). Molecular cloning and genomic organization of a second probable allatostatin receptor from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 273, 571–577 10.1006/bbrc.2000.3062 [DOI] [PubMed] [Google Scholar]
- Li X., Wu N., Ra N., Forte M. (1992). Cloning, functional expression, and developmental regulation of a neuropeptide Y receptor from Drosophila melanogaster. J. Biol. Chem. 267, 9–12 [PubMed] [Google Scholar]
- Li X. J., Wolfgang W., Wu Y. N., North R. A., Forte M. (1991). Cloning, heterologous expression and developmental regulation of a Drosophila receptor for tachykinin-like peptides. EMBO J. 10, 3221–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemans M., Janssen T., Husson S. J., Meelkop E., Temmerman L., Clynen E., et al. (2009). A neuromedin-pyrokinin-like neuropeptide signaling system in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 379, 760–764 10.1016/j.bbrc.2008.12.121 [DOI] [PubMed] [Google Scholar]
- Lingo P. R., Zhao Z., Shen P. (2007). Co-regulation of cold-resistant food acquisition by insulin- and neuropeptide Y-like systems in Drosophila melanogaster. Neuroscience 148, 371–374 10.1016/j.neuroscience.2007.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 10.1126/science.290.5494.1151 [DOI] [PubMed] [Google Scholar]
- Maestro J. L., Aguilar R., Pascual N., Valero M. L., Piulachs M. D., Andreu D., et al. (2001). Screening of antifeedant activity in brain extracts led to the identification of sulfakinin as a satiety promoter in the German cockroach. Are arthropod sulfakinins homologous to vertebrate gastrins-cholecystokinins? Eur. J. Biochem. 268, 5824–5830 10.1046/j.0014-2956.2001.02527.x [DOI] [PubMed] [Google Scholar]
- Maier W., Adilov B., Regenass M., Alcedo J. (2010). A neuromedin U receptor acts with the sensory system to modulate food type-dependent effects on C. elegans lifespan. PLoS Biol. 8:e1000376. 10.1371/journal.pbio.1000376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinissen M. J., Gutkind J. S. (2001). G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol. Sci. 22, 368–376 10.1016/S0165-6147(00)01678-3 [DOI] [PubMed] [Google Scholar]
- Matteri R. (2001). Overview of central targets for appetite regulation. J. Anim. Sci. 79(suppl.), E148–E158 [Google Scholar]
- Mayoral J. G., Nouzova M., Brockhoff A., Goodwin M., Hernandez-Martinez S., Richter D., et al. (2010). Allatostatin-C receptors in mosquitoes. Peptides 31, 442–450 10.1016/j.peptides.2009.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott M. T., Kidd G. S. (1987). The role of calcitonin in the development and treatment of osteoporosis. Endocr. Rev. 8, 377–390 10.1210/edrv-8-4-377 [DOI] [PubMed] [Google Scholar]
- McKay R. M., McKay J. P., Suh J. M., Avery L., Graff J. M. (2007). Tripeptidyl peptidase II promotes fat formation in a conserved fashion. EMBO Rep. 8, 1183–1189 10.1038/sj.embor.7401086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon L. R., Wellman P. J. (1997). Decreased intake of a liquid diet in nonfood-deprived rats following intra-PVN injections of GLP-1 (7-36) amide. Pharmacol. Biochem. Behav. 58, 673–677 10.1016/S0091-3057(97)90017-4 [DOI] [PubMed] [Google Scholar]
- McMahon L. R., Wellman P. J. (1998). PVN infusion of GLP-1-(7-36) amide suppresses feeding but does not induce aversion or alter locomotion in rats. Am. J. Physiol. 274, R23–R29 [DOI] [PubMed] [Google Scholar]
- Melcher C., Pankratz M. J. (2005). Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 3:e305. 10.1371/journal.pbio.0030305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Wahlstrom G., Immonen T., Kolmer M., Tirronen M., Predel R., et al. (2002). The Drosophila hugin gene codes for myostimulatory and ecdysis-modifying neuropeptides. Mech. Dev. 117, 5–13 10.1016/S0925-4773(02)00175-2 [DOI] [PubMed] [Google Scholar]
- Mercer R. E., Chee M. J., Colmers W. F. (2011). The role of NPY in hypothalamic mediated food intake. Front. Neuroendocrinol. 32, 398–415 10.1016/j.yfrne.2011.06.001 [DOI] [PubMed] [Google Scholar]
- Mertens I., Clinckspoor I., Janssen T., Nachman R., Schoofs L. (2006). FMRFamide related peptide ligands activate the Caenorhabditis elegans orphan GPCR Y59H11AL.1. Peptides 27, 1291–1296 10.1016/j.peptides.2005.11.017 [DOI] [PubMed] [Google Scholar]
- Millington G. W. (2007). The role of proopiomelanocortin (POMC) neurones in feeding behaviour. Nutr. Metab. (Lond.) 4, 18. 10.1186/1743-7075-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitreva M., Jasmer D. P., Zarlenga D. S., Wang Z., Abubucker S., Martin J., et al. (2011). The draft genome of the parasitic nematode Trichinella spiralis. Nat. Genet. 43, 228–235 10.1038/ng.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier D., Colas J. F., Rosay P., Hen R., Borrelli E., Maroteaux L. (1992). NKD, a developmentally regulated tachykinin receptor in Drosophila. J. Biol. Chem. 267, 1298–1302 [PubMed] [Google Scholar]
- Montell C. (2009). A taste of the Drosophila gustatory receptors. Curr. Opin. Neurobiol. 19, 345–353 10.1016/j.conb.2009.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody T. W., Ito T., Osefo N., Jensen R. T. (2011). VIP and PACAP: recent insights into their functions/roles in physiology and disease from molecular and genetic studies. Curr. Opin. Endocrinol. Diabetes Obes. 18, 61–67 10.1097/MED.0b013e328342568a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley J. E., Horowitz M., Morley P. M., Flood J. F. (1992). Pituitary adenylate cyclase activating polypeptide (PACAP) reduces food intake in mice. Peptides 13, 1133–1135 10.1016/0196-9781(92)90019-Y [DOI] [PubMed] [Google Scholar]
- Murase T., Arima H., Kondo K., Oiso Y. (1996). Neuropeptide FF reduces food intake in rats. Peptides 17, 353–354 10.1016/0196-9781(95)02137-X [DOI] [PubMed] [Google Scholar]
- Murdoch C., Finn A. (2000). Chemokine receptors and their role in inflammation and infectious diseases. Blood 95, 3032–3043 [PubMed] [Google Scholar]
- Nagarathnam B., Kalaimathy S., Balakrishnan V., Sowdhamini R. (2012). Cross-Genome Clustering of Human and C. elegans G-Protein Coupled Receptors. Evol. Bioinform. Online 8, 229–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassel D. R., Winther A. M. (2010). Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 92, 42–104 10.1016/j.pneurobio.2010.04.010 [DOI] [PubMed] [Google Scholar]
- Nathoo A. N., Moeller R. A., Westlund B. A., Hart A. C. (2001). Identification of neuropeptide-like protein gene families in Caenorhabditiselegans and other species. Proc. Natl. Acad. Sci. U.S.A. 98, 14000–14005 10.1073/pnas.241231298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neary N. M., Goldstone A. P., Bloom S. R. (2004). Appetite regulation: from the gut to the hypothalamus. Clin. Endocrinol. (Oxf.) 60, 153–160 10.1046/j.1365-2265.2003.01839.x [DOI] [PubMed] [Google Scholar]
- Nichols R. (2007). The first nonsulfated sulfakinin activity reported suggests nsDSK acts in gut biology. Peptides 28, 767–773 10.1016/j.peptides.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Nichols R., Schneuwly S. A., Dixon J. E. (1988). Identification and characterization of a Drosophila homologue to the vertebrate neuropeptide cholecystokinin. J. Biol. Chem. 263, 12167–12170 [PubMed] [Google Scholar]
- Niswender C. M., Conn P. J. (2010). Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol. 50, 295–322 10.1146/annurev.pharmtox.011008.145533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom K. J., Sallman Almen M., Edstam M. M., Fredriksson R., Schioth H. B. (2011). Independent HHsearch, Needleman – Wunsch-based, and motif analyses reveal the overall hierarchy for most of the G protein-coupled receptor families. Mol. Biol. Evol. 28, 2471–2480 10.1093/molbev/msr061 [DOI] [PubMed] [Google Scholar]
- Ohki-Hamazaki H., Watase K., Yamamoto K., Ogura H., Yamano M., Yamada K., et al. (1997). Mice lacking bombes in receptor subtype-3 develop metabolic defects and obesity. Nature 390, 165–169 10.1038/36568 [DOI] [PubMed] [Google Scholar]
- Paluzzi J. P., Park Y., Nachman R. J., Orchard I. (2010). Isolation, expression analysis, and functional characterization of the first antidiuretic hormone receptor in insects. Proc. Natl. Acad. Sci. U.S.A. 107, 10290–10295 10.1073/pnas.1003666107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y., Kim Y. J., Adams M. E. (2002). Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin, and AKH supports a theory of ligand-receptor coevolution. Proc. Natl. Acad. Sci. U.S.A. 99, 11423–11428 10.1073/pnas.152011799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pjetri E., Adan R. A., Herzog H., De Haas R., Oppelaar H., Spierenburg H. A., et al. (2012). NPY receptor subtype specification for behavioral adaptive strategies during limited food access. Genes Brain Behav. 11, 105–112 10.1111/j.1601-183X.2011.00732.x [DOI] [PubMed] [Google Scholar]
- Poels J., Birse R. T., Nachman R. J., Fichna J., Janecka A., Vanden Broeck J., et al. (2009). Characterization and distribution of NKD, a receptor for Drosophila tachykinin-related peptide 6. Peptides 30, 545–556 10.1016/j.peptides.2008.10.012 [DOI] [PubMed] [Google Scholar]
- Poels J., Verlinden H., Fichna J., Van Loy T., Franssens V., Studzian K., et al. (2007). Functional comparison of two evolutionary conserved insect neurokinin-like receptors. Peptides 28, 103–108 10.1016/j.peptides.2006.06.014 [DOI] [PubMed] [Google Scholar]
- Pollock V. P., McGettigan J., Cabrero P., Maudlin I. M., Dow J. A., Davies S. A. (2004). Conservation of capa peptide-induced nitric oxide signalling in Diptera. J. Exp. Biol. 207, 4135–4145 10.1242/jeb.01255 [DOI] [PubMed] [Google Scholar]
- Posnien N., Schinko J. B., Kittelmann S., Bucher G. (2010). Genetics, development and composition of the insect head – a beetle’s view. Arthropod Struct. Dev. 39, 399–410 10.1016/j.asd.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Prakash M., Steele M. (2010). The hungry fly: hydrodynamics of feeding in the common house fly. Phys. Fluids 23, 091110–091110 10.1063/1.3640023 [DOI] [Google Scholar]
- Pratt G. E., Farnsworth D. E., Fok K. F., Siegel N. R., Mccormack A. L., Shabanowitz J., et al. (1991). Identity of a second type of allatostatin from cockroach brains: an octadecapeptide amide with a tyrosine-rich address sequence. Proc. Natl. Acad. Sci. U.S.A. 88, 2412–2416 10.1073/pnas.88.6.2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinster S. C., Hague C., Hall R. A. (2005). Heterodimerization of g protein-coupled receptors: specificity and functional significance. Pharmacol. Rev. 57, 289–298 10.1124/pr.57.3.1 [DOI] [PubMed] [Google Scholar]
- Radford J. C., Davies S. A., Dow J. A. (2002). Systematic G-protein-coupled receptor analysis in Drosophila melanogaster identifies a leucokinin receptor with novel roles. J. Biol. Chem. 277, 38810–38817 10.1074/jbc.M203694200 [DOI] [PubMed] [Google Scholar]
- Richard D., Lin Q., Timofeeva E. (2002). The corticotropin-releasing factor family of peptides and CRF receptors: their roles in the regulation of energy balance. Eur. J. Pharmacol. 440, 189–197 10.1016/S0014-2999(02)01428-0 [DOI] [PubMed] [Google Scholar]
- Riediger T., Zuend D., Becskei C., Lutz T. A. (2004). The anorectic hormone amylin contributes to feeding-related changes of neuronal activity in key structures of the gut-brain axis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 286, R114–R122 10.1152/ajpregu.00333.2003 [DOI] [PubMed] [Google Scholar]
- Robertson H. M., Thomas J. H. (2006). “The putative chemoreceptor families of C. elegans,” in WormBook, ed. The C. elegans Research Community (WormBook) , Available at: http://www.wormbook.org 10.1895/wormbook.1.66.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha-Sousa A., Tavares-Silva M., Fonseca S., Falcão M., Falcão-Reis F., Leite- Moreira A. (2010). Diabetes, ghrelin and related peptides: from pathophysiology to vasculopathy. Open Circ. Vasc. J. 3, 17–29 10.2174/1874382601003010017 [DOI] [Google Scholar]
- Rogers C., Reale V., Kim K., Chatwin H., Li C., Evans P., et al. (2003). Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat. Neurosci. 6, 1178–1185 10.1038/nn1140 [DOI] [PubMed] [Google Scholar]
- Roller L., Yamanaka N., Watanabe K., Daubnerova I., Zitnan D., Kataoka H., et al. (2008). The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38, 1147–1157 10.1016/j.ibmb.2008.04.009 [DOI] [PubMed] [Google Scholar]
- Rosay P., Colas J. F., Maroteaux L. (1995). Dual organisation of the Drosophila neuropeptide receptor NKD gene promoter. Mech. Dev. 51, 329–339 10.1016/0925-4773(95)00382-7 [DOI] [PubMed] [Google Scholar]
- Saideman S. R., Ma M., Kutz-Naber K. K., Cook A., Torfs P., Schoofs L., et al. (2007). Modulation of rhythmic motor activity by pyrokinin peptides. J. Neurophysiol. 97, 579–595 10.1152/jn.00772.2006 [DOI] [PubMed] [Google Scholar]
- Sakurai T. (1999). Orexins and orexin receptors: implication in feeding behavior. Regul. Pept. 85, 25–30 10.1016/S0167-0115(99)00076-2 [DOI] [PubMed] [Google Scholar]
- Sakurai T., Amemiya A., Ishii M., Matsuzaki I., Chemelli R. M., Tanaka H., et al. (1998). Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell 92, 573–585 10.1016/S0092-8674(02)09256-5 [DOI] [PubMed] [Google Scholar]
- Sano H., Feighner S. D., Hreniuk D. L., Iwaasa H., Sailer A. W., Pan J., et al. (2004). Characterization of the bombesin-like peptide receptor family in primates. Genomics 84, 139–146 10.1016/j.ygeno.2004.01.008 [DOI] [PubMed] [Google Scholar]
- Schioth H. B., Fredriksson R. (2005). The GRAFS classification system of G-protein coupled receptors in comparative perspective. Gen. Comp. Endocrinol. 142, 94–101 10.1016/j.ygcen.2004.12.018 [DOI] [PubMed] [Google Scholar]
- Schoofs L., Clynen E., Cerstiaens A., Baggerman G., Wei Z., Vercammen T., et al. (2001). Newly discovered functions for some myotropic neuropeptides in locusts. Peptides 22, 219–227 10.1016/S0196-9781(00)00385-5 [DOI] [PubMed] [Google Scholar]
- Schulte G. (2010). International union of basic and clinical pharmacology. LXXX. The class frizzled receptors. Pharmacol. Rev. 62, 632–667 10.1124/pr.110.002931 [DOI] [PubMed] [Google Scholar]
- Shioda S., Takenoya F., Yagi M., Wang L., Hori Y., Kageyama H. (2008). Neural networks of several novel neuropeptides involved in feeding regulation. Nutrition 24, 848–853 10.1016/j.nut.2008.06.016 [DOI] [PubMed] [Google Scholar]
- Simoni M., Gromoll J., Nieschlag E. (1997). The follicle-stimulating hormone receptor: biochemistry, molecular biology, physiology, and pathophysiology. Endocr. Rev. 18, 739–773 10.1210/er.18.6.739 [DOI] [PubMed] [Google Scholar]
- Sommer R. J., Streit A. (2011). Comparative genetics and genomics of nematodes: genome structure, development, and lifestyle. Annu. Rev. Genet. 45, 1–20 10.1146/annurev-genet-110410-132417 [DOI] [PubMed] [Google Scholar]
- Stanley S., Wynne K., McGowan B., Bloom S. (2005). Hormonal regulation of food intake. Physiol. Rev. 85, 1131–1158 10.1152/physrev.00015.2004 [DOI] [PubMed] [Google Scholar]
- Stay B. (2000). A review of the role of neurosecretion in the control of juvenile hormone synthesis: a tribute to Berta Scharrer. Insect Biochem. Mol. Biol. 30, 653–662 10.1016/S0965-1748(00)00036-9 [DOI] [PubMed] [Google Scholar]
- Styer K. L., Singh V., Macosko E., Steele S. E., Bargmann C. I., Aballay A. (2008). Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322, 460–464 10.1126/science.1163673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwa M., Sugihara M., Ono Y. (2011). Functional and structural overview of G-protein-coupled receptors comprehensively obtained from genome sequences. Pharmaceuticals 4, 652–664 10.3390/ph4040652 [DOI] [Google Scholar]
- Suzuki K., Simpson K. A., Minnion J. S., Shillito J. C., Bloom S. R. (2010). The role of gut hormones and the hypothalamus in appetite regulation. Endocr. J. 57, 359–372 10.1507/endocrj.K10E-077 [DOI] [PubMed] [Google Scholar]
- Tachibana T., Cline M. A., Sugahara K., Ueda H., Hiramatsu K. (2009). Central administration of somatostatin stimulates feeding behavior in chicks. Gen. Comp. Endocrinol. 161, 354–359 10.1016/j.ygcen.2009.01.022 [DOI] [PubMed] [Google Scholar]
- Tang-Christensen M., Vrang N., Larsen P. J. (2001). Glucagon-like peptide containing pathways in the regulation of feeding behaviour. Int. J. Obes. Relat. Metab. Disord. 25(Suppl. 5), S42–S47 10.1038/sj.ijo.0801912 [DOI] [PubMed] [Google Scholar]
- Teng M. S., Shadbolt P., Fraser A. G., Jansen G., McCafferty J. (2008). Control of feeding behavior in C. elegans by human G protein-coupled receptors permits screening for agonist-expressing bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 14826–14831 10.1073/pnas.0803290105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhzaz S., Cabrero P., Robben J. H., Radford J. C., Hudson B. D., Milligan G., et al. (2012). Mechanism and function of Drosophila capa GPCR: a desiccation stress-responsive receptor with functional homology to human neuromedinU receptor. PLoS ONE 7:e29897. 10.1371/journal.pone.0029897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R., Nusse R. (2009). Towards an integrated view of Wnt signaling in development. Development 136, 3205–3214 10.1242/dev.033910 [DOI] [PubMed] [Google Scholar]
- Vashlishan A. B., Madison J. M., Dybbs M., Bai J., Sieburth D., Ch’Ng Q., et al. (2008). An RNAi screen identifies genes that regulate GABA synapses. Neuron 58, 346–361 10.1016/j.neuron.2008.02.019 [DOI] [PubMed] [Google Scholar]
- Waldhoer M., Bartlett S. E., Whistler J. L. (2004). Opioid receptors. Annu. Rev. Biochem. 73, 953–990 10.1146/annurev.biochem.73.011303.073940 [DOI] [PubMed] [Google Scholar]
- Wei Z., Baggerman G. R. J. N., Goldsworthy G., Verhaert P., De Loof A., Schoofs L. (2000). Sulfakinins reduce food intake in the desert locust, Schistocerca gregaria. J. Insect Physiol. 46, 1259–1265 10.1016/S0022-1910(00)00046-9 [DOI] [PubMed] [Google Scholar]
- Wilding J. P. (2002). Neuropeptides and appetite control. Diabet. Med. 19, 619–627 10.1046/j.1464-5491.2002.00790.x [DOI] [PubMed] [Google Scholar]
- Wilson R., Ainscough R., Anderson K., Baynes C., Berks M., Bonfield J., et al. (1994). 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature 368, 32–38 10.1038/368032a0 [DOI] [PubMed] [Google Scholar]
- Wong K. K., Ng S. Y., Lee L. T., Ng H. K., Chow B. K. (2011). Orexins and their receptors from fish to mammals: a comparative approach. Gen. Comp. Endocrinol. 171, 124–130 10.1016/j.ygcen.2011.01.001 [DOI] [PubMed] [Google Scholar]
- Woodhead A. P., Khan M. A., Stay B., Tobe S. S. (1994). Two new allatostatins from the brains of Diploptera punctata. Insect Biochem. Mol. Biol. 24, 257–263 10.1016/0965-1748(94)90005-1 [DOI] [PubMed] [Google Scholar]
- Woods S. C., Lutz T. A., Geary N., Langhans W. (2006). Pancreatic signals controlling food intake; insulin, glucagon and amylin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1219–1235 10.1098/rstb.2006.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods S. C., Seeley R. J., Cota D. (2008). Regulation of food intake through hypothalamic signaling networks involving mTOR. Annu. Rev. Nutr. 28, 295–311 10.1146/annurev.nutr.28.061807.155505 [DOI] [PubMed] [Google Scholar]
- Wu Q., Wen T., Lee G., Park J. H., Cai H. N., Shen P. (2003). Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron 39, 147–161 10.1016/S0896-6273(03)00396-9 [DOI] [PubMed] [Google Scholar]
- Xia Q., Zhou Z., Lu C., Cheng D., Dai F., Li B., et al. (2004). A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306, 1937–1940 10.1126/science.1102210 [DOI] [PubMed] [Google Scholar]
- Xu Y. L., Jackson V. R., Civelli O. (2004). Orphan G protein-coupled receptors and obesity. Eur. J. Pharmacol. 500, 243–253 10.1016/j.ejphar.2004.07.029 [DOI] [PubMed] [Google Scholar]
- Yamada K., Wada E., Santo-Yamada Y., Wada K. (2002). Bombesin and its family of peptides: prospects for the treatment of obesity. Eur. J. Pharmacol. 440, 281–290 10.1016/S0014-2999(02)01435-8 [DOI] [PubMed] [Google Scholar]
- Yona S., Lin H. H., Siu W. O., Gordon S., Stacey M. (2008). Adhesion-GPCRs: emerging roles for novel receptors. Trends Biochem. Sci. 33, 491–500 10.1016/j.tibs.2008.07.005 [DOI] [PubMed] [Google Scholar]
- Zamanian M., Kimber M. J., McVeigh P., Carlson S. A., Maule A. G., Day T. A. (2011). The repertoire of G protein-coupled receptors in the human parasite Schistosoma mansoni and the model organism Schmidtea mediterranea. BMC Genomics 12:596. 10.1186/1471-2164-12-596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. V., Ren P. G., Avsian-Kretchmer O., Luo C. W., Rauch R., Klein C., et al. (2005). Obestatin, a peptide encoded by the ghrelin gene, opposes ghrelin’s effects on food intake. Science 310, 996–999 10.1126/science.1117255 [DOI] [PubMed] [Google Scholar]
- Zorrilla E. P., Brennan M., Sabino V., Lu X., Bartfai T. (2007). Galanin type 1 receptor knockout mice show altered responses to high-fat diet and glucose challenge. Physiol. Behav. 91, 479–485 10.1016/j.physbeh.2006.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence of the nematodes GPCR transmembrane (TM) domains from non-model nematodes within each receptor family were extracted by sequence homology using the roundworm C. elegans TM regions. To facilitate visualization the TM1, 3, 5, and 7 were annotated in gray.
Sequence of the Arthropod GPCR transmembrane (TM) domains used in for phylogenetic analysis. TM domains from non-model arthropods within each receptor family were extracted by sequence homology using the D. melanogaster TMs. To facilitate visualization the TM1, 3, 5 and 7 were annotated in gray.