Background: Polo-like kinase 1 (Plk1) has multiple functions and substrates in human mitosis.
Results: Plk1 functions are separable via distinct thresholds of activity, and partial functional loss leads to chromosome missegregation.
Conclusion: Plk1 has several distinct mitotic roles that are separable by chemical genetics.
Significance: It is possible to dissect individual functions of a multifunctional enzyme using activity thresholds.
Keywords: Cancer Therapy, Cell Division, Chemical Biology, DNA Topology, Kinetochore, Mitosis, Chemical Genetics, Polo Box Domain, Separation of Function, Spindle
Abstract
Protein kinases play key roles in regulating human cell biology, but manifold substrates and functions make it difficult to understand mechanism. We tested whether we could dissect functions of a pleiotropic mitotic kinase, Polo-like kinase 1 (Plk1), via distinct thresholds of kinase activity. We accomplished this by titrating Plk1 activity in RPE1 human epithelial cells using chemical genetics and verifying results in additional lines. We found that distinct activity thresholds are required for known functions of Plk1 including (from low to high activity) bipolar spindle formation, timely mitotic entry, and formation of a cytokinesis cleavage furrow. Subtle losses in Plk1 activity impaired chromosome congression and produced severe anaphase dysfunction characterized by poor separation of chromosome masses. These two phenotypes were separable, suggesting that they stem from distinct phosphorylation events. Impaired chromosome segregation in anaphase was the most sensitive to modest loss in Plk1 activity. Mechanistically, it was associated with unpaired sister chromatids with stretched kinetochores, suggestive of merotelic attachments. The C-terminal Polo box domain of Plk1 was required for its anaphase function, although it was dispensable for forming a bipolar spindle. The ultimate effect of partial inhibition of Plk1 was the formation of micronuclei, an increase in tetraploid progeny, and senescence. These results demonstrate that different thresholds of Plk1 activity can elicit distinct phenotypes, illustrating a general method for separating pleiotropic functions of a protein kinase even when these are executed close in time.
Introduction
Polo-like kinase 1 (Plk1)2 is a key regulator of human mitosis. Its function is essential for multiple mitotic processes including centrosome maturation, formation of a bipolar mitotic spindle, chromosome attachment, loss of cohesion between sister chromatids, and cytokinesis (1, 2). On a molecular level, Plk1 interacts with many of its substrates via its C-terminal Polo box domain (PBD), a phosphopeptide binding domain. Binding via the PBD requires substrates to be previously phosphorylated (“primed”) by protein kinases such as cyclin-dependent kinase 1 and can elicit positive feedback when Plk1 itself generates the PBD binding site (3, 4). The PBD binding provides a high local activity of Plk1 that allows it to phosphorylate substrate proteins and elicit its biologic functions.
Despite the importance of Plk1 in cell division, it is difficult to reconcile its mitotic functions with an expanding list of binding partners and substrates. Although hundreds of Plk1 interactors and substrates have been identified (5–7), strong links between function and substrates are established for only a handful. Compounding this difficulty, its activities occur almost exclusively within the ∼60 min of mitosis, making it difficult to isolate individual functional events. A comprehensive understanding of Plk1 will require reconciliation of each mitotic function with its cognate phosphorylation event. However, this has been a challenging goal because the many functions of Plk1 are executed in a short interval of time.
We previously reported a chemical genetic system for specific interrogation of Plk1 in immortalized human retinal pigment epithelial (RPE1) cells (8). In this system, loxP sites were introduced flanking exon 3 in one copy of PLK1 and deleted from the other, generating a PLK1flox/Δ conditional knock-out cell line. An EGFP-tagged Plk1 construct was reintroduced followed by Cre-mediated excision of the flox-PLK1 locus. In this manner, cells were reconstituted with wild-type Plk1 to create Plk1WT RPE1 cells or by analog-sensitive Plk1 (C67V/L130G) to generate Plk1as RPE1 cells. In these cell lines, EGFP-Plk1WT and EGFP-Plk1as transgenes were expressed at levels similar to that of endogenously expressed Plk1; however, Plk1as was modestly elevated possibly due to its decreased catalytic efficiency (8). In the analog-sensitive cell line, Plk1as can be selectively inhibited by the bulky ATP analog 3-MB-PP1, resulting in loss of Plk1 functions with the expected phenotypes. Importantly, wild-type Plk1 is unaffected by 3-MB-PP1, allowing explicit controls for on- versus off-target effects (8, 9). Moreover, BI-2536, an inhibitor of wild-type Plk1, does not affect activity of Plk1as, allowing these two functional alleles to be orthogonally controlled independently with these chemicals (10).
We previously used this system to identify the role of Plk1 in triggering cytokinesis concordant with late mitosis and identified the cognate molecular substrate HsCyk4/RacGAP1 (2, 8). This work demonstrates the power of chemical biology to rapidly and specifically inactivate an enzyme to resolve functions that are separable in time.
Here we seek to extend the chemical genetic system to allow dissection of discrete kinase functions that are not separable in time. We hypothesized that kinase functions that occur close together may be separable by activity thresholds. In this model, distinct enzyme functions may be interrupted at different Plk1 thresholds due to differences in local kinase activity, counteracting phosphatases, or different phosphorylation requirements to elicit downstream functions. Indeed, we show here that careful titration of Plk1 activity in human cells undergoing mitosis revealed several discrete functions that were separable at thresholds of Plk1 activity, and in one testable case, this threshold matched that of the cognate molecular phosphorylation.
We demonstrate the power of this method for dissecting pleomorphic enzymatic functions that are close in time and space. Moreover, the phenotypes observed with partial loss of function presage in vivo effects of Plk1-targeted drugs that will cross these thresholds of inhibition in human subjects at times dictated by pharmacology.
EXPERIMENTAL PROCEDURES
Cell Culture Procedures
All cell lines were propagated at 37 °C in 5% CO2 in media supplemented with 10% fetal bovine serum and 100 units/ml penicillin-streptomycin except MCF-10A cells, which were propagated as reported previously (11). T47D, hTert-RPE1 (ATCC, Manassas, VA), and RPE1-derived cell lines were propagated in a 1:1 mixture of DMEM and Ham's F-12 medium supplemented with 2.5 mm l-glutamine. MCF-7 cells were propagated in DMEM supplemented with 4 mm l-glutamine, 4500 mg/liter glucose, and 10 μg/ml insulin.
EGFP-Plk1as and EGFP-Plk1WT RPE1 cell lines were reported previously (8). Plk1as cell lines stably expressing mCherry-Plk1WT constructs with a wild-type (AS/WT) or “pincer mutant” (AS/AA) PBD were derived as reported (2), and clones were obtained by limiting dilutions using 0.4 μg/ml G418 for selection.
To assay cell proliferation with prolonged Plk1 inhibition, 25,000 Plk1as cells were plated in individual wells of 12-well plates, allowed to acclimate for 24 h, and then challenged with DMSO or 0.25 μm 3-MB-PP1 for up to 12 days. Medium was replaced every 4 days. Adherent and non-adherent cells were collected every 24 h, incubated with trypan blue to exclude dead cells, and counted with a hemacytometer. Counts were performed in duplicate and averaged. Cellular senescence was assayed using a pH-dependent β-galactosidase staining kit (Cell Signaling Technology) according to the manufacturer's instructions.
Immunoblotting, Immunoprecipitation, and Kinase Assays
For immunoblotting, cells were lysed in buffer (50 mm HEPES, pH 7.5, 100 mm NaCl, 0.5% Nonidet P-40, 10% glycerol) containing phosphatase inhibitors (10 mm sodium pyrophosphate, 5 mm β-glycerol phosphate, 50 mm NaF, 0.3 mm Na3VO4), 1 mm PMSF, 1× protease inhibitor mixture (Thermo Scientific), and 1 mm dithiothreitol. Proteins were separated by SDS-PAGE, transferred to Immobilon PVDF membrane (Millipore), and blocked for 30 min in 4% milk and 0.1% Tween 20 in Tris-buffered saline, pH 7.4 (TBST + milk). Membranes were incubated with gentle agitation for 2 h at room temperature with primary antibodies diluted in TBST + milk, washed three times with TBST, and incubated for 1 h at room temperature in secondary antibodies conjugated to horseradish peroxidase in TBST + milk. Membranes were washed and developed with luminol/peroxide (Millipore) and visualized with film.
For immunoprecipitation and in vitro kinase assays, RPE1 cells expressing EGFP-Plk1as, EGFP-Plk1WT, or untagged Plk1 were incubated with nocodazole for 16 h. Whole cell extracts were incubated with EGFP antibody (Invitrogen) at a concentration of 1 μg of antibody/1 ml of total protein in lysis buffer for 1 h on ice. EGFP-bound Plk1 was immunoprecipitated using protein A- and protein G-Sepharose beads (GE Healthcare) in a 1:1 ratio for 2 h at 4 °C with gentle rotation. Beads were washed three times with lysis buffer and once with kinase buffer (20 mm Tris, pH 7.4, 10 mm MgCl2, 50 mm KCl, 1 mm dithiothreitol) and then incubated in kinase buffer plus 5 μg of casein, 1 μm ATP, 50 μCi of [γ-32P]ATP, and 3-MB-PP1 for 30 min at 30 °C. 32P incorporation was observed by SDS-PAGE and a Typhoon TRIO imager (GE Healthcare) and quantified using ImageJ (12).
Immunofluorescence (IF) and Microscopy
For IF, cells were seeded on glass coverslips in 24-well plates and fixed with 100% ice-cold methanol (centrosomal and central spindle epitopes) for 15 min to overnight or 4% paraformaldehyde in PBS (all other epitopes) for 10 min. Fixed cells were then blocked for 30 min in 3% bovine serum albumin (BSA) and 0.1% Triton X-100 in PBS (PBSTx + BSA). Primary antibodies were incubated in PBSTx + BSA for 1 h at room temperature and washed three times in PBSTx followed by secondary antibody incubation in PBSTx + BSA for 30 min at room temperature and two washes with PBSTx. Cells were counterstained with DAPI, mounted on glass slides with Prolong Gold antifade medium (Invitrogen), and allowed to cure overnight.
Image acquisition was performed on a Nikon Eclipse Ti inverted microscope equipped with 10×, 20×, 40×, and 100× objectives; a temperature-controlled motorized stage with 5% CO2 support (In Vivo Scientific); and CoolSNAP HQ2 charge-coupled device camera (Photometrics). Optical sections were taken at 0.2-μm intervals and deconvolved using Nikon Elements. Where appropriate, the observer was blinded to treatment condition during image acquisition and analysis. For live cell imaging, cells were seeded in 35-mm glass bottom plates and incubated with media containing DMSO, 3-MB-PP1, or BI-2536 for 1 h prior to imaging in 5% CO2 at 37 °C. Images were processed and analyzed using Nikon Elements. Panels were cropped using Photoshop CS5 (Adobe) and assembled with overlays using Illustrator CS5 (Adobe).
Calculations and Statistics
IC50 calculations were performed using BioDataFit 1.02 (Chang Bioscience) using an exponential decay model. Replicate experiments were performed, and standard errors are reported as indicated below. Statistical evaluations were performed using Excel (Office 2008, Microsoft) or Mstat (McArdle Laboratory for Cancer Research). Two-tailed t tests or Wilcoxon rank sum tests were used to estimate the likelihood that the observed difference was obtained by chance. No corrections were made for multiple hypothesis testing.
Chemicals and Antibodies
Chemicals used in this study include monastrol (100 μm; Tocris), nocodazole (0.2 mg/ml; EMD Biosciences), thymidine (2.5 mm; EMD Biosciences), ICRF-193 (Enzo Life Sciences), 3-MB-PP1 (Toronto Research Chemicals), and BI-2536 (a gift from P. Jallepalli). Antibodies used in this study were β-actin (AC-15, Abcam; 1:15,000), Bloom helicase (C-18, Santa Cruz Biotechnology; 1:500); CREST (detects centromeres; Immunovision; 1:2500), HsCyk4/RacGAP1 (Genetex; 1:375), DsRed (detects mCherry; Clontech; 1:1000), Plk1 (F-8, Santa Cruz Biotechnology; 1:500), α-tubulin (YL1/2, Millipore; 1:1000), and γ-tubulin (GTU-88, Thermo Scientific; 1:250). Antibodies against Ser(P)-170 of Cyk4 were developed as reported previously (2). Alexa Fluor (Invitrogen) secondary antibodies were used at 1:350.
RESULTS
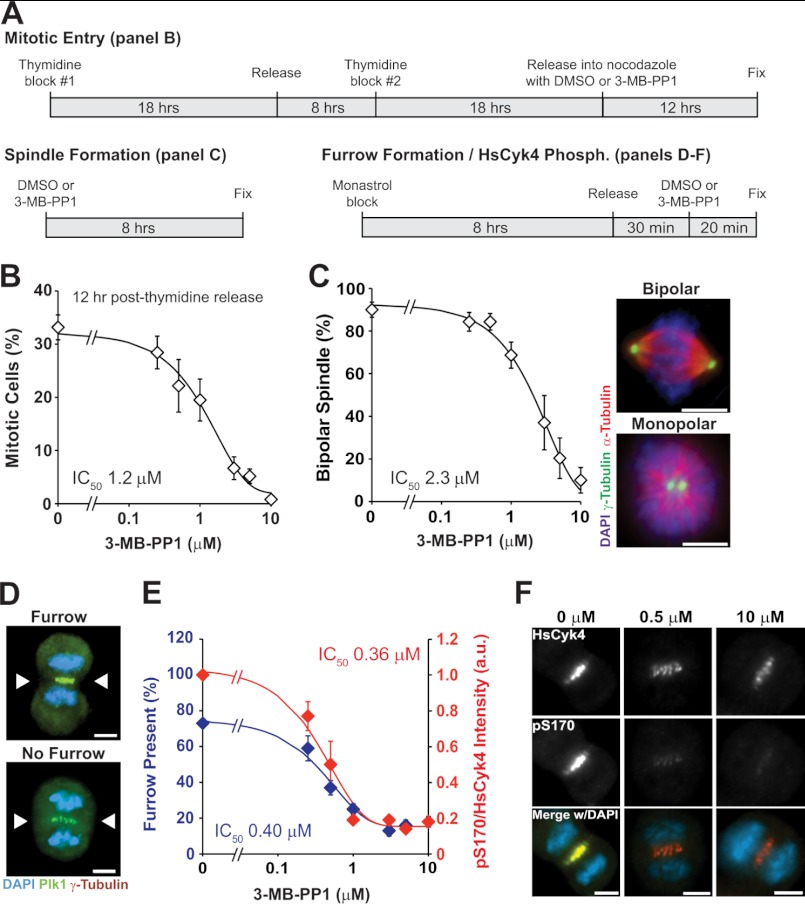
We surveyed the thresholds of activity required for several known functions of Plk1 using synchronization strategies (Fig. 1A) and a range of 3-MB-PP1 concentrations up to 10 μm. This maximum concentration was chosen because the observed phenotype matches the effects of knockdown, knock-out, and antibody microinjection (8). The survey of Plk1-dependent phenotypes revealed a graded effect in Plk1 function on mitotic entry (Fig. 1B), bipolar spindle formation (Fig. 1C), and formation of a cleavage furrow in anaphase (Figs. 1, D and E). Each Plk1-dependent phenotype had a distinct 50% inhibitory concentration (IC50) ranging from 400 nm to 2.3 μm (Table 1). These results demonstrate that individual Plk1 functions are separable as they have distinct thresholds for inactivation.
FIGURE 1.
Individual Plk1 phenotypes have distinct thresholds of inactivation. A, experimental synchronization schemes used to assess phenotypes. B–F, Plk1as RPE1 cells were challenged with DMSO (0 μm) or 3-MB-PP1 (0.25, 0.5, 1, 3, 5, or 10 μm) to determine the IC50 for mitotic entry (B), bipolar spindle formation (C), and cleavage furrow formation and HsCyk4 phosphorylation (Phosph.) (D–F). B, mitotic entry. Cells were synchronized in S phase using a double thymidine block. Upon release, DMSO or 3-MB-PP1 was added to the media with nocodazole (to prevent mitotic exit). After 12 h, cells were fixed and stained with Hoescht 33258 to visualize nuclei. The percentage of mitotic cells was plotted for each concentration (n = 600 cells; two independent experiments) to determine the IC50. Data represent averages ± S.E. (error bars). C, bipolar spindle formation. Asynchronously growing cells were challenged with DMSO or 3-MB-PP1 for 8 h and fixed for IF. The percentage of preanaphase mitotic cells exhibiting a normal bipolar spindle was plotted for each concentration (n = 300 cells; three independent experiments) to determine the IC50. Data represent averages ± S.E. (error bars). Representative images of cells exhibiting a normal bipolar or monopolar spindle are displayed. Other abnormal spindle structures (i.e. multipolar and unfocused bipolar spindles) were observed infrequently and are not depicted here. D–F, cleavage furrow formation and HsCyk4 phosphorylation. Cells were synchronized in prometaphase with a monastrol block, released into fresh media for 30 min to allow cells to enter anaphase, and then challenged with DMSO or 3-MB-PP1 for 20 min prior to fixation. D, late anaphase cells were scored for the presence (top) or absence (bottom) of a cleavage furrow (arrowheads). E, graph indicating the percentage of cells exhibiting a cleavage furrow (blue plot; n = 300 cells/concentration) and the ratio of phosphorylated (Ser(P)-170) to total HsCyk4 intensity (red plot; n = 100 cells/concentration). Data represent averages from three independent experiments ±S.E. (error bars). F, representative images of single optical z-sections through cells demonstrating decreased serine 170 phosphorylation (pS170) with increasing Plk1 inhibition as compared with total HsCyk4 intensity. Scale bars, 5 μm.
TABLE 1.
IC50 values for various phenotypes associated with loss of Plk1 activity
| Phenotype | 50IC 3-MB-PP1 | Residual activitya |
|---|---|---|
| μm | % | |
| Bipolar spindle formation | 2.3 | 0.4 |
| Mitotic entry | 1.2 | 0.7 |
| Chromosome congression | 0.50 | 11 |
| Formation of cytokinesis furrow | 0.40 | 16 |
| Chromosome segregation | 0.21 | 38 |
a In vitro inhibition with 1 μm total ATP. For higher intracellular ATP concentrations, absolute residual activities will be greater but the order will be the same.
We hypothesized that the inhibitory concentration for molecular phosphorylation events will match that of the associated phenotypic events. To test this, we measured the known Plk1-dependent phosphorylation of HsCyk4/RacGAP1 at the spindle midzone, an event known to trigger cytokinesis (Fig. 1, E and F) (2, 13). Indeed, we found that the IC50 for HsCyk4 phosphorylation closely matches that seen for furrow formation (0.36 versus 0.40 μm). We conclude that graded inhibition of a kinase such as Plk1 can resolve and link individual sets of cognate molecular and biologic events.
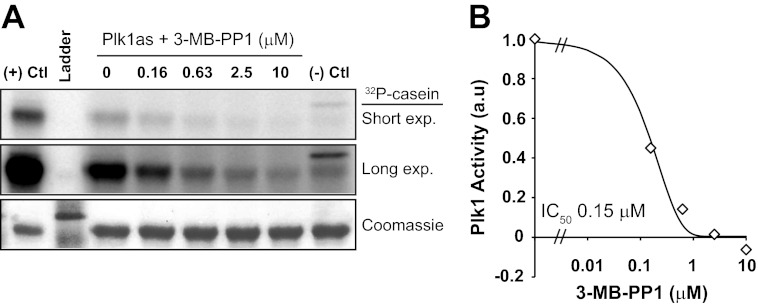
To calibrate these biologic IC50 values with biochemical inhibitory concentrations, we performed in vitro kinase assays using Plk1as. Using a range of 3-MB-PP1 concentrations and 1 μm total ATP, we measured a biochemical IC50 of 0.15 μm (Fig. 2). This allows us to estimate the residual kinase activity needed to elicit biologic functions (Table 1, right). The actual residual intracellular catalytic activity depends on local concentrations of Plk1, ATP, and the protein substrate, which may differ from that selected in our biochemical assay. However, if intracellular concentrations of these were uniform, then relative values would be as reported in Table 1.
FIGURE 2.
Plk1as activity is inhibited by 3-MB-PP1. A, Plk1 was immunoprecipitated from mitotic extracts of RPE1 cells expressing EGFP-Plk1as, EGFP-Plk1WT (positive control (Ctl)), or untagged Plk1 (negative control) using an EGFP antibody and then incubated with casein and [γ-32P]ATP. Where indicated, 3-MB-PP1 was added to the reactions. B, Plk1as activity was quantified by casein labeling relative to Plk1as reactions without inhibitor and plotted to determine the IC50. exp., exposure.
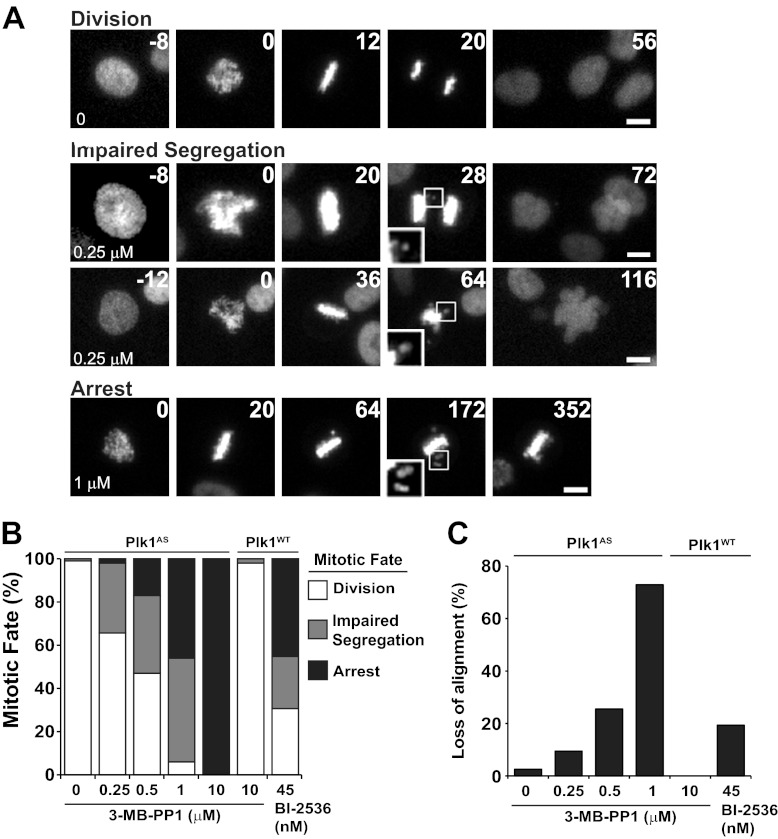
Our survey revealed that spindle polarity is relatively insensitive to loss of Plk1 function (IC50, 2.3 μm; Fig. 1B). This allowed us to evaluate detailed spindle-independent phenotypes associated with loss of Plk1 function. Previous work has demonstrated the presence of such spindle-independent Plk1 functions using other approaches (14, 15). To evaluate these in detail, we inhibited Plk1 at levels below this threshold and observed the effect on division of live cells by time lapse videomicroscopy (Fig. 3A and supplemental Movies S1–S4). Three phenotypes were observed. (i) Normal-appearing divisions (top), typically completed within an hour, were seen frequently with 0–0.25 μm 3-MB-PP1. (ii) Mitotic arrest (bottom) was seen frequently at high levels of 3-MB-PP1 even when there was evidence of bipolar spindle formation. (iii) At intermediate levels of Plk1 inhibition, we observed a distinct phenotype that we term “impaired segregation.” These phenotypes were observed with frequencies that depended on the level of Plk1 inhibition (Fig. 3B). Importantly, these phenotypes were not due to an off-target effect of 3-MB-PP1 or associated solely with Plk1as cells; a similar phenotype was observed with RPE1 cells harboring wild-type Plk1 when interrogated with low levels of BI-2536, a pharmacologic inhibitor of wild-type Plk1 (10, 16), but no effect in this cell line was observed with the Plk1as-specific inhibitor, 3-MB-PP1 (Fig. 3B, right). In addition to impaired chromosome segregation, we also found that chromosomes frequently aligned at the metaphase plate followed by loss of alignment (Fig. 3A, third and fourth rows, insets). This loss of alignment occurred with increasing frequency as 3-MB-PP1 was escalated. Again, controls demonstrated that this phenotype was specific to loss of Plk1 function as it was recapitulated with BI-2536 but not 3-MB-PP1 in cells with wild-type Plk1 (Fig. 3C). We conclude that partial loss of Plk1 catalytic activity impairs chromosome segregation during mitosis in human cells.
FIGURE 3.
Live cell imaging reveals that partial loss of Plk1 activity results in impaired chromosome alignment and segregation. To visualize chromosome movements during mitosis, mCherry-tagged histone 2B was expressed in Plk1as and Plk1WT cells. Cells were challenged with DMSO, 3-MB-PP1, or BI-2536 and observed for 10 h at 4-min intervals using time lapse videomicroscopy. A, still frames from time lapse movies (supplemental Movies S1–S4) highlighting three observed mitotic fates: a clear separation of chromosomes during anaphase (“Division,” top row), anaphase with a few lagging chromosomes (second row, see inset in frame 28) or frank failure to separate chromosome masses (third row, see inset in frame 64) collectively termed “Impaired Segregation”, and a sustained (≥240 min) preanaphase mitotic arrest (“Arrest,” bottom row). 3-MB-PP1 concentrations are indicated in the first frame. Time in minutes after nuclear envelope breakdown is shown on the upper right. Scale bars, 5 μm. B, cumulative frequency of mitotic fates observed in A (n = 30–80 cells/concentration). Note that Plk1WT cells are resistant to the effects of 3-MB-PP1 (>90% division with 10 μm exposure) but sensitive to the pharmacological inhibitor BI-2536 (<40% division with 45 nm exposure). C, chromosomes frequently lost alignment after initial midline congression (compare inset from frame 172 with frame 20 in Arrest in A). The frequency of cells exhibiting loss of chromosome alignment after establishing midline congression was recorded for each concentration (as in B). Plk1as cells challenged with 10 μm 3-MB-PP1 are excluded because initial midline congression did not occur.
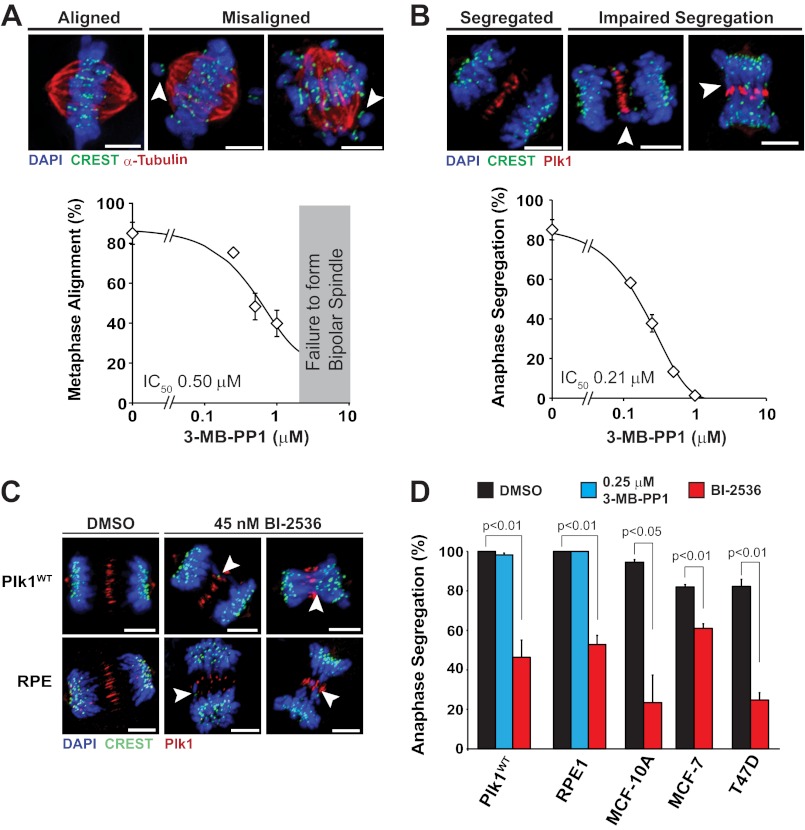
Careful observation of cells with impaired segregation revealed additional abnormalities. In the most striking example, chromosomes congressed at the metaphase plate followed by loss of alignment and onset of anaphase with poor separation of chromosome masses (Fig. 3A, third row, frame 64 and supplemental Movie S3). To characterize the misalignment and anaphase segregation defects, we analyzed cells by IF (Fig. 4). We found that chromosomes frequently were misaligned (distinctly separated from midline-oriented chromosomes) on bipolar spindles at 0.5 and 1 μm 3-MB-PP1, but this was less common at concentrations <0.5 μm (IC50, 0.5 μm; Fig. 4A). The number of misaligned chromosomes and their spindle position varied, but the severity increased with degree of Plk1 inhibition. In many cases, chromosomes appeared detached from microtubules, judged by α-tubulin staining, and remained paired as indicated by the presence of two distinct CREST signals (Fig. 4A, arrowheads). We considered that this chromosome misalignment could simply reflect a delay in chromosome congression during prometaphase; however, time lapse videomicroscopy revealed that a significant fraction of cells exhibited loss of chromosome alignment after initial midline congression (Fig. 3C and supplemental Movie S4). We conclude that maintenance of chromosome alignment requires high activity of Plk1.
FIGURE 4.
Anaphase chromosome segregation is most sensitive to Plk1 inhibition, and impairment is associated with shortened spindles and lagging chromosomes. A and B, to determine the IC50 for metaphase chromosome alignment and anaphase chromosome segregation, asynchronously growing Plk1as cells were challenged with DMSO (0 μm) or 3-MB-PP1 (0.125, 0.25, 0.5, or 1 μm) for 8 h and fixed for IF. A, for chromosome alignment, preanaphase cells exhibiting distinct midline chromosome alignment (n = 300 cells/condition; three independent experiments) were scored as having midline alignment of all chromosomes (“Aligned,” top left) or misalignment of a few (top center) or many (top right) chromosomes (indicated by arrowheads) collectively termed “Misaligned.” The percentage of metaphase cells with aligned chromosomes was plotted for each inhibitor concentration to determine the IC50. Data represent averages ± S.E. (error bars). B, for chromosome segregation, anaphase cells (n = 75–100 cells/condition from three independent experiments) were identified by central spindle localization of Plk1 (in red) and scored as having separated chromosome masses without midline-localized chromosomes (“Segregated”) or lagging/poorly segregated chromosomes (“Impaired Segregation,” arrowheads). The percentage of anaphase cells with segregated chromosomes was plotted for each inhibitor concentration to determine the IC50. Data represent averages ± S.E. (error bars). C and D, impaired chromosome segregation is specific to Plk1 inhibition but independent of cell type. C, Plk1WT and RPE1 cells were challenged with DMSO or BI-2536 for 8 h prior to fixation for IF, revealing defects in anaphase chromosome segregation with lagging chromosomes (arrowheads). D, anaphase cells (n = 120–150/condition; three independent experiments) were identified and scored by IF for anaphase segregation after 8 h of treatment as shown. Data represent averages ± S.E. (error bars). Significance was determined by t test. Scale bars, 5 μm.
Compared with chromosome misalignment in metaphase, impaired segregation in anaphase was more sensitive to loss of Plk1 function (Fig. 4B). Anaphase chromosomes frequently lagged in as little as 0.125 μm 3-MB-PP1. Consistent with time lapse observations, impaired segregation was sometimes severe with a large number of chromosomes remaining at the midline spindle (Fig. 4B, top right panel). We were nevertheless able to confirm that such cells were in anaphase due to both polar-oriented CREST signals and relocalization of Plk1 at the spindle midzone. Thus, among the phenotypes observed with loss of Plk1 function, the anaphase chromosome segregation was most sensitive to partial loss of function, suggesting the existence of a required substrate that either is not readily accessible to this kinase or is influenced by high local phosphatase activity.
To ensure that the anaphase segregation defect could be directly attributed to Plk1, we again analyzed cells with wild-type Plk1. As expected, BI-2536 but not 3-MB-PP1 elicited the segregation defect in cells with wild-type Plk1 (Fig. 4, C and D). This was not specific to any one subline as similar observations were obtained with both wild-type RPE1 cells (with endogenous Plk1) and Plk1WT cells (PLK1−/− knock-out cells complemented with EGFP-Plk1WT). Moreover, these findings were not specific to RPE1 cells as they were also observed in MCF-10A, MCF-7, and T47D cells (Fig. 4D). We conclude that anaphase chromosome missegregation is directly related to loss of Plk1 activity.
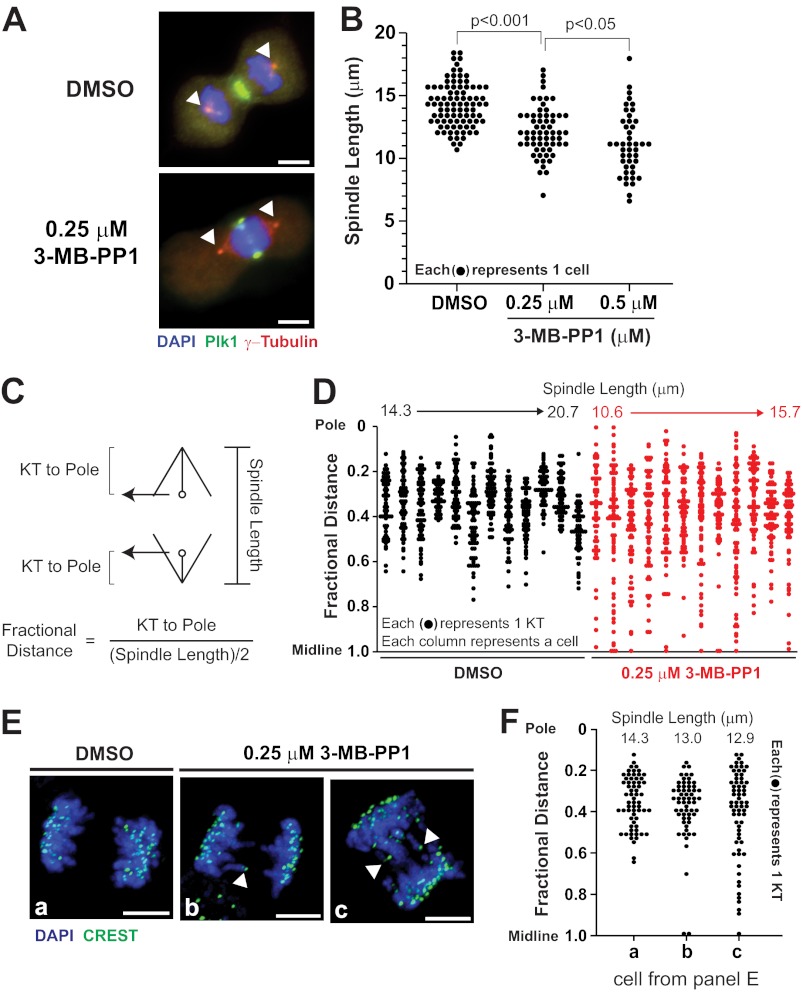
We reasoned that impaired chromosome segregation could be attributable either to poor chromosome migration in anaphase A or to dysfunction of spindle elongation in anaphase B, which is known to require Plk1 function (17). To distinguish these, we performed a detailed analysis of spindle elongation and chromosome segregation after partial impairment of Plk1 activity (Fig. 5). Under these conditions, we were able to confirm anaphase and visualize other Plk1-dependent processes including its own localization to the spindle midzone and initiation of cytokinesis, consistent with the higher IC50 associated with these processes. As expected, we observed a shortening in average spindle length as was reported previously with complete inhibition of Plk1 at anaphase onset (Fig. 5, A and B) (17). We next surveyed kinetochore position relative to poles (Fig. 5C). Using this scheme, we evaluated kinetochore position and spindle length in 12 Plk1as cells for each condition (DMSO control and 0.25 μm 3-MB-PP1) (Fig. 5D). As illustrated, chromosome laggards (0.8–1.0 fractional distance) were absent in control cells but present in the majority of cells after partial loss of Plk1 activity; no correlation was seen between the presence of laggards and spindle length. We confirmed this finding by comparing three cells with similar spindle lengths (Fig. 5, E and F). This demonstrated that, even controlling for spindle length, there was impaired chromosome segregation to poles with Plk1 inhibition. We conclude that both chromosome segregation and spindle elongation are sensitive to modest losses of Plk1 activity.
FIGURE 5.
Spindle elongation and chromosome segregation are impaired with partial loss of Plk1 activity. To characterize impaired chromosome segregation, asynchronously growing Plk1as cells were challenged with DMSO or 3-MB-PP1 (0.25 or 0.5 μm) for 8 h and fixed for IF. A, anaphase spindle lengths were determined by measuring the distance between the peak γ-tubulin intensities at each centrosome (arrowheads). B, spindle lengths of individual cells were plotted for each inhibitor concentration. p values were determined by Wilcoxon rank sum test. C–F, spindle position of individual chromosomes was determined using a fractional distance calculation adapted from Lampson and Kapoor (42) where the distance of the chromosome's kinetochore (KT) from the spindle pole is divided by half the length of the spindle axis. Fractional distance is then plotted to visualize kinetochore positions relative to the spindle midline (1.0) and pole (0). D, fractional distance plots in 12 control cells (black) and 12 3-MB-PP1-treated Plk1as cells (red). Cells are ordered by spindle length from shortest to longest (top). E and F, fractional distance plots of kinetochores for three cells (a–c) with similar spindle lengths. E, images of cells presented in F (arrowheads identify midline positioned kinetochores). Scale bars, 5 μm.
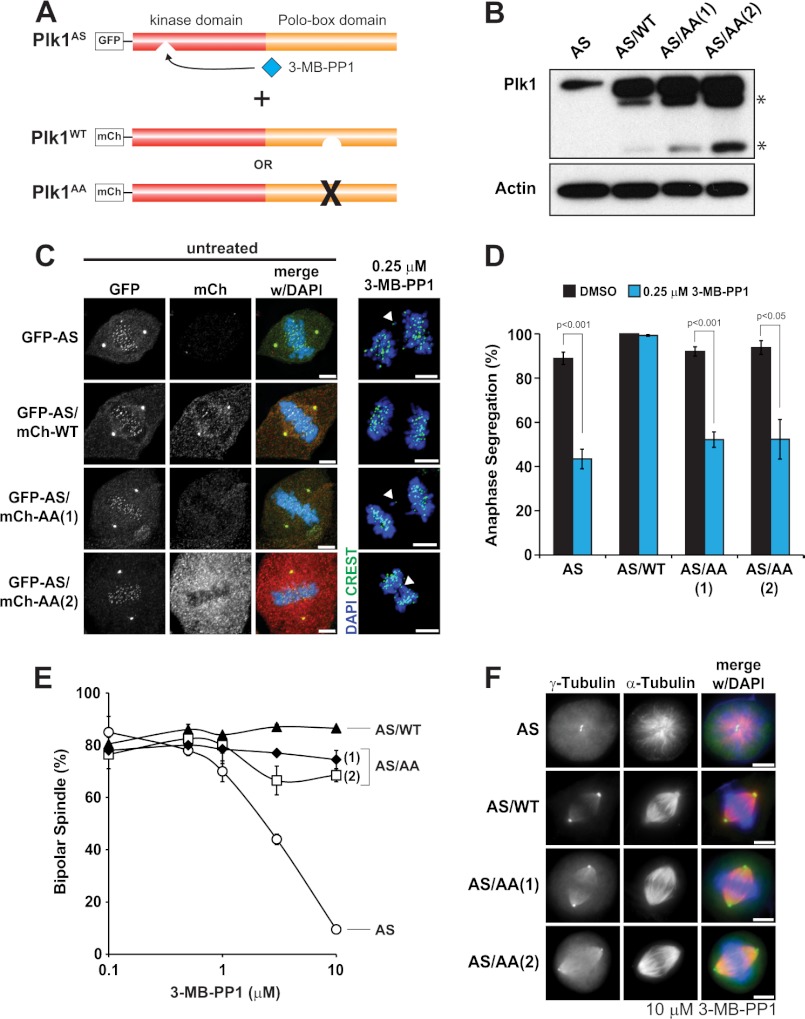
Most known functions of Plk1 are dependent on a functional C-terminal PBD, which localizes the kinase activity to specific locales or substrates that are primed by phosphorylation, often by cyclin-dependent kinase 1 or by Plk1 itself (3, 5, 6, 15). In the prevailing model, the PBD binds to substrates to generate a high local Plk1 concentration (3), although some kinase functions may not require the cis-acting PBD (15). We considered that, in principle, PBD-dependent binding could buffer the loss of Plk1 activity by providing a high local concentration. If so, the PBD-independent substrates may be exquisitely sensitive to partial Plk1 kinase inhibition because of the absence of this buffering activity. We therefore tested whether the PBD is dispensable for chromosome segregation. To do this, we used a chemical genetic complementation assay using either wild-type or a mutant PBD that is nonfunctional (PBDAA; Fig. 6A) (2, 3). We developed cell lines in which Plk1as is complemented by a separate Plk1 allele with a wild-type kinase domain and either wild-type PBD or nonfunctional PBDAA. We confirmed that both were expressed at similar levels and that the Plk1AA failed to localize to kinetochores and centrosomes as expected (Fig. 6, B and C). Next, we performed the rescue experiment. Whereas Plk1 with wild-type PBD readily rescued the anaphase missegregation phenotype, the allele with PBDAA failed to rescue this activity (Fig. 6, C, right, and D). We conclude that the anaphase functions of Plk1 require its cis-acting PBD-localizing function and reject the hypothesis that its buffering activity is dispensable for sensitive substrates.
FIGURE 6.
The PBD is required for chromosome segregation but dispensable for bipolar spindle formation. A, GFP-Plk1as (AS) cells were transduced with retroviruses expressing mCherry-tagged wild-type Plk1 (WT) or Plk1 with a mutated PBD (AA), and clonal populations (two for AS/AA) were selected for experiments. B, total Plk1 expression was determined by immunoblotting mitotic cell lysates for Plk1 and β-actin (loading control). * indicates proteolysis products. C, clonal cells were challenged with 3-MB-PP1 or left untreated for 8 h and fixed for IF. Localization of each Plk1 allele was determined by GFP (AS) or mCherry (mCh) (WT or AA) in untreated metaphase cells. Images were scaled identically for each channel. Note that the Plk1 AA allele fails to localize to kinetochores and centrosomes. Representative images of anaphase chromosome segregation in clonal cells challenged with 3-MB-PP1 are shown. Arrowheads indicate lagging chromosomes. D, the percentage of cells with segregated chromosomes was determined (as in Fig. 3B) for each clone challenged with DMSO or 3-MB-PP1 (n = 100 cells/condition; three independent experiments). Data represent averages ± S.E. (error bars). p values were determined by t test. E and F, for bipolar spindle formation, clonal cells were challenged with DMSO (0 μm) or 3-MB-PP1 (0.5, 1, 3, or 10 μm) for 8 h and fixed for IF. E, the percentage of cells with normal bipolar spindles was determined (as in Fig. 1C) for each clone (n = 200 cells/concentration; two independent experiments). Data represent averages ± S.E. (error bars). F, representative images of preanaphase mitotic spindles observed for each clone challenged with 10 μm 3-MB-PP1. Scale bars, 5 μm.
Hanisch et al. (15) reported that the PBD is not required for formation of bipolar spindles. We confirmed this result using complementation: Plk1 with the mutant PBDAA was capable of preserving bipolar spindles (Fig. 6, E and F) even though this mutant failed to localize to centrosomes (Fig. 6C). These results demonstrate that a functional PBD is required to execute Plk1 functions that require high catalytic activities but is dispensable for at least one function that requires only a small amount of activity.
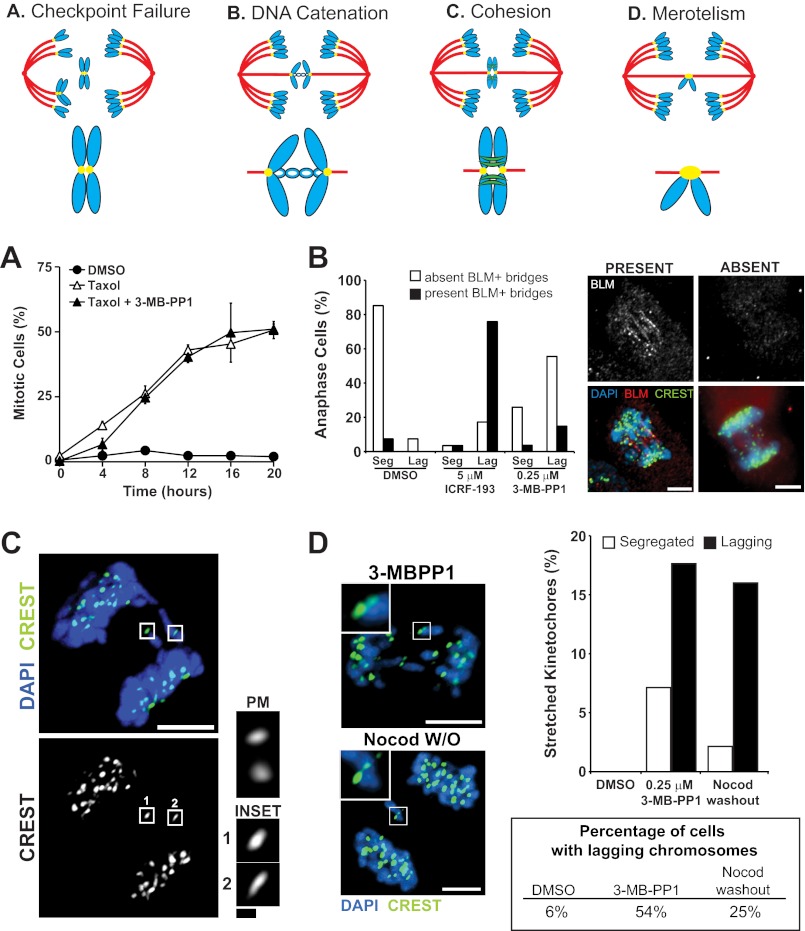
We next considered four mechanisms by which loss of Plk1 function could impair chromosome segregation in anaphase (Fig. 7, top). We first considered the possibility that anaphase was triggered prematurely due to an inactive mitotic checkpoint with partial loss of Plk1 activity (Fig. 7A). The mitotic checkpoint has been proposed to detect chromosome attachment to microtubules and possibly interkinetochore tension (18). We tested whether partial inhibition of Plk1 could interrupt the checkpoint by challenging cells with nocodazole, which precludes both attachment and tension. This revealed that loss of Plk1 activity did not impair activation of the mitotic checkpoint (not shown). Similarly, the checkpoint was active with Taxol, which activates the mitotic checkpoint by a more subtle effect of loss of attachment and/or tension on kinetochores (Fig. 7A). We conclude that the mitotic checkpoint is intact with partial loss of Plk1 activity.
FIGURE 7.
Impaired chromosome segregation is not caused by a mitotic checkpoint failure or persistent topologic linkages, but includes merotelic attachments. Top, schematic illustrating four potential sources of impaired segregation with Plk1 inhibition. A, checkpoint failure. Plk1as cells were challenged with Taxol (to activate the mitotic checkpoint) and 0.5 μm 3-MB-PP1 and then collected at 4-h intervals for 20 h. The percentage of mitotic cells was determined at each time point. Data points represent averages (three sets of 100 cells) ± S.E. (error bars). B, DNA catenation. Plk1as cells were synchronized for 8 h with monastrol, released into fresh media for 30 min, and then challenged with DMSO or ICRF-193 (topoisomerase II inhibitor) for 20 min before fixation. For 3-MB-PP1 exposure, asynchronous Plk1as cells were challenged with 0.25 μm 3-MB-PP1 for 8 h prior to fixation. Anaphase cells (n = 25 cells/condition) were identified (as in Fig. 3B) and scored for Bloom helicase (BLM)-positive (+) bridges in cells exhibiting segregated or missegregated chromosomes. Scale bars, 5 μm. C, persistent cohesion. Plk1as cells were challenged with 0.25 μm 3-MB-PP1 for 8 h and fixed. CREST signals (boxes 1 and 2), magnified 300X in insets, depict single lagging chromosomes during anaphase instead of paired sisters as seen during prometaphase (PM). Scale bars, 5 μm. Inset, 0.5 μm. D, merotelism. Asynchronous Plk1as cells were challenged with DMSO, 3-MB-PP1 (8 h), or nocodazole (Nocod) 7 h + 60-min washout (W/O) prior to fixation. Anaphase cells were identified (as in Fig. 3B). CREST-positive kinetochores of segregated (Seg) and lagging (Lag) chromosomes were measured along their longest dimension. Images illustrate cells with stretched CREST signals, magnified 300× in insets. Scale bars, 5 μm. The graph depicts the percentage of segregated and lagging kinetochores in each condition that exceeded the maximal segregated kinetochore width (0.74 μm) observed in DMSO-treated cells. The table indicates the frequency of cells exhibiting lagging chromosomes for each condition (n = 120 cells/condition; three independent experiments).
We turned to the possibility that lagging chromosomes arise from impaired DNA decatenation on anaphase onset. During replication, sister chromatids become intertwined, and these topologic links are sundered in mitosis by topoisomerase II (19, 20). When these links are not resolved, protein-coated, DNase-sensitive ultrafine bridges can be observed in anaphase. Plk1 has a possible function in resolving DNA catenation because it is known to regulate both topoisomerase II and Plk1-interacting helicase, which is found on ultrafine bridges (21, 22). To test this, we treated cells with either 3-MB-PP1 or ICRF-193, an inhibitor of topoisomerase II (Fig. 7B). For each, we quantified anaphase cells with ultrafine bridges, detected through staining of the Bloom helicase (23). As expected, ICRF-193 frequently yielded lagging anaphase chromosomes with Bloom helicase-coated bridges. In contrast, 3-MB-PP1-induced laggards rarely harbored ultrafine bridges. Similar results were obtained when visualizing ultrafine bridges with an antibody detecting Plk1-interacting helicase (not shown). We conclude that partial loss of Plk1 activity does not impair resolution of DNA catenation.
In budding yeast, the Polo kinase homolog, Cdc5, phosphorylates cohesin subunits at kinetochores, promoting its removal upon anaphase onset (24). We considered the possibility that loss of Plk1 activity impairs cohesin removal for some sister chromatid pairs. To test this, we evaluated CREST signals from kinetochores to determine whether they were singlet or doublet by comparing them with paired signals seen in prometaphase (Fig. 7C). We found that signals on lagging chromosomes were single, demonstrating successful separation of sister chromatids in anaphase. This is consistent with the finding that phosphorylation of cohesin subunits by Plk1 is dispensable for its cleavage by separase in human cells (25).
Finally, we considered the possibility that partial inhibition of Plk1 induces merotelic attachments of a single kinetochore to both spindle poles. Merotelic attachments arise when a single kinetochore is simultaneously linked to both spindle poles, precluding its migration to either pole at anaphase onset. Although difficult to observe directly, merotelic attachments can be identified by stretching of a kinetochore, which occurs in anaphase as it is pulled simultaneously to both spindle poles (26). To test this, we measured the width of CREST signals on lagging kinetochores and compared them with the effect of nocodazole washout, which increases lagging chromosomes and merotelic attachments (27) (Fig. 7D). We observed an increased number of stretched kinetochores on lagging chromosomes with 0.25 μm 3-MB-PP1, matching the effect of nocodazole washout. We conclude that Plk1 function may be important for resolving merotelic chromosome attachments.
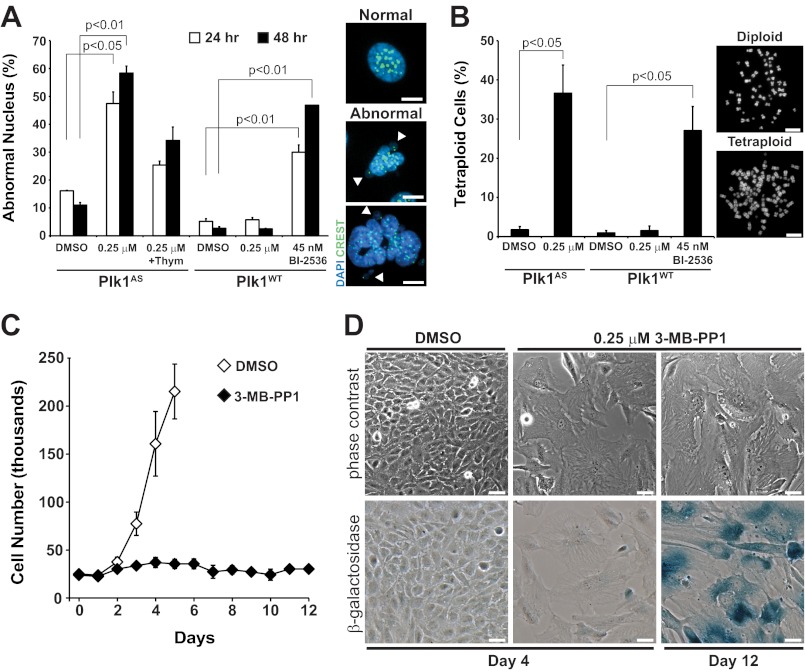
Several Plk1 inhibitors are in clinical development for anticancer therapy (28, 29). In clinical studies, mitotic arrest and abnormal spindle structure are classically used to determine whether Plk1 is actually inhibited in vivo. However, treatment doses will frequently result in partial inhibition of Plk1; even at high drug doses, this is virtually unavoidable given pharmacologic drug metabolism and washout. We therefore sought to determine the ultimate fate of human cells that encounter partial loss of Plk1 activity. To do this, we challenged Plk1as cells or matched Plk1WT cells with 24–48-h treatment with 0.25 μm 3-MB-PP1 and 45 nm BI-2536 (which provides similar effects of inhibition; Fig. 3B). We immediately identified a large number of cells with abnormal nuclear morphologies when Plk1 was inhibited (Fig. 8A). This included not only micronuclei as noted previously (14) but also lobed, binucleate, and multinucleate cells (Fig. 8A, right, and not shown). Again, this phenotype was specific to inhibition of Plk1 rather than an off-target effect: similar nuclear morphologies were seen in Plk1WT cells when challenged with BI-2536 but not with 3-MB-PP1. Moreover, blocking cell cycle progression with thymidine reduced the effect, consistent with it being the result of an abnormal prior mitosis.
FIGURE 8.
Impaired chromosome segregation produces progeny that exhibit abnormal nuclear morphologies, are tetraploid, and fail to proliferate. A, Plk1as and Plk1WT cells were challenged with DMSO, 3-MB-PP1, or BI-2536 for 24 or 48 h prior to fixation. Interphase cells were scored for normal or abnormal nuclear morphology. The graph represents the average percentage ±S.E. (error bars) of interphase cells (n = 300 cells/time point/condition; three independent experiments) with abnormal nuclei. p values were determined by t test. Thymidine (Thym) was added in one condition to verify that effects require cell cycle progression. Representative images of normal and abnormal nuclei with arrowheads indicating micronuclei are shown. Scale bars, 5 μm. B, Plk1as and Plk1WT cells were challenged with DMSO, 3-MB-PP1, or BI-2536 for 48 h; released; and incubated with nocodazole for 5 h. Mitotic cells were collected by shake-off, and chromosome spreads were performed to determine the incidence of tetraploidy. The graph represents the average percentage ±S.E. (error bars) of tetraploid cells (>80 chromosome pairs) for each condition (n = 120 cells/condition; three independent experiments). Representative images of diploid or tetraploid chromosome spreads are shown. Scale bars, 10 μm. C and D, 25,000 Plk1as cells were seeded in 12-well plates and challenged with DMSO or 0.25 μm 3-MB-PP1 up to 12 days to determine the effects of prolonged Plk1 inhibition. C, live cells (excluding trypan blue) were counted daily and plotted to determine cell proliferation. Data represent average cell counts ±S.E. (error bars) from two independent experiments. D, imaging of DMSO- and 3-MB-PP1-challenged cells at days 4 and 12 indicates that Plk1-inhibited cells become enlarged, flattened (top), and senescent as evidenced by pH-dependent β-galactosidase activity (bottom, blue staining). Scale bars, 50 μm.
Based on the findings above, we suspected a loss in genomic integrity in normal cells that traverse mitosis with partial loss of Plk1. To evaluate this directly, we made chromosome spreads of cells that experienced reduced Plk1 during proliferation for 48 h. This revealed a large increase in tetraploid cells (Fig. 8B), consistent with the observation of lobed and binucleated cells and the prior observations of failed cytokinesis. Thus, modest losses of Plk1 activity result in loss of genomic integrity with formation of tetraploid progeny, consistent with failed cytokinesis.
To observe the ultimate effect of persistent partial loss of Plk1 function, we treated Plk1as cells for up to 12 days with 3-MB-PP1 (Figs. 8, C and D). This revealed interruption of cell proliferation and stable cell number over the entire time observed (Fig. 8C). Morphologically, cells were often enlarged, granular, and binucleate, and demonstrated pH-dependent β-galactosidase activity, consistent with senescence (Fig. 8D). Thus, partial loss of Plk1 function can have a significant effect on the viability of nonmalignant human cells, leading to impaired chromosome segregation, secondary failure of cytokinesis, loss of genomic integrity, and senescence. This can occur despite normal spindle structure and minor mitotic delays. We conclude that mitotic arrest and spindle morphologies are insensitive biomarkers of Plk1 inhibition.
DISCUSSION
It is difficult to decode complex intracellular signaling networks that regulate homeostasis and proliferation of human cells. However, such a comprehensive understanding is required to rationally utilize enzymatic inhibitors for medical therapy. In cancer, antimicrotubule antimitotics represent a validated therapeutic paradigm, but less success has been realized to date with newer inhibitors of mitotic protein kinases. One proposed reason for this is that newer antimitotic drugs have an all-or-none effect rather than a graded effect as observed with antimicrotubule agents such as Taxol (30). This is not true for Plk1; we demonstrate here that Plk1 inhibition, like Taxol, exhibits a graded effect. Indeed, such concentration-dependent effects of a Plk1 inhibitor have been reported with differences in spindle structure and mitotic index as a function of inhibitor concentration (14). This may support the hypothesis that a graded effect is desirable for anticancer therapy because promising results have been reported in a recent Phase I clinical study of the Plk1 inhibitor BI-6727 (29).
Activity Thresholds
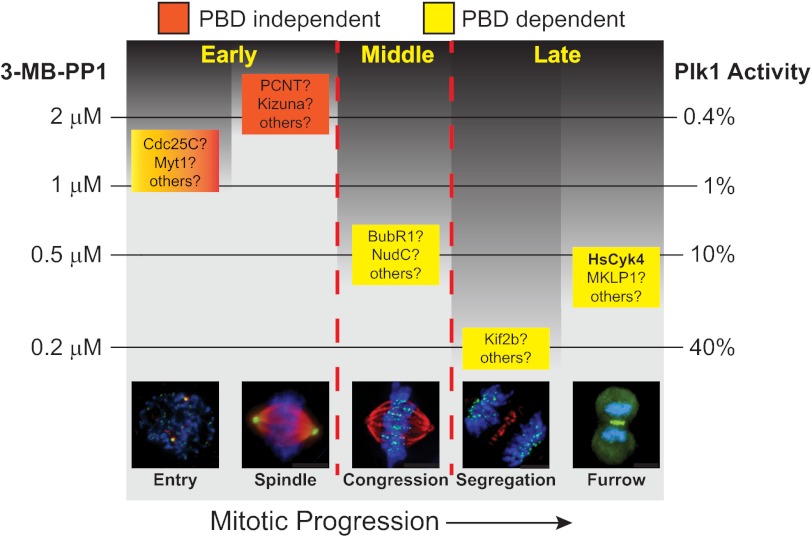
Our results support a model in which different thresholds of Plk1 activity are required to execute discrete functions that occur throughout mitosis (Fig. 9). Accurate chromosome segregation in anaphase was the most sensitive function related to loss of Plk1 activity. Additional functions required lower levels of residual Plk1 activity for proper execution.
FIGURE 9.
Separation of Plk1 function by activity threshold reveals its most sensitive functions and may help link these to substrates. Each box represents an observable phenotype seen at a specific time in mitosis (horizontal axis) and the IC50 for this phenotype (vertical axis). The box is colored by dependence on the PBD of Plk1 (mixed colors for mitotic entry, not tested). Within each box, substrates that may mediate the observed function are shown. HsCyk4 is shown in bold because we confirmed here that molecular and biologic effects have similar IC50 thresholds. The dark background represents regions in the schematic in which additional kinase functions and substrates may exist but are not readily isolated by activity thresholds. Residual Plk1 kinase activity estimated from the in vitro assay is displayed on the right.
Isolating kinase functions with thresholds may assist identification of substrates that elicit these functions. One current strategy to isolate Plk1 functions utilizes cell synchronization to separate mitosis into early, middle, and late stages. However, synchronization has limitations. For example, releasing cells from a nocodazole block generates lagging chromosomes (27), potentially obscuring effects of Plk1 inhibition.
Our findings indicate that Plk1 functions are separable by careful inhibitor titration (Fig. 9 and Table 1) to permit isolation of functions in an asynchronous cell population. Notably, we observed concordance between the Plk1 activity required for cleavage furrow formation and phosphorylation of HsCyk4 (Fig. 1E), suggesting a link between biologic function and cognate molecular phosphorylation. Considering the numerous substrates reported for individual Plk1 functions (some are shown in Fig. 9), we propose to use activity thresholds to identify the relevant substrate associated with each.
Using thresholds to isolate functional kinase activities has limitations. First, it will not be able to isolate mitotic functions that require minimal kinase activity (Fig. 9, darkly shaded regions). Second, it is possible that the activity threshold for some phenotypes could depend on the assay selected. For example, an assay that detects more subtle errors in anaphase chromosome segregation might yield a lower IC50 for this phenotype than we report. This observer effect may be lesser for robust assays of readily detected dichotomous phenotypes (e.g. spindle polarity). Finally, although we identified cell senescence as a late effect of partial loss of Plk1 activity, we cannot definitively ascribe this to the observed mitotic defect as a more subtle effect not directly visualized could lead to loss of cell viability.
The Role of the PBD
It is interesting to speculate on why nature would link a kinase to a C-terminal phosphopeptide binding module, the PBD. The PBD detects an upstream signal, allowing the kinase activity to be recruited at high local concentration. This recruited kinase, in turn, can phosphorylate multiple adjacent proteins although usually at sites different from the PBD docking site. In effect, Plk1 is a mitotic signal transducer and amplifier, which allow weak upstream molecular signals to be converted into substantial increases in local phosphorylation to mediate major events in cell division. This can even be autoamplification if the upstream signal is generated by Plk1 itself as occurs in late mitosis (2, 4). Given this view, it is perhaps unsurprising that transducer/amplifier Plk1 is involved in a multitude of mitotic functions or that many Plk1 functions require a functional PBD. Moreover, our observation that Plk1 functions required different activity thresholds suggests that different degrees of signal amplification are required for each functional event.
However, not all Plk1 functions are mediated through a functional PBD (see Fig. 9, orange versus yellow boxes). Prior studies with dominant-negative PBD expression (15) or competitive PBD binding (31) demonstrated abnormal chromosome congression but normal spindle formation. Here we confirmed that the PBD is dispensable for spindle function, suggesting that the Plk1-dependent substrate required to form a bipolar spindle is readily accessible and is isolated from phosphatases or requires little phosphorylation to elicit spindle bipolarity. Moreover, it suggests that the spindle function of Plk1 might not be mediated by a substrate localized at the centrosome. The substrate could be, for example, a plus-end-directed motor required to maintain spindle polarity. If so, the centrosome-localized pool of Plk1 may have a more obscure function or may simply represent a reservoir of kinase; this idea is supported by the recent discovery that Plk1 function requires dynamic relocalization from the centrosome (32).
In contrast to the effect on spindle polarity, a functional PBD is essential for cells to accurately congress and segregate chromosomes even though these functions can be readily blocked with modest inhibition of Plk1. Thus, the PBD appears to provide a high local activity of Plk1 to overcome phosphatases and saturate substrates. We speculate that PBD-dependent functions of Plk1 on chromosome congression and segregation emanate from kinetochores, a complex zone of activities of overlapping kinases and phosphatases in which high local kinase activity may be required to elicit effects.
Anaphase Function of Plk1
We demonstrate here that Plk1 has a function in chromosome segregation in anaphase. Indeed, this function is distinct from the spindle elongation defect described previously (17). Moreover, we and others did not observe impaired chromosome segregation when Plk1 was inhibited specifically upon anaphase onset (8, 17, 33). This suggests that the observed anaphase phenotype is related to an earlier mitotic event that requires a high level of Plk1 activity. This is not related to the effect on the mitotic checkpoint or impaired resolution of sister chromatid cohesion or DNA topology. However, the stretched CREST signals at kinetochores suggests merotelic attachment as a likely mechanism (26, 34). Intriguingly, Aurora B, a known mediator of resolving merotelic attachments, has recently been described as a direct activator of Plk1 at the centromere/inner kinetochore (35). Moreover, improper syntelic attachments due to kinetochore dysfunction have been reported with Polo depletion in Drosophila S2 cells (36). These observations suggest a number of intriguing possibilities, but it remains unclear how Plk1 might mediate resolution of merotelic microtubule attachments.
Aside from merotely, weak chromosome attachment is an important alternative explanation for the observed missegregation of chromosomes seen in anaphase. Plk1 is known to mediate stable kinetochore-microtubule attachment in part through phosphorylation of BubR1 (37, 38). Consistent with this, misaligned chromosomes were often seen with partial inhibition of Plk1 (Figs. 3, A and C, and 4A) as expected from the known function of Plk1 promoting stable end-on attachments (37, 39). However, we do not believe that poor kinetochore-microtubule attachment is sufficient to explain poorly segregated anaphase chromosomes. First, residual Plk1 activity was sufficient to silence the mitotic checkpoint and allow cells to proceed into anaphase, suggesting adequate attachment of all kinetochores. Second, poor chromosome congression at the metaphase plate is a characteristic feature of poor kinetochore-spindle attachment, but the threshold for this phenotype is distinct from that of impaired anaphase segregation (0.5 versus 0.2 μm; Fig. 4, A and B). For these reasons, the anaphase phenotype is unlikely to be a product of unstable kinetochore-microtubule attachment.
Clinical Implications
We demonstrate here that low levels of Plk1 inhibition lead to loss of genomic integrity. This occurs with the development of micronuclei, which can lead to chromosome pulverization and chromothripsis (40). However, the primary abnormality observed at the chromosome was development of tetraploid progeny, which are known to lead to subsequent chromosome instability (41). Loss of Plk1 activity can therefore allow progression through a bipolar mitosis, albeit abnormally, and results in loss of genomic integrity and cell senescence.
These results have important implications in the clinical development of Plk1 inhibitors. First, pharmacokinetic end-points for Plk1 inhibitors should include chromosome segregation errors, which appear to be the most sensitive phenotype related to partial loss of Plk1 function. Second, under some conditions, Plk1 inhibitors may lead to a loss of genomic integrity in proliferating healthy cells similar to classic chemotherapies and radiation, and (unless prevented by senescence) latent second malignancies are a consideration when Plk1 inhibition is used as a component of curative therapy. Third, the therapeutic window for cancer therapy could depend markedly on drug pharmacology as complete Plk1 loss of function for short times (e.g. intravenous dosing with a short half-life drug) may produce remarkably different effects than achieved with partial Plk1 loss of function for extended times (e.g. low dose continual oral dosing of a long half-life drug).
Conclusions
A major strength of our study is the use of chemical genetics with explicit controls to ensure that phenotypes are linked to Plk1 inhibition. Although we confirmed similar phenotypes in other cell lines, we cannot rule out the possibility that the relative order of Plk1 activity thresholds may be cell line-dependent. This may occur both because of various levels of Plk1 expression, differences in activity of wild-type versus analog-sensitive Plk1 alleles, and different intracellular phosphatase activities. Despite limitations, we were able to demonstrate that it is possible to dissect the multiple functions of a kinase through activity thresholds.
In sum, we demonstrated that different thresholds of Plk1 activities are required to execute each specific biologic function of this kinase. We anticipate that this will allow dissection of Plk1 functions and ultimately provide a detailed understanding of how the many substrates of Plk1 mediate its functions in human cell division. Chemical genetics can separate functions in both time and concentration threshold, allowing dissection and mechanistic interrogation of discrete functions of a multifunctional kinase.
Supplementary Material
Acknowledgments
We thank B. Weaver and R. Blank for critical reading of the manuscript, P. Jallepalli for sharing BI-2536, and A. Mastrocola and members of the Burkard and Weaver laboratories for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM097245 (to M. E. B.) and P30 CA014520 (for R. L.) and Clinical and Translational Science Award 9U54TR000021 from the National Center for Advancing Translational Sciences (K Award to M. E. B.) This work was also supported by Flight Attendant Medical Research Institute Grant 062541_YCSA.

This article contains supplemental Movies S1–S4.
- Plk1
- Polo-like kinase 1
- 3-MB-PP1
- 3-methylbenzylpyrazolopyrimidine
- DMSO
- dimethyl sulfoxide
- EGFP
- enhanced green fluorescent protein
- IF
- immunofluorescence
- PBD
- Polo box domain
- Plk1as
- analog-sensitive mutant of Plk1, C67V/L130G
- RPE
- retinal pigment epithelial
- CREST
- Calcinosis, Raynaud's, Esophageal dismotility, Sclerodactyly, Telangiectasia.
REFERENCES
- 1. Barr F. A., Silljé H. H., Nigg E. A. (2004) Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5, 429–440 [DOI] [PubMed] [Google Scholar]
- 2. Burkard M. E., Maciejowski J., Rodriguez-Bravo V., Repka M., Lowery D. M., Clauser K. R., Zhang C., Shokat K. M., Carr S. A., Yaffe M. B., Jallepalli P. V. (2009) Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol. 7, e1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Elia A. E., Rellos P., Haire L. F., Chao J. W., Ivins F. J., Hoepker K., Mohammad D., Cantley L. C., Smerdon S. J., Yaffe M. B. (2003) The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 115, 83–95 [DOI] [PubMed] [Google Scholar]
- 4. Neef R., Gruneberg U., Kopajtich R., Li X., Nigg E. A., Sillje H., Barr F. A. (2007) Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat. Cell Biol. 9, 436–444 [DOI] [PubMed] [Google Scholar]
- 5. Liu F., Park J. E., Qian W. J., Lim D., Scharow A., Berg T., Yaffe M. B., Lee K. S., Burke T. R., Jr. (2012) Identification of high affinity Polo-like kinase 1 (Plk1) Polo-box domain binding peptides using oxime-based diversification. ACS Chem. Biol. 7, 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lowery D. M., Clauser K. R., Hjerrild M., Lim D., Alexander J., Kishi K., Ong S. E., Gammeltoft S., Carr S. A., Yaffe M. B. (2007) Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J. 26, 2262–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oppermann F. S., Grundner-Culemann K., Kumar C., Gruss O. J., Jallepalli P. V., Daub H. (2012) Combination of chemical genetics and phosphoproteomics for kinase signaling analysis enables confident identification of cellular downstream targets. Mol. Cell. Proteomics 11, O111.012351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burkard M. E., Randall C. L., Larochelle S., Zhang C., Shokat K. M., Fisher R. P., Jallepalli P. V. (2007) Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc. Natl. Acad. Sci. U.S.A. 104, 4383–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Randall C. L., Burkard M. E., Jallepalli P. V. (2007) Polo kinase and cytokinesis initiation in mammalian cells: harnessing the awesome power of chemical genetics. Cell Cycle 6, 1713–1717 [DOI] [PubMed] [Google Scholar]
- 10. Burkard M. E., Santamaria A., Jallepalli P. V. (2012) Enabling and disabling Polo-like kinase 1 inhibition through chemical genetics. ACS Chem. Biol. 7, 978–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Debnath J., Muthuswamy S. K., Brugge J. S. (2003) Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 [DOI] [PubMed] [Google Scholar]
- 12. Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wolfe B. A., Takaki T., Petronczki M., Glotzer M. (2009) Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 7, e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gilmartin A. G., Bleam M. R., Richter M. C., Erskine S. G., Kruger R. G., Madden L., Hassler D. F., Smith G. K., Gontarek R. R., Courtney M. P., Sutton D., Diamond M. A., Jackson J. R., Laquerre S. G. (2009) Distinct concentration-dependent effects of the polo-like kinase 1-specific inhibitor GSK461364A, including differential effect on apoptosis. Cancer Res. 69, 6969–6977 [DOI] [PubMed] [Google Scholar]
- 15. Hanisch A., Wehner A., Nigg E. A., Silljé H. H. (2006) Different Plk1 functions show distinct dependencies on Polo-box domain-mediated targeting. Mol. Biol. Cell 17, 448–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steegmaier M., Hoffmann M., Baum A., Lénárt P., Petronczki M., Krssák M., Gürtler U., Garin-Chesa P., Lieb S., Quant J., Grauert M., Adolf G. R., Kraut N., Peters J. M., Rettig W. J. (2007) BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol. 17, 316–322 [DOI] [PubMed] [Google Scholar]
- 17. Brennan I. M., Peters U., Kapoor T. M., Straight A. F. (2007) Polo-like kinase controls vertebrate spindle elongation and cytokinesis. PLoS One 2, e409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maresca T. J., Salmon E. D. (2010) Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. J. Cell Sci. 123, 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clarke D. J., Johnson R. T., Downes C. S. (1993) Topoisomerase II inhibition prevents anaphase chromatid segregation in mammalian cells independently of the generation of DNA strand breaks. J. Cell Sci. 105, 563–569 [DOI] [PubMed] [Google Scholar]
- 20. Spence J. M., Phua H. H., Mills W., Carpenter A. J., Porter A. C., Farr C. J. (2007) Depletion of topoisomerase IIα leads to shortening of the metaphase interkinetochore distance and abnormal persistence of PICH-coated anaphase threads. J. Cell Sci. 120, 3952–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baumann C., Körner R., Hofmann K., Nigg E. A. (2007) PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128, 101–114 [DOI] [PubMed] [Google Scholar]
- 22. Li H., Wang Y., Liu X. (2008) Plk1-dependent phosphorylation regulates functions of DNA topoisomerase IIα in cell cycle progression. J. Biol. Chem. 283, 6209–6221 [DOI] [PubMed] [Google Scholar]
- 23. Chan K. L., North P. S., Hickson I. D. (2007) BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26, 3397–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alexandru G., Uhlmann F., Mechtler K., Poupart M. A., Nasmyth K. (2001) Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105, 459–472 [DOI] [PubMed] [Google Scholar]
- 25. Hauf S., Roitinger E., Koch B., Dittrich C. M., Mechtler K., Peters J. M. (2005) Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 3, e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cimini D., Howell B., Maddox P., Khodjakov A., Degrassi F., Salmon E. D. (2001) Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 153, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cimini D., Moree B., Canman J. C., Salmon E. D. (2003) Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J. Cell Sci. 116, 4213–4225 [DOI] [PubMed] [Google Scholar]
- 28. Medema R. H., Lin C. C., Yang J. C. (2011) Polo-like kinase 1 inhibitors and their potential role in anticancer therapy, with a focus on NSCLC. Clin. Cancer Res. 17, 6459–6466 [DOI] [PubMed] [Google Scholar]
- 29. Schöffski P., Awada A., Dumez H., Gil T., Bartholomeus S., Wolter P., Taton M., Fritsch H., Glomb P., Munzert G. (2012) A phase I, dose-escalation study of the novel Polo-like kinase inhibitor volasertib (BI 6727) in patients with advanced solid tumours. Eur. J. Cancer 48, 179–186 [DOI] [PubMed] [Google Scholar]
- 30. Gascoigne K. E., Taylor S. S. (2009) How do anti-mitotic drugs kill cancer cells? J. Cell Sci. 122, 2579–2585 [DOI] [PubMed] [Google Scholar]
- 31. Yun S. M., Moulaei T., Lim D., Bang J. K., Park J. E., Shenoy S. R., Liu F., Kang Y. H., Liao C., Soung N. K., Lee S., Yoon D. Y., Lim Y., Lee D. H., Otaka A., Appella E., McMahon J. B., Nicklaus M. C., Burke T. R., Jr., Yaffe M. B., Wlodawer A., Lee K. S. (2009) Structural and functional analyses of minimal phosphopeptides targeting the polo-box domain of polo-like kinase 1. Nat. Struct. Mol. Biol. 16, 876–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kishi K., van Vugt M. A., Okamoto K., Hayashi Y., Yaffe M. B. (2009) Functional dynamics of Polo-like kinase 1 at the centrosome. Mol. Cell. Biol. 29, 3134–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Petronczki M., Glotzer M., Kraut N., Peters J. M. (2007) Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev. Cell 12, 713–725 [DOI] [PubMed] [Google Scholar]
- 34. Cimini D., Fioravanti D., Salmon E. D., Degrassi F. (2002) Merotelic kinetochore orientation versus chromosome mono-orientation in the origin of lagging chromosomes in human primary cells. J. Cell Sci. 115, 507–515 [DOI] [PubMed] [Google Scholar]
- 35. Carmena M., Pinson X., Platani M., Salloum Z., Xu Z., Clark A., Macisaac F., Ogawa H., Eggert U., Glover D. M., Archambault V., Earnshaw W. C. (2012) The chromosomal passenger complex activates Polo kinase at centromeres. PLoS Biol. 10, e1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moutinho-Santos T., Conde C., Sunkel C. E. (2012) POLO ensures chromosome bi-orientation by preventing and correcting erroneous chromosome-spindle attachments. J. Cell Sci. 125, 576–583 [DOI] [PubMed] [Google Scholar]
- 37. Elowe S., Hümmer S., Uldschmid A., Li X., Nigg E. A. (2007) Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 21, 2205–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paschal C. R., Maciejowski J., Jallepalli P. V. (2012) A stringent requirement for Plk1 T210 phosphorylation during K-fiber assembly and chromosome congression. Chromosoma doi: 10.1007/s00412-012-375-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lénárt P., Petronczki M., Steegmaier M., Di Fiore B., Lipp J. J., Hoffmann M., Rettig W. J., Kraut N., Peters J. M. (2007) The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17, 304–315 [DOI] [PubMed] [Google Scholar]
- 40. Crasta K., Ganem N. J., Dagher R., Lantermann A. B., Ivanova E. V., Pan Y., Nezi L., Protopopov A., Chowdhury D., Pellman D. (2012) DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ganem N. J., Godinho S. A., Pellman D. (2009) A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lampson M. A., Kapoor T. M. (2005) The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 7, 93–98 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.