Abstract
Hybrid dendritic-linear block copolymers based on a 4-arm polyethylene glycol (PEG) core were synthesized using an accelerated AB2/CD2 dendritic growth approach through orthogonal amine/epoxy and thiol-yne chemistries. The biological activity of these 4-arm and the corresponding 2-arm hybrid dendrimers revealed an enhanced, dendritic effect with an exponential increase in cell internalization concomitant with increasing amine end-groups and low cytotoxicity. Furthermore, the ability of these hybrid dendrimers to induce endosomal escape combined with their facile and efficient synthesis makes them attractive platforms for gene transfection. The 4-arm-based dendrimer showed significantly improved DNA binding and gene transfection capabilities in comparison with the 2-arm derivative. These results combined with the MD simulation indicate a significant effect of both the topology of the PEG core and the multivalency of these hybrid macromolecules, on their DNA binding and delivery capablities.
Keywords: Dendrimer, polyethylene glycol, click chemistry, fluorescence microscopy, drug delivery, gene delivery
INTRODUCTION
The modular structure, plurality of functional end-groups and monodispersity of dendrimers make them appealing scaffolds for biomedicine,1-7 enabling a broad spectrum of applications including visualization of sub-cellular processes,8 as delivery agents9-10and as scaffolds for vaccines/anti-viral agents.11–13 While well-suited for these architectures, the synthesis of dendrimers is still considered to be a time-consuming process requiring rigorous purification processes.14 Furthermore, partial functionalization with bioactive moieties, which are often hydrophobic, leads to low loading and results in polydisperse materials.15
To address these challenges and simplify their preparation, several groups have reported the use of polyethylene glycol (PEG) as a difunctional or monofunctional polymeric core for the divergent synthesis of dendrimers.16–18 These strategies rely on the solubility of the PEG core to simplify purification of the PEG-dendrimer hybrids by dialysis or precipitation. Additionally, Park and coworkers demonstrated increased colloidal stability and reduced toxicity for PAMAM-PEG-PAMAM copolymers when compared to commercial PAMAM dendrimers.19 Recently, we reported the accelerated synthesis of internally-functionalized PEG-dendrimer hybrids and their application as a dual-functional delivery platform.20 Based on a bi-functional PEG core, a 4th generation dendrimer with 32 orthogonal surface groups and 20 internal functionalities was synthesized in only four steps. The success of this approach prompted the exploration of alternative hybrid dendritic architectures in order to understand their structure-property relationship with respect to biological activity.
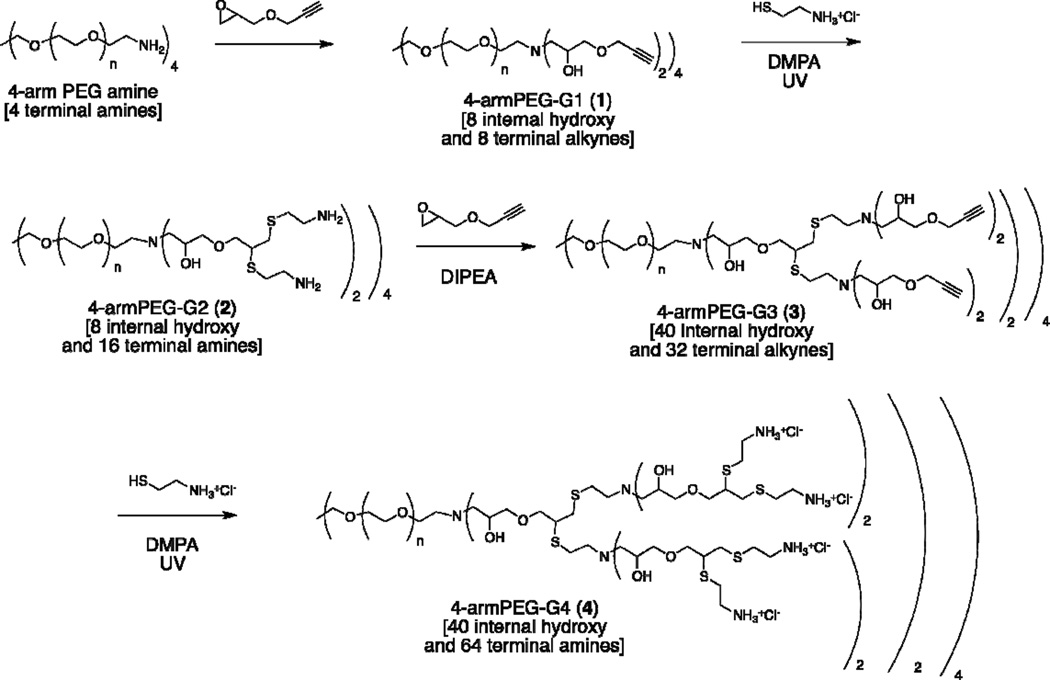
Herein we report the synthesis of amine-terminated hybrid dendrimers, based on a 4-arm PEG star core, in only four steps, with precipitation and dialysis as the only means of purification. The facile synthesis, simple purification, and compatibility of the alkyne groups of the third generation with both thiol-yne21–23 and copper-catalyzed azide-alkyne24–26 click chemistries, make these dendrimers highly attractive scaffolds for various biomedical applications. The role of architecture and number of end groups in determining the interaction of these 2-arm and 4-arm based hybrid cationic dendrimers with living cells was evaluated with respect to toxicity, membrane affinity, and internalization pathway (Figure 1).
Figure 1.
Schematic representation of the structures of 2nd and 4th generations of 2-armPEG-Gn and 4-armPEG-Gn series with increasing number of cationic amines at the chain ends.
EXPERIMENTAL SECTION
Materials
All solvents (ACS grade, Fisher) were used as received. Tetra-amine PEG 10K was purchased from Laysan Bio. 2,2’-Azobis(2-methylpropionitrile) (98%), cysteamine hydrochloride (98%) and 2,2-dimethoxy-2-phenylacetophenone (99%) were purchased from Aldrich. Glycidyl propargyl ether (technical >90%, Santa Cruz Biotechnology), N,N-diisopropylethylamine (98%, TCI America), Alexa Fluor 488 and 647 (Invitrogen) and deuterated solvents for NMR (Cambridge Isotope Laboratories, Inc.) were used as received. Dialysis tubes (regenerated cellulose, spectra/por 6, MWCO 1K) were purchased from Spectrum Laboratories. Dendrimers 2-armPEG-G2 and 2-armPEG-G4 were synthesized as previously reported.20
Instrumentation
1H and 13C NMR spectra were recorded on Varian 500 or Bruker Avance III 800 spectrometers respectively. Chemical shifts are reported in ppm and referenced to the solvent signal. The molecular weights of the dendrimers were determined by comparison of the areas of the peaks corresponding to the PEG block (3.63 ppm) and the proton peaks of the dendrimers. Infrared spectra were recorded on a Perkin Elmer Spectrum 100 with a Universal ATR sampling accessory. Gel permeation chromatography (GPC) analysis was performed on a Waters Alliance instrument equipped with three columns: Eprogen Inc., CATSEC 100 Å, 300 Å and 1000 Å with 1 wt% acetic acid/0.10 M sodium sulfate (aq) as eluent (flow rate: 0.25 ml/min at 25 °C). Detection was achieved with Waters photodiode array (PDA) and refractive index (RI) detectors with linear polyethylene glycol standards used for calibration. MALDI-TOF samples were run on a Voyager DE Pro (Applied Biosystems Instruments) using a stainless steel sample plate. All matrices (2,5-dihydroxybenzoic acid, α-cyano-4-hydroxycinnamic acid, dithranol, and sinapinic acid) were purchased from Sigma Aldrich and used without further purification. Dynamic Light Scattering (DLS) was conducted with a Wyatt DynaPro NanoStar equipped with a 100 mW, 662 nm air-launched laser and a detector at a constant angle of 90°. Solutions of dendrimers 2-armPEG-G4 and 4-armPEG-G4 were prepared using PBS buffer pH 7.4 (buffer was filtered using a Nylon 0.45 µm filter prior to sample preparation). Samples were mixed using a vortex shaker for 1 min and then filtered through Nylon 0.45 µm filters. Measurements were carried at 25 °C using disposable cuvettes.
Synthesis of dendrimers
Dendrimer 4-armPEG-G1
10kD tetra-amine PEG, (1.00 g, 0.1 mmol) was dissolved in MeOH (5.0 ml), glycidyl propargyl ether (1.70 ml, 16.0 mmol) was added and the reaction was stirred overnight. The crude mixture was precipitated into ether and the product was collected by filtration and dried overnight in a vacuum oven at 40 °C, to give the tetraalkyne dendrimer 4-armPEG-G1 (960 mg, 88% yield) in the form of white powder. FT-IR, ν (cm−1): 3240, 2880, 1460, 1360, 1340, 1280, 1240, 1150, 1100, 1060, 960, 840; 1H NMR (500 MHz, CDCl3) δ: 4.19 – 4.14 (m, O-CH2-alkyne), 3.86 – 3.45 (CHO and CH2-O of dendrimer; CH2 of PEG backbone), 3.38 (s, C-(CH2-O)4 of the PEG core), 2.89 – 2.42 (m, CH2N and alkyne-H); 13C-NMR (200 MHz, CDCl3) δ: 79.7, 74.7, 71.7, 70.4, 68.4, 67.3, 59.3, 58.3, 58.2, 55.4, 54.4.
Dendrimer 4-armPEG-G2
Dendrimer 4-armPEG-G1 (500 mg, 0.046 mmol), cysteamine hydrochloride (1.7 g, 15 mmol) and DMPA (38 mg, 0.15 mmol) were dissolved in MeOH (5 ml). The mixture was purged with argon for 10 minutes and then irradiated under UV irradiation for 1 hour. MeOH (5 ml) was subsequently added and the crude mixture dialyzed against DI water (3 × 1.0 L) for 24 hours. The solution was then removed from the dialysis tube and the solvent was removed by freeze-drying to give the dendrimer 4-armPEG-G2 (464 mg, 80% yield) in the form of white solid. FT-IR, ν (cm−1): 3410, 2880, 1470, 1360, 1340, 1280, 1240, 1150, 1100, 1060, 960, 840. 1H-NMR (500 MHz, DMSO-d6) δ: 8.15 (br, NH3 +), 3.74-3.39 (m, CH-O and CH2-O of dendrimer; CH2 of PEG backbone; CH2NH3 + and C-(CH2-O)4 of the PEG core), 3.19 – 2.78 (m, CH2N; CHS and CH2S); 13C-NMR (200 MHz, DMSO-d6) δ: 72.7, 72.1, 70.7, 69.6, 68.9, 44.4, 38.6, 33.3, 28.7, 27.4.
Dendrimer 4-armPEG-G3
Dendrimer 4-armPEG-G2, (460 mg, 0.036 mmol) was dissolved in MeOH (3.0 ml), glycidyl propargyl ether (5 ml, 46 mmol) and DIPEA (0.2 ml, 1.2 mmol) were added and the reaction was stirred overnight at room temperature. The crude mixture was precipitated into ether and the product was collected by filtration and dried overnight in a vacuum oven at 40 °C, to give dendrimer 4-armPEG-G3 (549 mg, quantitative) in the form of very light yellow powder. FT-IR, ν (cm−1): 3240, 2920, 2850, 1460, 1350, 1300, 1250, 1090, 950, 850. 1H-NMR (500 MHz, CDCl3) δ: 4.19 (m, O-CH2-alkyne), 3.89 – 3.35 (m, CH-O and CH2-O of dendrimer; CH2 of PEG backbone and C-(CH2-O)4 of the PEG core), 3.10 – 2.41 (m, CHS; CH2S; CH2N and alkyne-H); 13C-NMR (200 MHz, CDCl3) δ: 79.7, 78.5, 77.9, 75.0, 73.6, 73.2, 71.8, 70.5, 70.3, 68.3, 58.6, 58.4, 55.0, 54.7, 45.8, 35.0, 31.5, 30.1.
Dendrimer 4-armPEG-G4
Dendrimer 4-armPEG-G3 (100 mg, 6.4 µmol), cysteamine hydrochloride (930 mg, 8.2 mmol) and DMPA (21 mg, 0.082 mmol) were dissolved in MeOH (2.0 ml). The mixture was purged with argon for 15 minutes and then irradiated under UV irradiation for 1 hour. The solvent was evaporated and the crude mixture was dissolved in DI water (10.0 ml) and dialyzed against DI water (3×1.0 L) for 24 hours, followed by lyophilization to yield amino-functionalized dendrimer 4-armPEG-G4 in the form of yellow solid (129 mg, 88% yield). FT-IR, ν (cm−1): 3380, 2880, 1610, 1470, 1340, 1280, 1240, 1100, 950, 840. 1H-NMR (500 MHz, DMSO-d6) δ = 8.24 (br, NH3 +), 3.80 – 3.21 (m, CH-O and CH2-O of dendrimer; CH2 of PEG backbone; CH2NH3 + and C-(CH2-O)4 of the PEG core), 3.19 – 2.56 (m, CH2N; CHS and CH2S); 13C-NMR (200 MHz, DMSO-d6) δ: 73.3, 72.0, 70.2, 69.6, 69.1, 67.4, 44.8, 38.7, 33.4, 28.5, 27.5.
General Procedure for Fluorophore Labeling of Dendrimers
Dendrimers were conjugated with Alexa488 or Alexa647 fluorophores to afford labeled dendrimers following previously reported procedure.27,28 Briefly, conjugation was carried out via amide bond formation between the primary amine of the dendrimer and the N-hydroxysuccinimide activated carboxyl of the fluorophores. Dendrimers (50 nmol) were dissolved in DMSO, the reactive dye (1eq) added and the solution stirred for 4 h at room temperature and then dialyzed against water (MWCO = 10 kDa) to afford the desired dendrimer-dye conjugates, with an estimated average of one dye per dendrimer. The fluorescence intensity of different dendrimers was evaluated and normalized to account for differences in their brightness.
Flow Cytometry Measurements
HeLa cells were grown in a six well plate and after treatment with Alexa488-labeled dendrimer, were detached using trypsin-EDTA, washed with PBS buffer and fixed with 4% paraformaldheyde (PFA). Cells were washed with PBS until complete removal of PFA and finally resuspended in 250 µL of PBS. Flow cytometry was performed on a MACSQuant system (Miltenyi) by counting 10,000 events.
Cell Culture and confocal imaging
HeLa (CCL-2) were purchased from ATCC and cultured following manufacturer’s instructions. For live cell microscopy cells were plated onto 35mm glass-bottom dishes (WillCo-dish GWSt-3522) and imaged at 37 °C 5% CO2.
Cell imaging was performed on a Leica TCS SP2 inverted confocal microscope (Leica Microsystems) equipped with a 40× 1.25 NA oil immersion objective (Leica Microsystems). Imaging was obtained illuminating the samples with the inline Ar and He-Ne lasers of the microscope and with a 403 nm pulsed diode laser (M8903-01; Hamamatsu) at 50 MHz repetition rate. Fluorescence emission was collected with the AOBS-based built-in detectors of the confocal microscope (Hamamatsu R6357).
For the toxicity assay, HeLa cells were incubated for 2 h at 37 °C with DMEM containing 8 µg/mL propidium iodide, 1 µg/mL of calcein AM and different concentrations of dendrimers. The medium was then discarded and the cells were washed with PBS buffer containing the same concentration of propidium iodide before confocal imaging.
Internalization Assay and Colocalization Studies
To monitor dendrimer internalization cells were incubated with 100 nM of labeled dendrimers in DMEM for 1 h at 37 °C. To remove unbound molecules from the medium, cells were rinsed two times with PBS. After the initial preloading and subsequent washing, cells were incubated again in DMEM and imaged at the indicated time point. In order to identify the endocytic vesicles involved in dendrimer internalization, we performed colocalization assays in living cells. For this, HeLa cells were coincubated with dendrimers (as described above) and different dyes: these include 1mg/mL 70kDa dextran-FITC conjugate at 37 °C for 30 min to label macropinosomes, 50 mM Lysosensor for 10 min to label lysosomes, and 2 µg/mL transferrin Alexa488 conjugate to label recycling and sorting endosomes. Images were analyzed using ImageJ software version 1.37 (NIH Image; http://rsbweb.nih.gov/ij/).
Endosomal escape assay
For a typical calcein assay, cells were seeded 24 hours before the experiment in WillCo-dishes to reach a 70% confluence. Medium was replaced with 1.0 mL of DMEM containing the dendrimer together with 250 µM of Calcein. After 2 h of incubation at 37 °C cells were washed three times with PBS and then analyzed by confocal microscopy.
Ethidium bromide intercalation assay
Ethidium bromide (1µg/ml) and DNA (3µg/ml) were dissolved in 0.05 mol/l Tris–HCl buffer with 50 mmol/l NaCl (pH = 7.4). The fluorescence spectra of ethidium bromide (EB) in the presence of DNA before and after addition of dendrimers were taken with a Cary Eclipse fluorimeter. EB was excited at 477 nm and the emission spectra were recorded from 490 to 850 nm. A sample of EB with dendrimer was studied to check that no interaction of the macromolecule with the dye is present.
Transfection experiments and confocal imaging
HeLa cells were grown in DMEM (Invitrogen) supplemented in 10 % FCS at 37°C and 5% CO2. For imaging experiments, cells were seeded in 8-well chambered ibiTreat µ-Slides (Ibidi GmbH, Munich) at a density of 10000 cells per well. Plasmid DNA (pCMVLuc) was labeled with Cy5 fluorophore using the Label IT kit (MIRUS, Madison, WI, USA) according to the manufacurer’s instructions. Dendriplexes were generated by incubating DNA (Cy5 labeled pCMVLuc for single particle imaging experiments, unlabeled peGFPNuc for gene expression) with different dendrimers at N/P = 4 for 30 minutes at room temperature in HBG buffer (20 mM Hepes pH 7.1, 5% glucose w/v). 20 µl of dendriplexes equivalent to 400 ng DNA, were administered to cells in a total volume of 240 µl serum-free CO2 independent cell medium (Invitrogen). Cells were washed three times in PBS buffer (either after 1h or 5h) and incubated in CO2 independent medium supplemented with 10 % FCS. Single cells were analyzed on a heated microscope stage by spinning disk confocal microscopy using a Nikon TE2000E microscope, the Yokogawa CSU10 spinning disk unit, an EMCCD camera (iXon DV884; Andor) and a Nikon 1.49 NA 100x Plan Apo oil immersion objective. Cells were imaged either after 1h or 22h. Z-stacks were imaged with 641 nm laser excitation at 300 ms per frame and with a spacing of 166 nm between two planes. Z-projections of the recorded image sequences were built in Image J. GFPNuc expression was detected by widefield microscopy on a custom build setup based on a Nikon Ti microscope equipped with a 10x or 20x objective. The total fluorescence signal per field of view was calculated in ImageJ by summing all pixel intensities above a set intensity threshold. To analyze the fluorescence intensity per dendriplex, single particles were identified in ImageJ using a defined size restriction and an intensity threshold criterion.
RESULTS AND DISSCUSION
Synthesis
4-armPEG-Gn dendrimers of generations 1 through 4 were synthesized using a sequence of amine/epoxy and thiol/yne reactions as presented in Scheme 1. The 10 kDa 4-arm PEG amine, which serves as a core, and an excess of commercially available glycidyl propargyl ether (GPE) were stirred in methanol overnight to give the hybrid dendrimer 4-armPEG-G1 (1) in 84% yield after precipitation in ether. Subsequent thiol-yne reaction with excess cysteamine hydrochloride gave 4-armPEG-G2 (2) in 80% yield after dialysis. Repetition of this 2-step epoxy ring-opening/thiol-yne growth strategy then gave 4-armPEG-G3 (3) in near quantitave yields with the fourth generation hybrid dendrimer 4- armPEG-G4 (4) being obtained in 88% after sequential thiol-yne reaction followed by dialysis.
Scheme 1.
Synthesis of 4-armPEG-G4 dendrimer, 4, with 40 internal OH groups and 64 terminal NH2 groups.
This accelerated dendritic growth strategy, coupled with a starting tetravalent PEG-core allows a fourth generation dendrimer, having a total of 64 chain end amino groups and 40 internal hydroxy functional groups, to be prepared in only four steps. Furthermore, the high molecular weight 4-arm PEG core (10 kDa) permits the use of precipitation or dialysis as the only means of purification, greatly simplifying the synthetic process.
Dendrimer-Cell interactions
Previous studies have shown that overall structure, generation, and number of functional groups have important implications for biological activity of various dendrimers.27–36 Thus, an extensive evaluation of the interactions of 2-arm and 4-arm PEG-dendrimer hybrids with living cells was carried out with particular attention to toxicity, cell internalization ability, and their capability to induce endosomal escape, which are three crucial factors relevant for intracellular delivery applications. In order to study the biological properties of these dendrimers by confocal microscopy and flow cytometry (FACS) the PEG-dendrimer hybrids were fluorescently labeled with Alexa-488.
Toxicity Assay
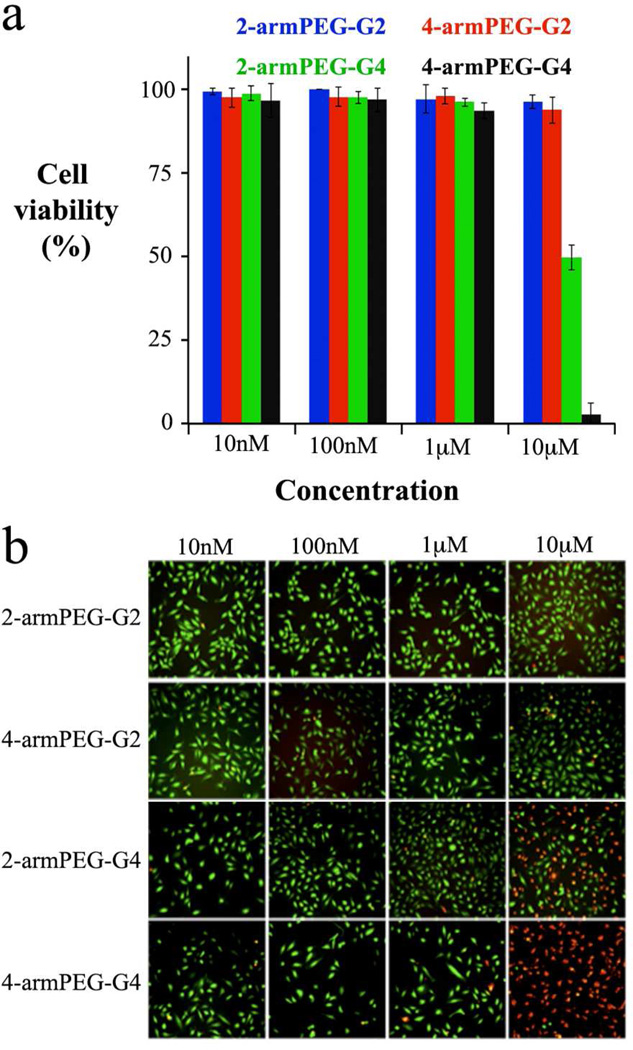
Cell viability with respect to dendrimer structure and concentration was assessed by a propidium iodide (PI) and calcein acetoxymethyl ester (AM) assay. PI dye cannot permeate intact cell membranes and it can be internalized only when the membrane is defective, which allows monitoring of cell viability as PI permeates only dead cells. On the other hand, the calcein AM dye readily passes through the cellular plasma membrane into the cytoplasm, where the acetyl groups are cleaved by esterases to yield the more hydrophilic calcein. Trapped inside the cell, the calcein can readily bind to intracellular calcium, resulting in a strong fluorescence. As dead cells lack cytoplasmatic esterases, fluorescence is observed only in live cells. Figure 2 shows images of PI (in red) and calcein AM (in green) in HeLa cells incubated with dendrimers at 10 nM, 100 nM, 1.0 µM and 10 µM concentrations. High cell viability was observed for all dendrimers up to a concentration of 1.0 µM and the compounds were found to be toxic only at a concentration of 10 µM. At this concentration, the dendrimers’ toxicity is correlated to the number of functional groups, as the higher generation dendrimers show higher toxicity, which agrees well with previous reports of cationic PAMAM dendrimers containing an equivalent number of end groups.27
Figure 2.
Dendrimer toxicity in HeLa cells. The histogram (a) reports the cell viability measured with PI/calcein AM assay after cells incubation with different amount of dendrimer. (b) Imaging of calcein AM (green = live cells) and PI (red = dead cells).
Cellular Uptake
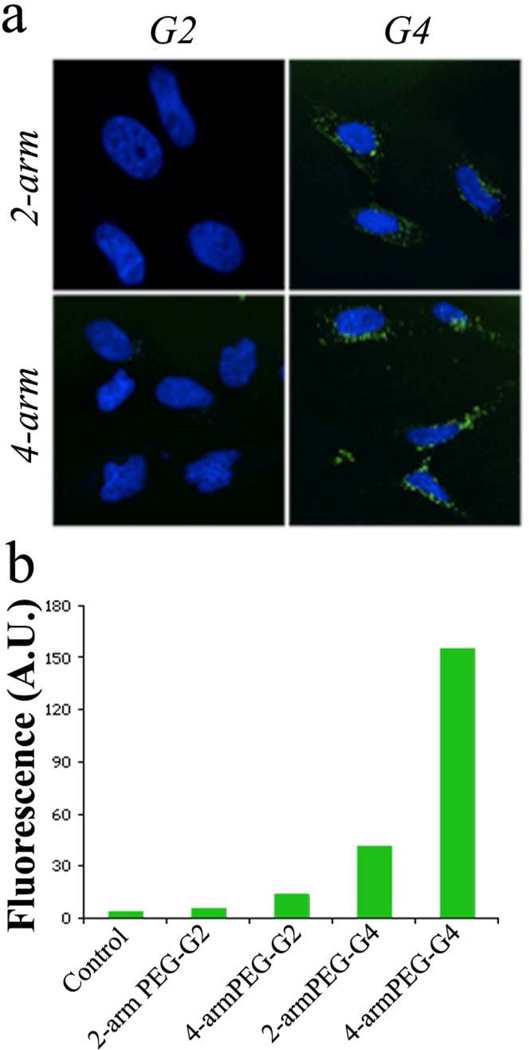
A flow cytometry assay was performed to quantify cell uptake of different dendrimers. Based on the toxicity results, we expected the hybrid dendrimers to show a similar trend in their membrane affinity, and consequently in cell internalization, when compared to cationic dendrimers where higher cells affinities typically correlate with higher cell toxicity. HeLa cells were incubated for 30 min with the dendrimers and the membrane binding was quantified by flow cytometry. The FACS (Figure 3) results indicate negligible binding for the smaller 2nd generation 2-armPEG and 4-armPEG dendrimers. In direct contrast, membrane affinity was observed to increase as the number of end groups increased with a notable exponential jump for the 4th generation 4-arm derivative. This non-linear trend cannot be explained a simple, increased number of charged ammonium moieties at the chain ends of the dendrimers and the pronounced difference in performance between the 2-arm and 4-arm hybrid dendrimers must be due to the influence of the core architecture. To further understand the origin of this promising balance between toxicity and cell affinity for the 4th generation, 4-arm derivative, an in-depth analysis of the solution structure and their biological performance as a gene delivery platform was undertaken.
Figure 3.
Cell binding and internalization. (a) Confocal imaging of HeLa cells treated with AF488-labeled dendrimers at 100nM concentration (nuclei were visualized with Hoechst (blue)) and (b) membrane binding quantification of HeLa cells treated for 30 min with PEG-dendrimer hybrids.
Internalization Pathways
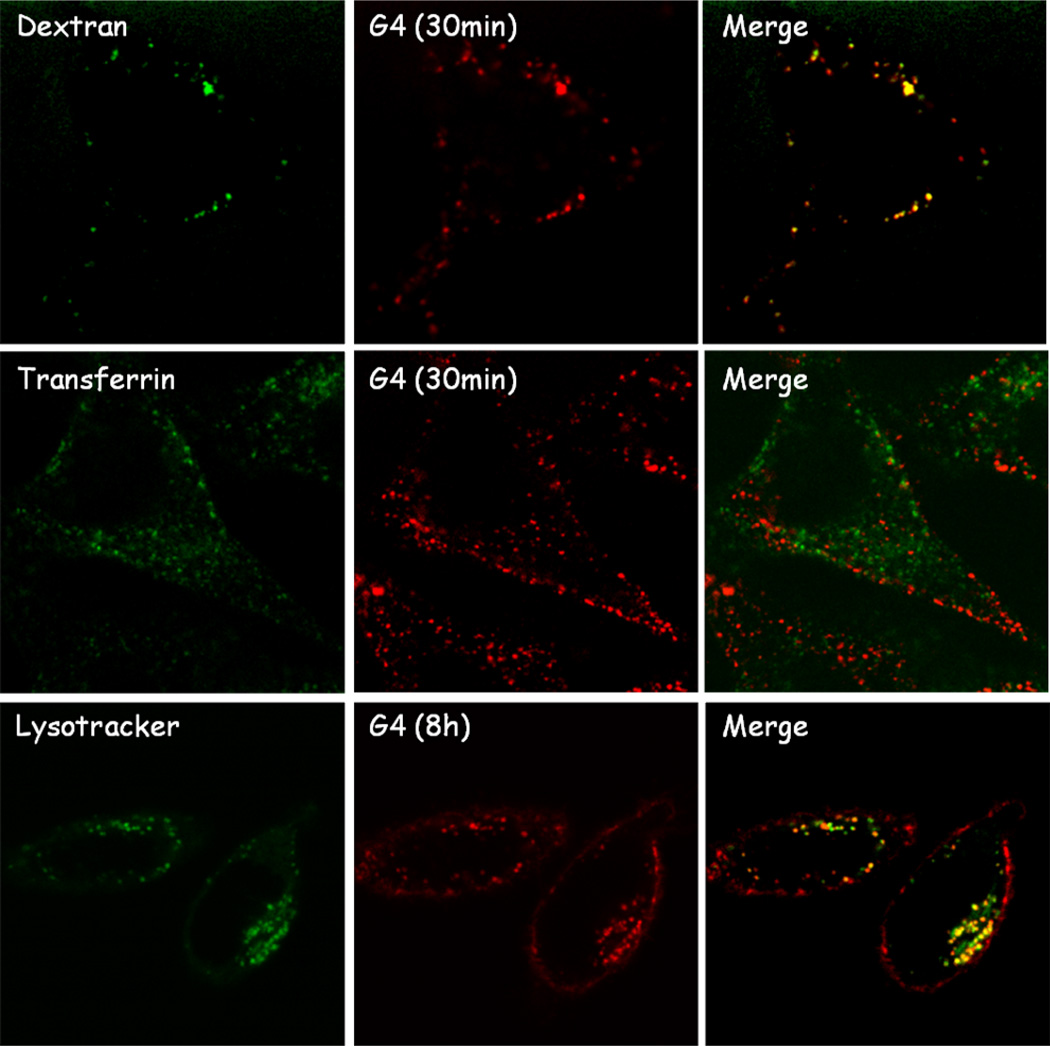
In order to identify the biological pathways involved in internalization of these hybrid dendritic macromolecules, colocalization assays with biomarkers for different endocytic vesicles were performed. Transferrin-Alexa 488 and 70kDa dextran-FITC were used to trace clathrin-dependent endocytosis and macropinocytosis, respectively. Preliminary studies showed the colocalization of the 4-arm, 4th generation PEG-dendrimer, 4, with the dextran marker (Figure 4), which suggests internalization by macropinocytosis with the clathrin-mediated pathway not playing a significant role. Interestingly these results differ from what observed for PAMAM dendrimers that are internalized both via micropinocytosis and clathrin-mediated endocytosis,27 but are in good agreement with recently reported studies of other PEG-dendron hybrids. 28,37 More detailed studies showed that the dendrimers reach their final destination in the perinuclear region in ca. 8h and a colocalization assay with lysotracker at this time point showed localization in the lysosomal compartment, similarly to other cationic dendrimers such as PAMAM.27 No further changes in localization have been observed after lysosomal delivery up to 48h suggesting the lysosome represent the final destination of dendrimers inside the cell. This lysosomal localization is of particular interest for delivery applications owing to the low pH and high activity of hydrolytic enzymes in this compartment which allows for the design of tailored dendritic platforms responsive to the specific biochemical properties of the lysosome.20,37
Figure 4.
Colocalization of 4-armPEG-G4 dendrimer (100mM, in red) with endocytic vesicle markers (in green). Strong colocalization signal was observed for the macropinocytosis marker (top) and lysosome marker (bottom).
Endosomal Escape
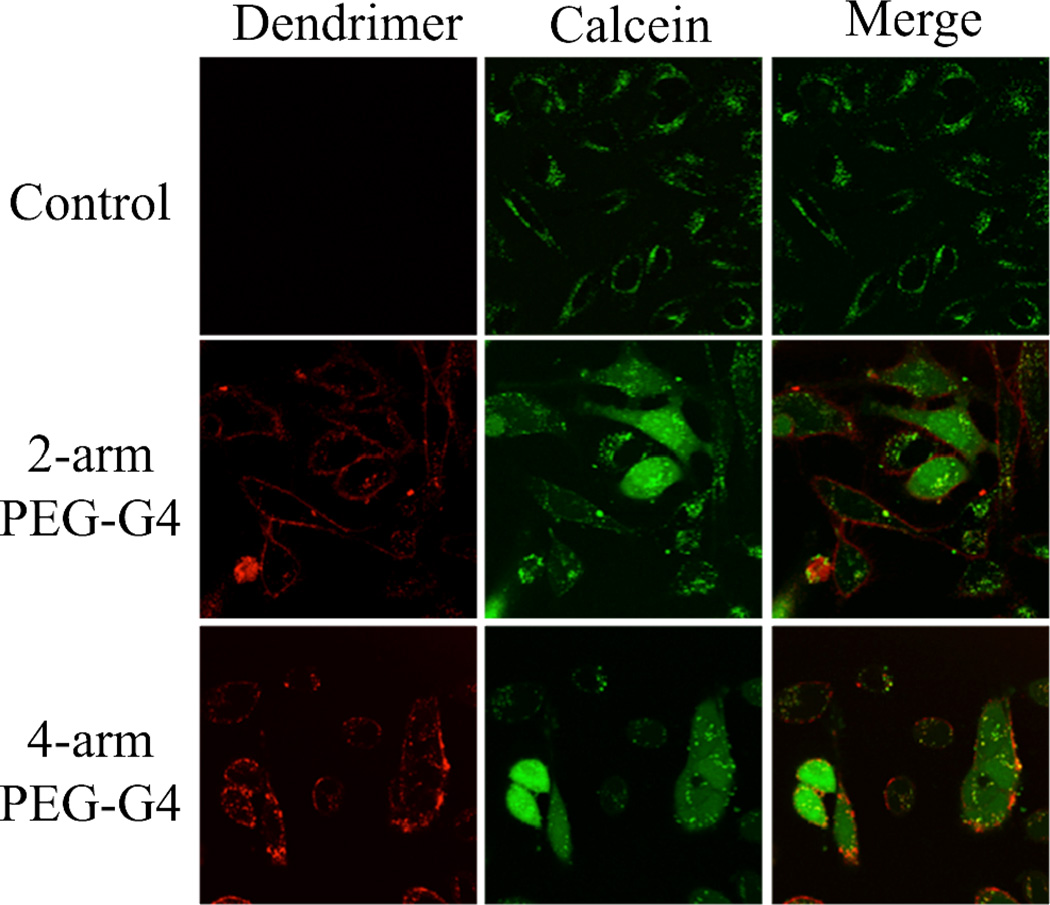
In a recent report, we showed the release of coumarin dyes from a 2-armPEG-G4 in B16 cells.20 After cleavage of covalently loaded coumarins from the dendritic carrier, the released dyes showed a cytoplasmic localization while the dendrimers remain in the endolysosomal system. To investigate the role of our hybrid dendrimers on the endosomal escape of covalently attached dye molecules, the ability of the dendrimers to induce endosomal escape of co-incubated calcein was investigated. Figure 5 shows confocal fluorescence images of HeLa cells after treatment with calcein alone or with a mixture of calcein and the 2-arm and 4-arm 4th generation dendrimers. As can be seen in the upper panel, cells incubated only with calcein show green fluorescence localized in endocytic vesicles with no signal coming from the cytoplasm. This is expected as the hydrophilicity of the calcein molecules does not allow them to penetrate the membrane of the vesicles, and hence, the molecules are locked within endosomal and lysosomal vesicles. In direct constrast, for cells incubated with the 2-arm and 4-arm 4th generation dendrimers, green fluorescence from calcein is observed in the cytoplasm, demonstrating that the dendrimers are able to destabilize vesicles and allow for the escape of small molecules. Notably the dendrimer still localizes in the endosome, indicating that the endolysosomal vesicles are destabilized but they still retain their shape and some degree of integrity. This is of great importance in order to reduce the toxicity associated to the leakage of lysosomal enzymes. The similar performance for both dendrimers may be related to the proton sponge effect, which is characteristic of dendrimers rich in tertiary amines, 3,10 which is the case for both the 2-arm and 4-arm derivatives. These cell-based studies are critical as they confirm the ability of our dendrimers to address two key issues in designing delivery platforms: cell internalization and endosomal escape.
Figure 5.
HeLa cells treated with calcein only (top), calcein with 200nM of 2-armPEG-G4 dendrimer (middle) and calcein with 200nM of 4-armPEG-G4 dendrimer (bottom). Calcein and dendrimer fluorescence is represented in green and red, respectively.
Molecular Simulations
The biological results presented in the previous sections suggest a direct correlation between performance and the macromolecular structure with the number of amine end-groups and architecture playing important roles. In order to investigate the details of these structural effects, Molecular Dynamics (MD) simulations were performed on the 2-armPEG-G4 and 4-armPEGG4 dendrimers.
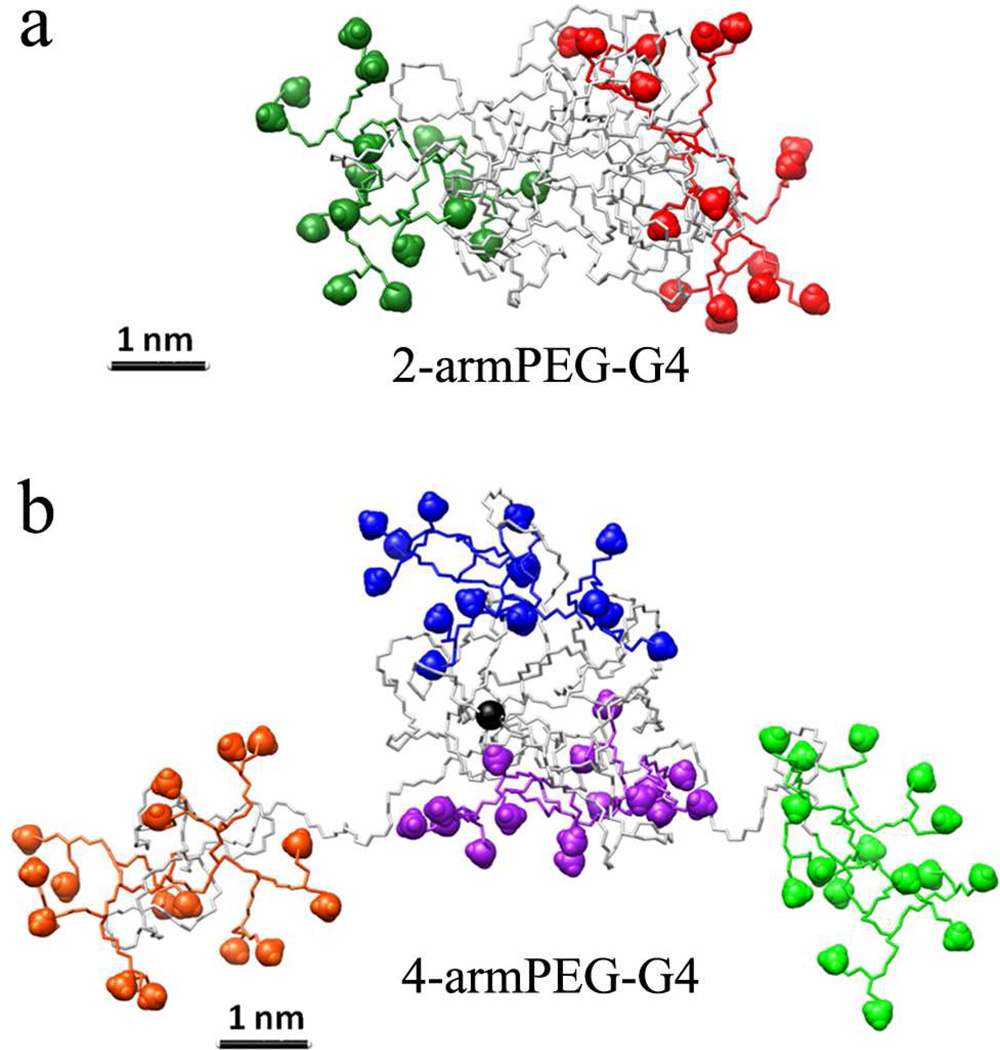
The structure of 2-armPEG-G4 was created and simulated according to the procedure described in the Computational Methods section (SI). Figure 6a reports snapshots taken from the equilibrated phase of the MD simulation and shows that the PEG core of 2-armPEG-G4 dendrimer collapses into a globular shape, exposing the dendrons at both sides. Despite its relative hydrophilicity, the collapse of the PEG core can be attributed to the tendency of the polymer to decrease the surface exposed to the surrounding aqueous solution. This is in good agreement with previous MD studies of dendrimers decorated with PEG chains.38,39 The swelling of the dendrons results from electrostatic repulsion between their charged amino end-groups groups with folding of the PEG core and stability of this collapsed structure being in good agreement with DLS measurements, which showed a diameter of 5 nm for the 2-armPEG-G4 and 9 nm for the 4-armPEG-G4 (SI).
Figure 6.
Molecular dynamics snapshot of the equilibrated structure for the 2-armPEG-G4 (a) and 4- armPEG-G4 (b) dendrimers. Simulations were carried out in water pH7.4 containing 150mM of NaCl.
Interestingly, molecular dynamics of the 4-armPEG-G4 (Figure 6b) showed significantly difference behavior with the PEG core of the 4-armPEG-G4 dendrimer being less dense than the corresponding 2- arm derivative (Figure 6a). To understand the influence of architecture and core branching on the dynamic difference between 2-arm PEG and 4-arm PEG hybrid dendrimers, the average distances between each dendron and the center of the PEG core were calculated over the duration of the MD simulation. The plots reported in Figure 7, together with the snapshots taken from different time points during the simulation, illustrate the differences in dynamic behavior between the two dendrimer-PEG hybrids. From the simulations, it is evident that the equilibrated configuration of the 2-armPEG-G4 remains stable during the entire 200 ns of MD simulation (Figure 7a). In direct contrast, the 4-armPEGG4 hybrid dendrimer assumes a much more dynamic structure with modeling showing that the 4-arm PEG core can accommodate only two of the four dendrons stably (purple and blue D3 and D4 in Figure 7b), while the other two dendrons fluctuate away from the center. The “oscillatory” behavior of dendrons D1 and D2 (Figure 7b, red and green) is most likely due to a competition between the characteristic tendency of the PEG to fold and the electrostatic repulsion between the charged amine end-groups. This leads to increased exposure of the dendrons to the surrounding aqueous environment and a reduced influence of the steric hindrance of the PEG core. It should be noted that this simulated phenomenon agrees well with the improved membrane-binding ability of the 4-arm based hybrid dendrimer, and with the transfection measurements, vide infra.
Figure 7.
Plots of the distances between the centers of each dendron (Dn) and that of the PEG core for 2-armPEG-G4 (a) and 4-armPEG-G4 (b).
DNA Binding and Gene Transfection
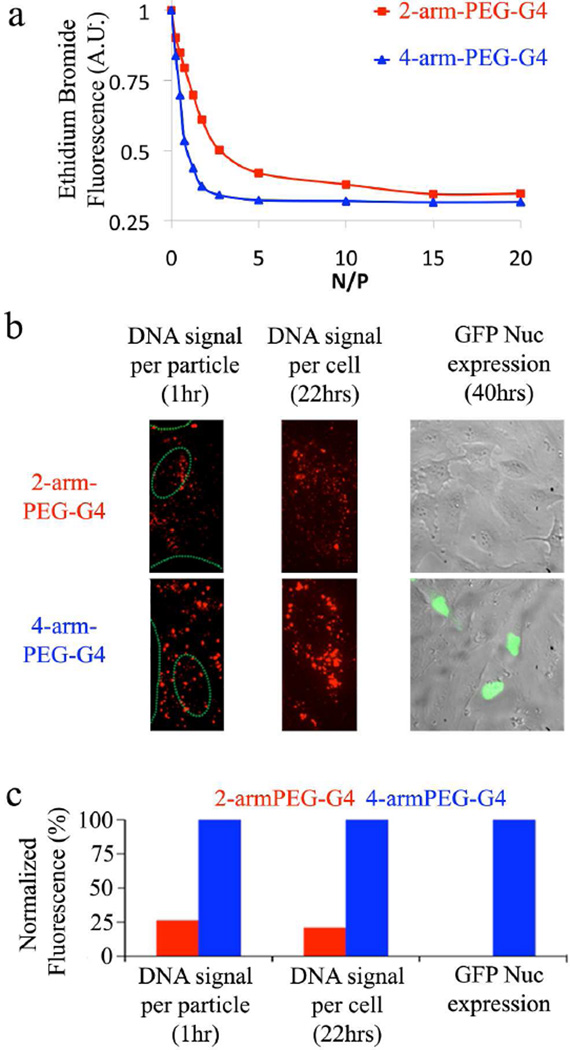
The unique structural dynamics of the 4-armPEG-G4 coupled with the numerous cationic surface groups, cell-internalization ability and endosomal destabilization suggests that these hybrid dendritic systems may be attractive candidates for binding of negatively charged DNA and subsequent delivery to the cytoplasm for gene therapy applications.8,19 In order to evaluate the DNA binding ability of the 2-armPEG-G4 and 4-armPEG-G4 dendrimers, an initial Ethidium Bromide (EB) displacement assay was performed. EB is known to emit red fluorescence upon intercalation into DNA. It has been reported that EB fluorescence is quenched by the presence of a second molecule that can bind the nucleic acid with higher affinity, displacing the dye. DNA-EB complexes were therefore titrated with 2-armPEG-G4 and 4-armPEG-G4 (Figure 8a). The assay demonstrates that 4-armPEG-G4 has significantly higher affinity for DNA when compared to 2-arm derivative with a significantly faster drop in EB fluorescence with the 4-armPEG-G4’s fluorescence signal plateauing at an N/P (the ratio between dendrimer amines versus DNA phosphates) of ca. 4, indicating a stable DNA-dendrimer complex, while the 2-armPEG-G4 requires a much higher N/P ratio (~ 15) to stabilize the dendriplex.
Figure 8.
Dendrimer-mediated gene delivery: (a) EB intercalation assay; (b) Confocal imaging (images widths are 30 µm for the left and center images and 205 µm for the image on the right) shows dendriplex formation (left), cellular accumulation of dendriplexes (center) and the nuclear accumulation of the expressed GFP Nuc (green) overlayed on a transmission light image of treated cells (right) and (c) Quantification of fluorescence signal (400 ngr of DNA, N/P = 4, 240 µl).
To further analyze the formation of 2-arm and 4-arm based dendriplexes and study their ability to bind cell membranes, both types of dendrimers were mixed with Cy5-labeled DNA to form Cy5-labeled dendriplexes. The 2-arm and 4-arm dendriplexes were then incubated for 1h with HeLa cells and single particles imaged using highly-sensitive confocal spinning disk microscopy at the single cell level. Both, 2-arm and 4-arm dendriplexes attached to the plasma membrane (Figure 8b), however significantly lower fluorescence intensity was detected in case of the 2-arm dendriplex particles (Figure 8c). This result supports the enhanced ability of 4-arm dendriplexes to bind DNA, as demonstrated in the EB intercalation assay. The amount of DNA delivered to HeLa cells after 22 hours incubation by the 2-arm and the 4-arm dendritic carriers was then quantified from the intensity of the fluorescence signal. Significantly, the amount of DNA that was delivered by the 4-armPEG-G4 was nearly an order of magnitude greater than for the 2-arm dendriplex (Figure 8c). This enhanced DNA delivery by the 4-arm dendriplex can be attributed to numerous property enhancements driven by changes due to the 4-arm architecture and higher number of amine end-groups, which leads to improved DNA-binding efficacy.
To evaluate the actual transfection potential of the dendriplexes, a plasmid encoding for green fluorescent protein bearing a nuclear localization signal (EGFP-Nuc) was then used in combination with either the 2-arm or 4-arm dendrimer. The fluorescence arising from GFP expression allows the transfection efficiency to be quantified by widefield-fluorescence microscopy. Figures 8b and 8c show the overlay of transmission light images for treated cells coupled with the green fluorescence signal for GFP expression followed by quantification of the GFP fluorescence signals, respectively. It is particularly noteworthy that the 2-armPEG-G4 shows no apparent transfection, while a strong fluorescence signal from GFP expression following transfection with the 4-arm dendriplexes was observed. As both dendrimers show endosome escaping abilities the significant difference is probably realted to the amount of DNA delivered to the cells. These results clearly show an enhanced ability for DNA delivery for the 4-armPEG-G4 platform compared to the corresponding 2-armPEG-G4 derivative. As higher N/P ratio can influence the trasnfection efficiency, in particular for the 2-arm dendrimer, the optimization of the trasnfection protocol in light of gene therapy is currently under further investigation.
CONCLUSIONS
Direct comparison between hybrid dendritic macromolecules based on 2-arm and 4-arm poly(ethylene glycol) cores has illustrated the influence of macromolecular architecture and core structure on their interactions with living cells. This leads to different biological properties which in turn can be related to a difference in solution structure with molecular dynamic studies showing that the 2-arm-PEG-G4 derivatives folds into a particle-like structure, with a compact, collapsed PEG core, while the 4-arm- PEG-G4 possesses a much less dense PEG core, resulting in a dynamic and oscillatory behavior in solution. While both dendrimers showed low toxicity and induced endosomal escape, the dynamic nature of the 4-arm hybride translates to enhanced biological performance, resulting in improved cell affinity and internalization. Significantly, the 4-arm-PEG based dendrimer also showed dramatically improved DNA binding and gene transfection capabilities in cell culture, demonstrating the potential of these synthetically accessible dendrimers as intracellular delivery platforms for a wide variety of applications.
Supplementary Material
Table 1.
Experimental and theoretical molecular weights of dendrimers 4-armPEG-Gn.
| Mn (GPC)a | PDI | Mpb(MALDI) | Theoretical Mn | |
|---|---|---|---|---|
| 4-armPEG-G0 | 12,000 | 1.21 | 10,700 | 10,000 |
| 4-armPEG-G1 | NAa | NAa | 11,300 | 10,900 |
| 4-armPEG-G2 | 17,600 | 1.25 | 12,100 | 12,700 |
| 4-armPEG-G3 | NAa | NAa | 13,200 | 15,700 |
| 4-armPEG-G4 | 23,800 | 1.19 | NDc | 23,000 |
Aqueous phase cationic GPC was used to determine Mn and PDI of the positively charged dendrimers. Data for the alkyne-terminated dendrimers is not available (NA) as they were not analyzed due to their poor solubility and aggregation in the mobile phase.
Mp: maximum of molecular ion peaks.
ND: not detectable
ACKNOWLEDGMENT
This material is based upon work supported by the National Institutes of Health as a Program of Excellence in Nanotechnology (HHSN268201000046C) (RJA, CJH). This work was partially supported by the Institute for Collaborative Biotechnologies through grant numbers W911NF-07-1-0279 and W911NF-09-D-0001 from the US Army Research Office (EA, KLK, CJH), MRSEC Program of the National Science Foundation through the MRL Central Facilities (LA, KLK, CJH) the Nanoinitiative Munich (NIM), the Elitenetzwerk Bayern, the SPP1313 and by fellowship support from Samsung (TK).
Footnotes
Supporting Information Available. MALDI-TOF MS, 1H-NMR spectra, dynamic light scattering results and computational methods. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Menjoge AR, Kannan RM, Tomalia DA. Drug Discovery Today. 2010;15:171–185. doi: 10.1016/j.drudis.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Bonduelle CV, Gillies ER. Pharmaceuticals. 2010;3:636–666. doi: 10.3390/ph3030636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calderón M, Quadir MA, Sharma SK, Haag R. Adv. Mater. 2010;22:190–218. doi: 10.1002/adma.200902144. [DOI] [PubMed] [Google Scholar]
- 4.Cheng Y, Zhao L, Li Y, Xu T. Chem. Soc. Rev. 2011;40:2673–2703. doi: 10.1039/c0cs00097c. [DOI] [PubMed] [Google Scholar]
- 5.Kaminskas LM, Boyd BJ, Porter CJH. Nanomedicine. 2011;6:1063–1084. doi: 10.2217/nnm.11.67. [DOI] [PubMed] [Google Scholar]
- 6.Soliman GM, Sharma A, Maysinger D, Kakkar A. Chem. Commun. 2011;47:9572–9587. doi: 10.1039/c1cc11981h. [DOI] [PubMed] [Google Scholar]
- 7.El Kazzouli S, Mignani S, Bousmina M, Majoral JP. New J. Chem. 2012;36:227–240. doi: 10.2174/0929867311209024995. [DOI] [PubMed] [Google Scholar]
- 8.Bonner DK, Leung C, Chen-Liang J, Chingozha L, Langer R, Hammond PT. Bioconjugate Chem. 2011;22:1519–1525. doi: 10.1021/bc200059v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mintzer MA, Simanek EE. Chem. Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 10.Turrin CO, Caminade AM. In: Dendrimers: Towards Catalytic, Material and Biomedical Uses. Caminade AM, Turrin CO, Laurent R, Ouali A, Delavaux-Nicot B, editors. Chichester, UK: John Wiley & Sons, Ltd; 2011. pp. 413–435. [Google Scholar]
- 11.Mumper RJ, Bell MA, Worthen DR, Cone RA, Lewis GR, Paull JRA, Moench TR. Drug Dev. Ind. Pharm. 2009;35:515–524. doi: 10.1080/03639040802488097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heegaard PMH, Boas U, Sorensen NS. Bioconjugate Chem. 2010;21:405–418. doi: 10.1021/bc900290d. [DOI] [PubMed] [Google Scholar]
- 13.Skwarczynski M, Zaman M, Urbani CN, Lin I, Jia Z, Batzloff MR, Good MF, Monteiro MJ, Toth I. Angew. Chem. Int. Ed. 2010;49:5742–5745. doi: 10.1002/anie.201002221. [DOI] [PubMed] [Google Scholar]
- 14.Antoni P, Robb MJ, Campos L, Montanez M, Hult A, Malmstorm E, Malkoch M, Hawker CJ. Macromolecules. 2010;43:6625–6631. [Google Scholar]
- 15.Duncan R, Izzo L. Adv. Drug Delivery Rev. 2005;57:2215–2237. doi: 10.1016/j.addr.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Megia E, Correa J, Riguera R. Biomacromolecules. 2006;7:3104–3111. doi: 10.1021/bm060580d. [DOI] [PubMed] [Google Scholar]
- 17.Clementi C, Miller K, Mero A, Satchi-Fainaro R, Pasut G. Mol. Pharmaceutics. 2011;8:1063–1072. doi: 10.1021/mp2001445. [DOI] [PubMed] [Google Scholar]
- 18.Sousa-Herves A, Riguera R, Fernandez-Megia E. New J. Chem. 2011;36:205–210. [Google Scholar]
- 19.Kim T, Seo HJ, Choi JS, Jang HS, Baek J, Kim K, Park JS. Biomacromolecules. 2004;5:2487–2492. doi: 10.1021/bm049563j. [DOI] [PubMed] [Google Scholar]
- 20.Amir RJ, Albertazzi L, Willis J, Khan A, Kang T, Hawker CJ. Angew. Chem. Int. Ed. 2011;50:3425–3429. doi: 10.1002/anie.201007427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe AB, Hoyle CE, Bowman CN. J. Mater. Chem. 2010;20:4745–4750. [Google Scholar]
- 22.Silvers AL, Chang CC, Emrick T. J. Polym. Sci. Part A: Polym. Chem. 2012;50:3517–3529. [Google Scholar]
- 23.Chen G, Kumar J, Gregory A, Stenzel MH. Chem. Commun. 2009;41:6291–6293. doi: 10.1039/b910340f. [DOI] [PubMed] [Google Scholar]
- 24.Kolb HC, Sharpless KB. Drug Discovery Today. 2003;8:1128–1137. doi: 10.1016/s1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- 25.Wang DK, Hill DJT, Rasoul FA, Whittaker AK. J. Polym. Sci., Part A: Polym. Chem. 2012;50:1143–1157. [Google Scholar]
- 26.Wu Z, Zeng X, Zhang Y, Feliu N, Lundberg P, Fadeel B, Malkoch M, Nyström AM. J. Polym. Sci., Part A: Polym. Chem. 2012;50:217–226. [Google Scholar]
- 27.Albertazzi L, Serresi M, Albanese A, Beltram F. Mol. Pharmaceutics. 2010;7:680–688. doi: 10.1021/mp9002464. [DOI] [PubMed] [Google Scholar]
- 28.Albertazzi L, Storti B, Marchetti L, Beltram F. J. Am Chem. Soc. 2010;132:18158–18167. doi: 10.1021/ja105689u. [DOI] [PubMed] [Google Scholar]
- 29.Reichert S, Welker P, Calderón M, Khandare J, Mangoldt D, Licha K, Kainthan RK, Brooks DE, Haag R. Small. 2011;7:820–829. doi: 10.1002/smll.201002220. [DOI] [PubMed] [Google Scholar]
- 30.Iliashevsky O, Amir L, Glaser R, Marks RS, Lemcoff NG. J. Mater. Chem. 2009;19:6616–6622. [Google Scholar]
- 31.Kelly CV, Liroff MG, Triplett LD, Leroueil PR, Mullen DG, Wallace JM, Meshinchi S, Baker JR, Orr BG, Banaszak Holl MM. ACS Nano. 2009;3:1886–1896. doi: 10.1021/nn900173e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones SP, Gabrielsen NP, Wong CH, Chow HF, Pack D, Posocco P, Fermeglia M, Pricl S, Smith DK. Mol. Pharmaceutics. 2011;8:416–429. doi: 10.1021/mp100260c. [DOI] [PubMed] [Google Scholar]
- 33.Bastings MMC, Helms BA, van Baal I, Hackeng TM, Merkx M, Meijer E. J. Am Chem. Soc. 2011;133:6636–6641. doi: 10.1021/ja110700x. [DOI] [PubMed] [Google Scholar]
- 34.Fox ME, Guillaudeu S, Fréchet JMJ, Jerger K, Macaraeg N, Szoka FC. Mol. Pharmaceutics. 2009;6:1562–1572. doi: 10.1021/mp9001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurtoglu YE, Navath RS, Wang B, Kannan S, Romero R, Kannan RM. Biomaterials. 2009;30:2112–2121. doi: 10.1016/j.biomaterials.2008.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lo Conte M, Robb MJ, Hed Y, Marra A, Malkoch M, Hawker CJ, Dondoni A. J. Polym. Sci Part A: Polym. Chem. 2011;49:4468–4475. doi: 10.1002/pola.24888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albertazzi L, Fernandez-Villamarin M, Giuera R, Fernandesz-Megia R. Bioconjugate. Chem. 2012;23:1059–1068. doi: 10.1021/bc300079h. [DOI] [PubMed] [Google Scholar]
- 38.Pavan GM, Mintzer MA, Simanek EE, Merkel OM, Kissel T, Danani A. Biomacromolecules. 2010;11:721–730. doi: 10.1021/bm901298t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albertazzi L, Brondi M, Pavan GM, Sulis Sato S, Signore G, Storti B, Ratto GM, Beltram F. PLoS ONE. 2011;6:e28450. doi: 10.1371/journal.pone.0028450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.