Abstract
The Diversity Outbred (DO) population is a heterogeneous stock derived from the same eight founder strains as the Collaborative Cross (CC) inbred strains. Genetically heterogeneous DO mice display a broad range of phenotypes. Natural levels of heterozygosity provide genetic buffering and, as a result, DO mice are robust and breed well. Genetic mapping analysis in the DO presents new challenges and opportunities. Specialized algorithms are required to reconstruct haplotypes from high-density SNP array data. The eight founder haplotypes can be combined into 36 possible diplotypes, which must be accommodated in QTL mapping analysis. Population structure of the DO must be taken into account here. Estimated allele effects of 8 founder haplotypes provide information that is not available in two-parent crosses and can dramatically reduce the number of candidate loci. Allele effects can also distinguish chance co-location of QTL from pleiotropy – which provides a basis for establishing causality in expression QTL studies. We recommended sample sizes of 200 to 800 mice for QTL mapping studies, larger than for traditional crosses. The CC inbred strains provide a resource for independent validation of DO mapping results. Genetic heterogeneity of the DO can provide a powerful advantage in our ability to generalize conclusions to other genetically diverse populations. Genetic diversity can also help to avoid the pitfall of identifying an idiosyncratic reaction that occurs only in a limited genetic context. Informatics tools and data resources associated with the CC, the DO, and their founder strains are developing rapidly. We anticipate a flood of new results to follow as our community begins to adopt and utilize these new genetic resource populations.
Keywords: QTL, mouse, genetics, association mapping, heterogeneous stock
Introduction
Genome-wide association studies in human populations have raised the bar for genetic mapping of complex traits and have motivated the development of new model organism resources that encompass high levels of genetic diversity and high mapping resolution to identify causal genetic variants. Unlike human studies, a model system can be designed to avoid the problems associated with environmental heterogeneity, population structure, and rare alleles, to increase power and reduce sample sizes. The Diversity Outbred (DO) mice have been developed with these goals in mind.
The DO (Svenson et al., 2012) is a heterogeneous stock derived from the same eight founder strains as the Collaborative Cross inbred strains (Collaborative Cross Consortium, 2012). In 2009, animals representing 144 independent lineages from the CC breeding colony at The Oak Ridge National Laboratory (Chesler et al., 2008) were used to seed the DO population, which was then maintained as a randomized breeding colony with a population size of 175 pairs. One female and one male from each first litter are randomly assigned to a new breeding pair to make the next generation. This mating scheme doubles the effective population size and minimizes the effects of drift and selection on allele frequencies in the DO (Rockman and Kruglyak, 2008; Unpublished simulations). Each DO mice is a unique individual with a high level of allelic heterozygosity and the DO population provides an effectively unlimited source of novel allelic combinations. The current generation, G10, is expected to have an average of 390 recombination events per animal, sufficient to provide sub-Mb mapping resolution and mapping resolution will continue to improve with each successive generation.
DO mice are robust and breed efficiently, with an average first litter size of 7 pups (+/− 2.4 SD) consistently observed from G4 through G9. Matings are allowed to produce two or more litters (second litters average 9 pups +/− 2.6 SD). Recently, the colony was expanded for production purposes. Two mating cages are set up from each first litter, with only one mating providing the next generation breeders. Hence, the colony size remains fixed at 175 lines while expansion allows for an effectively unlimited production capacity. Using this strategy, up to four generations can be produced annually and the production of mice can be adjusted to meet demand without altering the structure of the breeding population.
Genetic Mapping Populations
Inbred strain backcrosses and intercrosses have been widely used to map genetic loci associated with quantitative traits (quantitative trait loci; QTL). These study designs have high statistical power for detecting genetic loci but they have limited mapping resolution. As a result, QTL can span tens of Mb and extensive follow up studies are often required to identify causal variants. Another approach to genetic mapping is association mapping with inbred strain panels. The “classical” mouse inbred strains, such as C57BL/6J and DBA/2J, are most suitable for this purpose and have been extensively characterized by biomedical researchers. However, genetic variation in these strains is both limited and unevenly distributed across the genome (Yang et al., 2007; Yang et al., 2011). Furthermore, historical bottlenecks in the derivation of these strains have resulted in extensive long-range linkage disequilibrium (Petkov et al., 2005; Collaborative Cross Consortium, 2012), which confounds attempts to use these strains for association mapping. Heterogeneous stocks, such as the DO, hold promise for high-resolution mapping. A second, independent heterogeneous stock (HS-CC) has been generated from CC founders, as described in Iancu et al (2010). The HS-CC utilizes a circular mating design that effectively controls genetic drift but at the expense of reduced map expansion relative to random designs (Rockman and Kruglyak, 2008). Below we address the unique features and challenges of genetic mapping in the DO populations.
DO Phenotypes
Phenotypes described here were collected on 150 DO animals from generations G4 and G5. Mice of both sexes were maintained on either standard chow (n=100) or high fat diet (HFD; n=50) from weaning until 23 weeks of age. Details about husbandry and phenotyping methods have recently been described (Svenson et al., 2012). Briefly, animals were subject to a battery of noninvasive physiological measurements to obtain clinical parameters including early and late measurements of clinical plasma chemistries (lipids, glucose, insulin) and body composition (weight, percent fat, lean tissue mass).
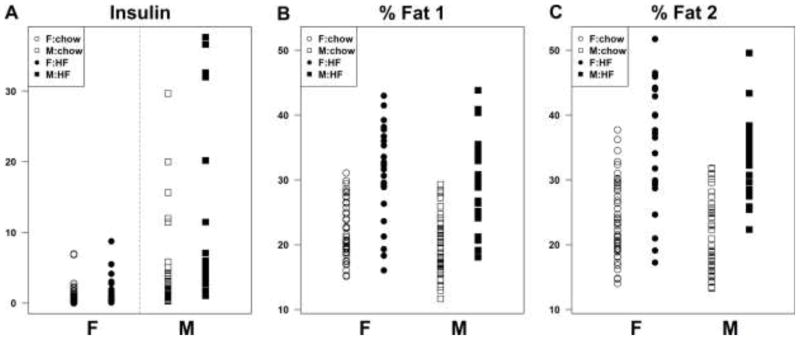
The eight founder strains vary widely in the phenotypes measured in this study (Svenson et al., 2007) and, as expected, we also observe a broad range of phenotypes among DO animals. The genetically heterogeneous DO can be used to investigate variation in response to environmental and dietary perturbations (for example, refer to the cholesterol QTL described below). In our study, insulin levels were observed to increase in both sexes in response to HFD, with much greater variation observed in males (Figure 1A). Weight gain was also greater on high fat diet and was generally due to an increase in fat mass in both sexes (Figure 1B & C). Insulin was more highly correlated with glucose and triglycerides in males who consumed the HFD (data not shown). Summary statistical analyses do not always capture the unique phenotypes that can be observed in the DO mice. While visually striking phenotypes including variations in coat color, body size and constitution are immediately obvious; underlying differences in disease-related phenotypes are also present. We have observed animals, mostly males, with coincident elevations of lipids, glucose, insulin and body fat after consuming the HFD, reflective of metabolic syndrome. As we expand our characterization of DO mice, we expect to see even more extreme ‘case studies’ that will provide a basis for investigation of how novel combinations of natural allelic variation can give rise to common and genetically complex diseases. Importantly, allele combinations predicted to cause outlier phenotypes in DO animals can be reproduced and validated in the CC strains (see “Validation in the CC” below).
Figure 1.
DO phenotypes by sex and diet groups are shown for insulin (A), Percent fat at time 1 (B), Percent fat time 2 (C).
Genotyping the DO
In an intercross mapping population, each locus has three possible diplotypes and the genotypes of diallelic SNPs correspond directly to parental haplotypes. In the DO there are eight founder haplotypes and 36 possible diplotypes (8 homozygotes + 28 heterozygotes). The problem of reconstructing founder haplotypes from binary SNP data is challenging but can be solved with a hidden Markov model (HMM) algorithm. High-density SNP data are required to capture small haplotype blocks and we have successfully used the Mouse Universal Genotyping Array (MUGA; Collaborative Cross Consortium, 2012), which includes 7,854 pairs of allele-specific probes to assay SNPs in the early generations of the DO. However as recombination events continue to accumulate, a higher density SNP platform will be required. Indeed, we observe that up to 20% of expected recombinations at G8 are being missed, which may reflect the limitation in marker density or re-use of recombination hot spots (unpublished data).
Genome Scans
A second challenge posed by the 36 diplotype states of the DO is the computation of suitable LOD scores for QTL mapping. Intercross LOD scores are computed (essentially) by a regression analysis with 3 free parameters. The corresponding regressions for DO mapping require 36 free parameters, which can result in reduced power and computational instability, especially with small sample sizes. One solution, which we have employed, is to reduce the complexity by regressing phenotype values on estimated allele dosage for each of the 8 founder alleles. This approach assumes that heterozygotes will have phenotypes that are intermediate between the corresponding homozygous genotypes. This approach does not account for dominance but it has proven to be effective in practice. Further developments can be expected and are the subject of ongoing investigation.
Allele Effects
The 8-state regression provides estimates of the additive genetic effects associated with each parental haplotype. In an intercross, estimated allele effects can indicate which parent has contributed the “high” allele. Allele effects in the DO provide much more information because they partition the 8 founder haplotypes into groups. Causal variants in the DO will most likely share the same grouping pattern as the estimated allele effects, which in some cases can dramatically reduce the number of candidate variants. Access to full genomic sequences of founders ensures that the screening of candidate SNPs is comprehensive (Keane, et al., 2011).
Allele effects can distinguish chance co-location of QTL from pleiotropy. This can be helpful for establishing causal relationships among traits, including both clinical and gene expression traits. If two traits share a common causal variant, they should have similar allele effects. QTL with distinct patterns of allele effects are most likely driven by different genetic variants.
Population structure in the DO
Intercross mice are all equally related to one another through a simple pedigree. In contrast, DO mice are related to one another in variable degree through an extended and complex pedigree. It is important to account for these relationships in the mapping analysis. This is achieved by including a “random effect” that reflects the degree of kinship between pairs of animals. Correlation between closely related animals will reduce power for QTL detection and the effect will be greater when many closely related animals are included in the study population. For this reason we try to avoid including siblings in a single study. The effect of kinship correction is minor for more distant relationships. Tracking the DO pedigree is impractical and therefore we use SNP genotypes to estimate kinship. The QTLRel software (Cheng et al., 2010) has been adapted for DO mapping and includes functions to estimate additive and dominance components of kinship.
Example: Mapping Cholesterol QTL
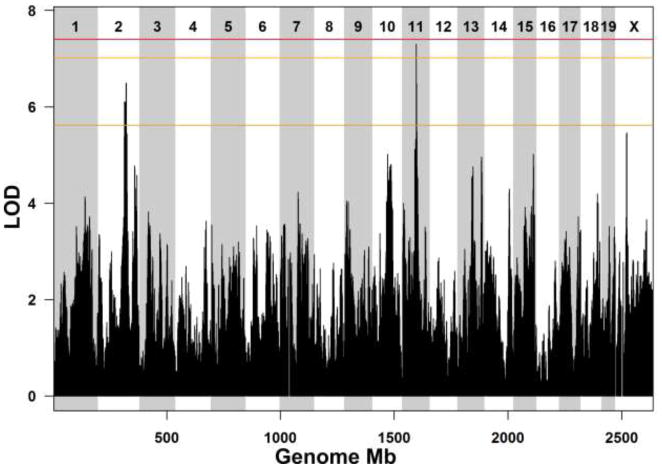
We illustrate QTL analysis in the DO by mapping 17-week plasma cholesterol in a subset of 141 of the mice from the (Svenson et al., 2012) study. A genome scan identifies suggestive QTL on chromosomes 2 and 11 (Figure 2). The Chr 11 QTL spans from 60.1 to 65.3 Mb and is within a larger interval that has been associated with HDL cholesterol in previous mapping studies (Machleder et al,1997; Wittenburg et al., 2006). There are 82 genes in this interval and 86,665 SNPs that are polymorphic among the 8 DO founder strains (Keane et al., 2011). The PWK/PhJ allele is associated with low cholesterol values (Figure 3A). However SNPs and indels that are private to PWK are abundant (18,690 private PWK SNPs and 93 private PWK indels) and distributed throughout the QTL interval, and do not help to narrow the candidate gene list.
Figure 2.
LOD score profile for week 17 plasma cholesterol levels. The horizontal axis shows genome location and the vertical axis shows the LOD score. Horizontal lines are permutation derived significance thresholds of 0.05 (red), 0.1 (orange) and 0.63 (yellow). The genome scan identifies two suggestive QTL loci, one on chromosome 2 and another on chromosome 11.
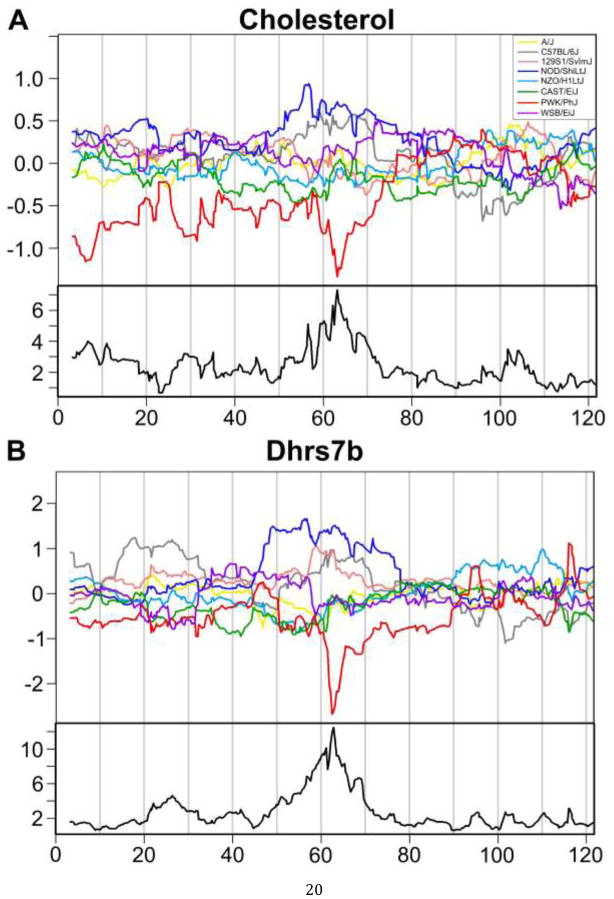
Figure 3.
Estimated allele effects along chromosome 11 are shown for week 17 cholesterol (A), and gene expression of Dhrs7b (B). The upper panel of each figure shows centered estimates of the effects of the eight founder alleles, with color code as indicated in the legend. The lower panel shows the LOD profile.
In addition to clinical phenotypes, we have obtained liver gene expression data by RNA-seq in 91 of the 141 DO animals in this study. Data and methods will be described elsewhere. Briefly, bar-coded cDNA libraries were prepared from total liver RNA with the TruSeq kit (Illumina) according to manufacturer’s protocols, and were sequenced on the HiSeq 2000 (Illumina). Paired-end 100bp reads were aligned to the mouse reference genome (NCBI37/mm9) with TopHat v1.3.2. Transcript abundance in Fragments Per Kilobase of Exon model per Million mapped fragments (FPKM) was calculated using TopHat and Cufflinks v1.0.3 without quartile normalization. Transcript abundance was transformed to normal scores and mapped using QTLRel.
We examined the allele effects for expression patterns of each of the 82 genes in the QTL region and identified a single gene (Dhrs7b) that closely matched the cholesterol QTL (Figure 3B). By itself, this is not conclusive evidence for a causal role of Dhrs7b. However this gene is now a strong candidate for additional studies to clarify its potential function in the regulation of plasma cholesterol.
Investigation of additional mouse resources revealed that Dhrs7b was knocked out by Lexicon-Genentech and included in the KOMP Phenotyping Pilot project (http://www.kompphenotype.org/). Mice homozygous for the null mutation are not viable, and therefore heterozygotes were phenotyped. No significant difference in plasma total cholesterol was observed in mutants compared to wild-type controls. The knockout strain background is B6;129S5, and therefore a direct phenotypic comparison to the DO is not necessarily the right one. Our results suggest that a PWK allele drives a lower plasma cholesterol level, and that PWK has lower expression of Dhrs7b. Further, if Dhrs7b is in fact the causal gene underlying the cholesterol QTL, it is not clear by what mechanism the PWK allele is influencing cholesterol levels, or that the effect is due to a loss of function.
Sample Size for QTL mapping studies
In genetic mapping studies, sample size together with variability due to measurement, environment and background genetic effects, the effect size and allele frequency of the causal variant will determine power to detect a QTL. Multiple test adjustment and kinship structure may also affect power and sample sizes. With so many variables to consider, a universal recommendation for sample size is not practical, but some general guidelines can be offered. In the best case -- a QTL with additive effects, allele frequency of 50%, and a mapping population with no siblings -- we can expect the DO to perform essentially like an intercross. While studies with smaller sample sizes can be successful, samples of N > 200 have proven to be most effective. It is helpful to consider a worst-case scenario of a private allele (frequency 1/8) with a recessive effect. The relevant genotype will occur in 1/64 of the study population. A recessive effect that shifts a trait mean by 1 standard deviation would likely be biologically important. To ensure that we will detect such an effect using a statistical test of size alpha = 0.05 (a “suggestive” QTL after genome-wide multiple test adjustment) and power 1-beta = 0.80, we should aim to obtain 8 animals with the homozygous recessive genotype (van Belle, 2002). Let n be number of animals with the recessive genotype at a given locus and let N be the total size of the cross. A binomial calculation indicates that at N=400 we will achieve n>= 8 at 22.8% of all loci; at N=640 we can expect to achieve n>= 8 at 67.9% of loci; and at N=800, 87.7% of potential QTL loci will achieve the desired level of coverage. These sample sizes are large but not unrealistic. It is worth noting that a recessive QTL effect can be validated quickly and easily on a small set of CC inbred strains.
Other applications of the DO
The DO can be useful in applications that do not require genetic mapping, including toxicology screens. Although genetic heterogeneity of the DO will introduce additional variability compared to an inbred strain screen, the ability to generalize conclusions from a genetically diverse population provides a powerful advantage. Genetic diversity can help to avoid the pitfall of identifying an idiosyncratic reaction that occurs only in a limited genetic context, or of missing an effect due to failure to test it in a sensitive genetic background. Generalization from animal studies to humans is always an extrapolation but the risk is magnified when we fail to include natural genetic diversity in the study design. The greater the diversity of the screening population, the more likely we are to encounter sensitive subpopulations.
Sample size is also an important consideration for screening studies. A study that aims to detect a compound with adverse effects in 20% or more of subjects against a spontaneous background rate of 2% would require n = 48 to achieve the usual size (alpha=0.05) and power (1-beta= 0.80). If the adverse reaction rate is 10%, the required sample size increases to n=140. When adverse reactions are rare, such that none are expected to occur, we can place an upper bound of 3/N on the true rate. Thus, to ensure an adverse reaction has a true rate of less than 1%, we would require that no events occur in a sample of 300 animals. These examples illustrate that reasonable sample sizes can provide powerful screening designs using a genetically heterogeneous population.
Validation in the CC
Individual DO animals cannot be replicated. However, genetic loci that are associated with phenotypic effects in DO mice can in principle be reproduced in CC strains or in the F1 progeny of two CC strains. In this way, the CC provides a resource for independent validation of mapping results, including complex multiple locus models, that are detected in the DO. The DO can be viewed as a hypothesis generator for genetic models that can be tested in the CC.
Resources
Informatics resources will extend the power and scope of QTL analysis in the DO. Databases of DO phenotypes and genotypes are still in early stages of development but will soon be available to provide a basis for combined data analysis and integration with the CC. We have listed some resources in Box 1 but we expect these to evolve as use of the DO population increases.
Box 1. Resource Links.
http://cgd.jax.org/datasets/datasets.shtml
Published data and software tools described in this paper are available here.
http://jaxmice.jax.org/strain/009376.html
DO mice are available from The Jackson Laboratory.
A newly established web site (under construction) that will host data and resources to support analysis of DO data.
Acknowledgments
Development and maintenance of the DO has benefitted from the effort and expertise of Lisa Somes, Marge Strobel, Elissa Chesler and Darla Miller and was supported with funding provided by The Jackson Laboratory. Riyan Cheng, Abraham Palmer, and William Valdar contributed to the development of analytical methods described here. Fernando Pardo-Manuel de Villena, Leonard McMillan, and Catherine Welsh at the University of North Carolina at Chapel Hill developed the MUGA array and associated software. Additional support was provided by NIH grants GM076468, GM070683.
References
- Cheng R, Lim JE, Samocha KE, Sokoloff G, Abney M, Skol AD, Palmer AA. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics. 2010;1851033(3):1044. doi: 10.1534/genetics.110.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, Churchill GA, Johnson DK, Manly KF. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome. 2008;19(6):382–389. doi: 10.1007/s00335-008-9135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Cross Consortium. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics. 2012;190(2):389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu OD, Darakjian P, Walter NA, Malmanger B, Oberbeck D, Belknap J, McWeeney S, Hitzemann R. Genetic diversity and striatal gene networks: focus on the heterogeneous stock-collaborative cross (HS-CC) mouse. BMC Genomics. 2010;11:585. doi: 10.1186/1471-2164-11-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellåker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assunção JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477(7364):289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machleder D, Ivandic B, Welch C, Castellani L, Reue K, Lusis AJ. Complex genetic control of HDL levels in mice in response to an atherogenic diet. Coordinate regulation of HDL levels and bile acid metabolism. J Clin Invest. 1997;99(6):1406–1419. doi: 10.1172/JCI119300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov PM, Graber JH, Churchill GA, DiPetrillo K, King BL, Paigen K. Evidence of a large-scale functional organization of mammalian chromosomes. PLoS Genet. 2005;1(3):e33. doi: 10.1371/journal.pgen.0010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. Breeding designs for recombinant inbred advanced intercross lines. Genetics. 2008;179(2):1069–1078. doi: 10.1534/genetics.107.083873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson KL, Von Smith R, Magnani PA, Suetin HR, Paigen B, Naggert JK, Li R, Churchill GA, Peters LL. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J Appl Physiol. 2007 Jun;102(6):2369–78. doi: 10.1152/japplphysiol.01077.2006. [DOI] [PubMed] [Google Scholar]
- Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, Chesler EJ, Palmer AA, McMillan L, Churchill GA. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics. 2012;190(2):437–447. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belle G. Statistical Rules of Thumb. Chapter 2 Wiley; NY: 2002. [Google Scholar]
- Wang JR, Pardo-Manuel de Villena F, Lawson HA, Cheverud JM, Churchill GA, McMillan L. Imputation of single-nucleotide polymorphisms in inbred mice using local phylogeny. Genetics. 2012;190(2):449–458. doi: 10.1534/genetics.111.132381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenburg H, Lyons MA, Li R, Kurtz U, Wang X, Mossner J, Churchill GA, Carey MC, Paigen B. QTL mapping for genetic determinants of lipoprotein cholesterol levels in combined crosses of inbred mouse strains. J Lipid Res. 2006;47(8):1780–1790. doi: 10.1194/jlr.M500544-JLR200. [DOI] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat Genet. 2007;39(9):1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH, Nachman MW, Pialek J, Tucker P, Boursot P, McMillan L, Churchill GA, Pardo-Manuel de Villena F. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet. 2011;43(7):648–655. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]