Background: mARC1 is a novel molybdenum enzyme functioning in pro-drug metabolism with unknown subcellular localization.
Results: mARC1 localizes to the outer mitochondrial membrane with its catalytic domain facing the cytosol. Import is dependent on two N-terminal motifs (mitochondrial targeting signal, transmembrane domain) resulting in high oligomeric complexes.
Conclusion: mARC1 is a novel signal-anchored protein of the outer mitochondrial membrane.
Significance: We identify mARC protein maturation.
Keywords: Drug Metabolism, Metalloenzymes, Mitochondria, Mitochondrial Transport, Molybdenum, mARC1, Molybdenum Cofactor, Outer Mitochondrial Membrane, Signal-anchored Protein
Abstract
The mitochondrial amidoxime-reducing component (mARC) was recently discovered as the fifth eukaryotic molybdenum cofactor-containing enzyme. The human genome encodes two mARC proteins, mARC1 and mARC2, sharing significant homologies with respect to sequence and function. Whereas mARC2 was identified as a mitochondrial enzyme, the subcellular localization of mARC1 has remained uncharacterized, although the similarity of both proteins suggested identical subcellular localizations. In addition, neither mARC1 nor mARC2 could be attributed unambiguously to one of the four mitochondrial subcompartments. Accordingly, mechanisms triggering the subcellular distribution of both enzymes have been unexplored so far. Here, we shed light on the subcellular localization of mARC1 and demonstrate that it is integrated into the outer mitochondrial membrane. The C-terminal catalytic domain of the protein remains exposed to the cytosol and confers an N(in)-C(out) membrane orientation of mARC1. This localization is triggered by the N terminus of the enzyme, being composed of a weak N-terminal mitochondrial targeting signal and a downstream transmembrane helix. We demonstrate the transmembrane domain of mARC1 to be sufficient for mitochondrial targeting and the N-terminal targeting signal to function as a supportive receptor for the outer mitochondrial membrane. According to its localization and targeting mechanism, we classify mARC1 as a novel signal-anchored mitochondrial protein. During mitochondrial import, mARC1 is not processed, and membrane integration proceeds membrane potential independently but requires external ATP, which finally results in the assembly of mARC1 into high oligomeric protein complexes.
Introduction
Molybdenum (Mo) constitutes an essential micronutrient for most eukaryotes and prokaryotes, where it is involved in the catalysis of a large variety of redox reactions. Molybdenum gains biological activity by its chelation and activation through its insertion into the molybdenum cofactor (Moco).2 In eukaryotes, Moco is synthesized from GTP by a multistep reaction cascade, with the final steps taking place in the cytosol (1). The biological relevance of Moco is fulfilled by its integration into a large number of Mo enzymes catalyzing a multitude of different cellular redox reactions (2). Most of these proteins only occur in prokaryotes, whereas five Moco-dependent enzymes are known in eukaryotes.
One of the Mo enzymes, nitrate reductase, is present solely in plants, algae, and fungi, whereas animals comprise a remaining subset of four different Moco-dependent enyzmes (3). The collection of animal Moco enzymes can be classified into two groups, which also reflect their respective subcellular localizations. One group is composed of the cytosolic proteins aldehyde oxidase and xanthine oxidase, which are involved in the oxidation of a large number of aldehydes and the metabolism of purines, respectively (1). Apart from its functional relevance in the cytosol, Moco is biologically active in mitochondria, where it is an essential component of the second group of Moco enzymes. Sulfite oxidase is a soluble protein of the mitochondrial intermembrane space and catalyzes the fundamental oxidation of toxic sulfite to nonhazardous sulfate (4). The second mitochondrial Moco enzyme was recently discovered and purified from mitochondria based on its function in the reduction of N-hydroxylated pro-drugs (5). The corresponding gene had been annotated as MOSC2 based on sequence similarities to the C-terminal domain of Moco sulfurase, an enzyme essential for the sulfuration of Moco in aldehyde and xanthine oxidases. Based on its first observed catalytic activity, the protein was termed the mitochondrial amidoxime-reducing component 2 (mARC2) according to its identified function (5). Apart from the conversion of amidoxime pro-drugs to the respective active amidine drugs, the only known physiological function of mARC2 accounts for its involvement in the regulation of nitric oxide synthesis (6). The catalytic activity of mARC2 requires its integration into a three-component enzyme system, in which electrons are transferred from NADH to cytochrome b5 reductase and via cytochrome b5 to mARC2, where the substrate is finally reduced at the Mo active site.
The human genome encodes an additional MOSC gene, which was identified based on sequence similarities to MOSC2 and the tandem orientation of both genes on chromosome 1 (7). According to similar enzymatic properties in the reduction and activation of N-hydroxylated pro-drugs, the corresponding protein was termed mARC1 (7). In analogy to mARC2, mARC1 is active in the same N-reductive enzyme system and was also found to contribute to the regulation of nitric oxide synthesis (6).
Initially, mARC2 was isolated from outer mitochondrial membranes, and consistently, its localization was later confirmed within mitochondria (8, 9). However, the exact localization to one of the four mitochondrial subcompartments has not been unambiguously addressed so far, as the evidence in the literature is contradictory in respect to the submitochondrial localization (5, 9, 10). In addition, the subcellular localization of mARC1 has remained completely unexplored.
EXPERIMENTAL PROCEDURES
Expression Constructs
The human MOSC1 coding sequence was purchased from ImaGenes and amplified by PCR. Sequencing revealed two reproducible polymorphisms resulting in substitution of threonine 165 by alanine and methionine 187 by lysine. However, these polymorphisms also appear in the databases, e.g. as protein accessions NP073583, AAH10619, and EAW93921 and hence obviously represent naturally occurring polymorphisms. GFP-tagged versions of human mARC1 and all truncated variants were achieved by cloning into the pEGFP-N1 vector (Clontech) using HindIII and KpnI restriction sites. mARC1 was cloned into pcDNA3.1 Myc/HisA (Invitrogen) using HindIII and EcoRI restriction sites to obtain myc-tagged and untagged mARC1.
Determination of Moco Content by the nit-1 Assay
Harvested human fibroblasts were homogenized by sonication on ice for 30 s and subsequently centrifuged at 21,000 × g for 15 min. Fibroblast extract was incubated anaerobically with nit-1 extract for 12 h. Nitrate reductase activity was subsequently determined as described (11).
Cell Culture and Transfection
HEK-293 cells, HEP-G2 cells, HeLa cells, and human fibroblasts were cultured in 10-cm dishes at 37 ºC and 5% CO2 in DMEM (PAA Laboratories). For confocal laser-scanning microscopy, 1 × 105 HEK-293 cells and 3 × 104 human fibroblasts were seeded on collagenized coverslips in 12-well plates. After 24 h, transfection of HEK-293 cells and human fibroblasts was conducted with polyethylenimine (1 mg/ml, diluted in H2O, pH 7.0). For each well, 3.4 μl of polyethylenimine was added to 66 μl of DMEM in the absence of fetal calf serum. Following 5 min of incubation, 0.85 μg of DNA was added, followed by another 20 min of incubation. This mixture was added to each well, and cells were grown for another 48 h. For biochemical studies using Western blotting, 1.45 × 106 HEK-293 cells were seed on 10-cm plates and transfected with polyethylenimine as described above but scaled linearly to the increased cell numbers. Cells were harvested 48 h after transfection.
Antibody Staining of Cell Culture Preparations and Confocal Microscopy
For mitochondrial staining, cells were incubated with MitoTracker Red CMXRos (Invitrogen) according to the instructions of the manufacturer. Cells were fixed with 4% paraformaldehyde for 20 min and either mounted directly on coverslips in the case of GFP-mediated detection or subsequently permeabilized with 0.2% Triton X-100 (diluted in PBS) for 20 min in the case of antibody-mediated detection. After incubation with 1% BSA for 2 min, the primary anti-myc antibody (9E10) was applied for 1 h at 37 ºC. Afterward, cells were incubated with 5 μg/ml secondary antibody (Alexa Fluor 488 goat anti-mouse; Invitrogen) for 1 h. Cells were finally washed two times with PBS and mounted on a microscope slide. Images of cells were taken with a Nikon Eclipse Ti confocal laser-scanning microscope and processed with the EZ-C1 software (Nikon).
Isolation of Mitochondria
Mitochondria were obtained from HEK-293 cells, HEP-G2 cells, or HeLa cells as described previously (12).
Na2CO3 Extraction
Enriched mitochondria were resuspended in 0.1 m Na2CO3, pH 11.5, to a concentration of 0.3 mg/ml. Mitochondria were incubated 30 min on ice and centrifuged at 100,000 × g for 1 h at 4 ºC. The pellets were washed with H2O and resuspended in SDS loading buffer. The supernatants were precipitated with TCA, and protein pellets were resuspended in SDS loading buffer.
Antibody Detection of mARC1
mARC1 was detected by Western blotting using anti-MOSC1 polyclonal (Abcam) or anti-myc monoclonal (9E10) antibodies.
In Vitro Translation of mARC1 and Mitochondrial in Vitro Imports
In vitro translation of human mARC1 occurred by means of the troponin T-coupled reticulocyte lysate system (Promega) using T7 RNA polymerase and [35S]methionine (Hartmann Analytic). For mitochondrial import assays, mitochondria were enriched from HEK-293 cells and concentrated to 1 mg/ml in import buffer (250 mm sucrose, 5 mm magnesium acetate, 80 mm potassium acetate, 10 mm sodium succinate, 1 mm DTT, 0.1 mm ATP, 20 mm Hepes-KOH, pH 7.4). Import reactions were conducted with 100 μl of mitochondria and 5 μl of reticulocyte lysate. Membrane potential inhibition occurred by application of 20 μg/ml valinomycin prior to the import reaction, and ATP dependence was determined using an import buffer lacking ATP. The import reactions were incubated at 37 ºC and 400 rpm for 5, 15, and 45 min. Mitochondria were pelleted and resuspended in 100 μl of solubilization buffer (1% digitonin, 20 mm Bis-Tris, 50 mm NaCl, 10% glycerol). Samples were incubated 30 min on ice and centrifuged at maximum speed. Supernatants were analyzed by Blue Native PAGE (BN-PAGE) and Western blotting. Blots were exposed to an x-ray cassette for 1 week and signals were analyzed by a Typhoon Trio Variable Mode Imager (Amersham Biosciences).
BN-PAGE
BN-PAGE was conducted as described previously (12).
RESULTS
The N Terminus of mARC1 Contains a Predicted Mitochondrial Targeting Signal and a Downstream Hydrophobic Domain
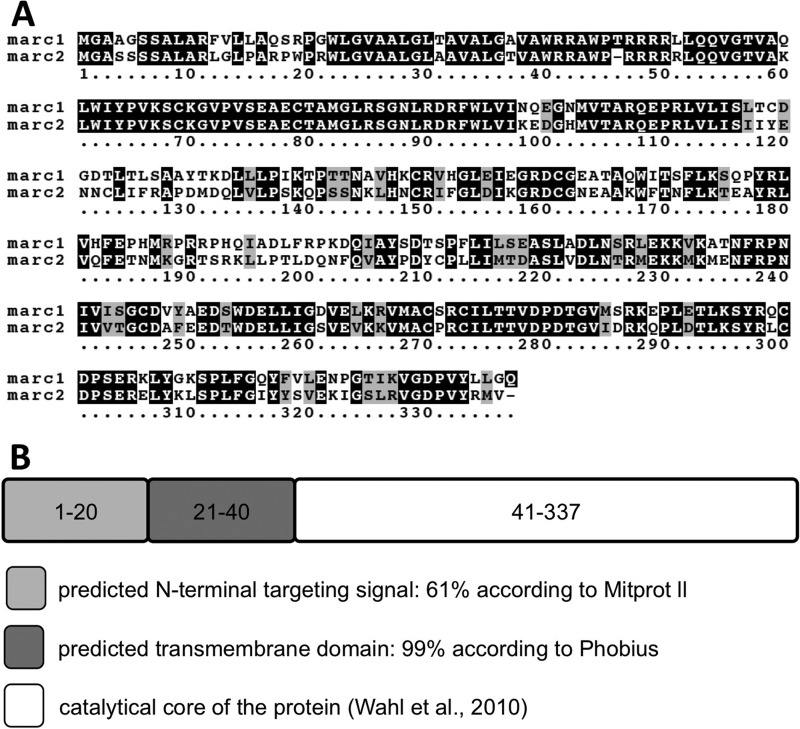
The mitochondrial activity of mARC2 (5) suggested a similar localization of mARC1, as both proteins are part of the same three-component enzyme system and reduce a similar subset of substrates. Consequently, human mARC1 and mARC2 are characterized by a high degree of sequence conservation (Fig. 1A). Altogether, the functional and primary sequence homologies of mARC1 and mARC2 suggested identical subcellular localizations and similar mechanisms of cellular trafficking. Therefore, we studied human mARC1 as a representative member of the family of mARC proteins.
FIGURE 1.
Analysis of mARC1 primary structure and sequence homology to mARC2. A, sequence alignment of human mARC1 and mARC2 using the Boxshade tool. Residues are framed black (high conservation), gray (moderate conservation), or white (no conservation). Protein accession numbers are NP_073583 (human mARC1) and NP_060368 (human mARC2). B, peptide sequence of human mARC1 analyzed by MITOPROT II and Phobius. Sketch represents the predicted motifs with numbers illustrating the involved residues.
First, we analyzed the primary structure of human mARC1 by means of in silico prediction tools (13, 14) to obtain first indications for its subcellular localization (Fig. 1B), which revealed the presence of a potential N-terminal mitochondrial targeting signal and a downstream hydrophobic domain with high probability scores. The remaining part of the protein was shown to be catalytically active in vitro in absence of residues 1–40 (8) but was not predicted to contain any further mitochondrial targeting or transmembrane motifs and was hence termed the catalytic core of the enzyme.
mARC1 Is Associated with a Mitochondrial Membrane
Considering the mitochondrial localization of mARC2 and the predictions stated above, we expected mARC1 to be directed to mitochondria. Therefore, mARC1 was C-terminally fused to GFP and expressed in HEK-293 cells. The exclusive co-localization of mARC1-GFP with mitochondria stained by MitoTracker Red, as depicted by confocal laser-scanning microscopy, demonstrated an efficient mitochondrial localization of mARC1 (Fig. 2A). Next, we aimed to identify to which of the four mitochondrial subcompartments mARC1 is localized. Given the sequence analysis of mARC proteins, a membrane association was predicted, which could be either the outer or inner mitochondrial membrane. However, many mitochondrial proteins contain hydrophobic regions in their precursor forms, which are lost during maturation by proteolytic processing, resulting in soluble mitochondrial proteins (15). To evaluate whether mARC1 is either bound to a mitochondrial membrane or appears as a soluble protein, we purified mitochondria from HEP-G2 cells and separated mitochondrial membranes from soluble fractions by alkaline treatment with 0.1 m Na2CO3 (16). Successful separation of both fractions was achieved by ultracentrifugation and demonstrated the exclusive detection of the respective marker proteins voltage dependent anion channel (outer membrane) and COX4 (inner membrane) in the pellet fractions and the exclusive accumulation of the soluble proteins SMAC (intermembrane space) and HSP60 (matrix) in the supernatant fractions. Following this separation protocol, endogenous mARC1 protein was only detected in the pellet fraction, demonstrating that mARC1 is membrane-bound in its mature form (Fig. 2B).
FIGURE 2.
Subcellular localization of human mARC1. A, mARC1-GFP (green) was expressed in HEK-293 cells for 48 h and visualized by confocal laser-scanning microscopy. Mitochondria were stained with MitoTracker Red CMXRos (mitoTR). Scale bar, 10 μm. Sketch illustrates the expression construct with the predicted mitochondrial targeting signal (light gray), the transmembrane domain (dark gray), the catalytic core (white), and GFP (green). B, mitochondria were enriched from HEP-G2 cells by differential centrifugation and resuspended in 0.1 m Na2CO3, pH 11.5. Fractions were separated by centrifugation at 100.000 × g, and the pellet (P) as well as supernatant (SN) were loaded on a 12% SDS-PAGE. Distribution of marker proteins and mARC1 was analyzed by Western blotting. C and D, mitochondria were enriched from HeLa cells and treated with 100 μg/ml (C) PK or (D) trypsin for 10 min. Swelling occurred by incubation of mitochondria in 10 mm Hepes, pH 7.4, for 10 min prior to protease application. After protease inactivation and TCA precipitation, mitochondrial proteins were loaded on a Tris/Tricine gradient SDS-PAGE (8–17.5%) followed by Western blotting.
mARC1 Is Integrated into the Outer Mitochondrial Membrane with an N(in)-C(out) Orientation
Because inner and outer mitochondrial membranes were not separated during Na2CO3 extraction, the subcellular localization of mARC1 was not entirely resolved. In addition, the topology of mARC1 within the respective membrane was not clarified. We therefore purified mitochondria from HeLa cells and assessed the accessibility of mARC1 to externally added proteases to purified mitochondria and interpreted protein stability against or susceptibility to degradation in light of protein localization. First, we purified mitochondria from HeLa cells and examined the stability of endogenous mARC1 to proteinase K (PK) exposure. MFN2, a protein of the outer mitochondrial membrane exposing a large cytosolic domain, was efficiently degraded upon PK treatment of mitochondria, whereas proteins of the intermembrane space (SMAC) and matrix (HSP60) were protected against the protease (Fig. 2C). Incubation of mitochondria in a hypotonic solution allows selective disruption of the outer mitochondrial membrane, whereas the inner membrane remains intact under these conditions (17). Consistently, MFN2 and SMAC were degraded upon osmotic swelling of mitochondria and PK treatment, whereas HSP60 residing in the mitochondrial matrix was unaffected. When we tested the stability of endogenous mARC1 to PK treatment of intact mitochondria, we found that mARC1, but not SMAC or HSP60, was degraded upon PK treatment, demonstrating that it is integrated into the outer mitochondrial membrane (Fig. 2C).
Notably, a proteolytic fragment of ∼30 kDa of mARC1 was formed upon protease treatment of intact mitochondria (Fig. 2C), which allowed us to examine the topology of mARC1 in the outer membrane. Considering the predictions in Fig. 1B, two orientations of mARC1 in the outer membrane are possible. Either, the C terminus may be exposed to the intermembrane space and the 20 residues upstream of the transmembrane domain are facing the cytosol (N(out)-C(in) orientation), or the N terminus may point toward the intermembrane space while the core of the protein remains cytosolic (N(in)-C(out) orientation). Compared with the intensity of full-length mARC1 in absence of PK, the truncated fragment following PK treatment was hardly detectable. This finding together with the size of the sequence preceding the transmembrane domain (2 kDa), which would be degraded in the event of an outside orientation, suggests that the weak signal at 30 kDa is derived from an incomplete degradation of the large C-terminal core facing the cytosol.
To confirm the proposed N(in)-C(out) orientation of mARC1, trypsin was chosen as a second protease. Assuming the C-terminal core of mARC1 to be directed toward the intermembrane space, a similar protease-protected fragment as seen upon PK treatment should appear following trypsin application. Because no smaller fragment was detected after trypsin exposure (Fig. 2D), we conclude that the truncated fragment seen in Fig. 2C was due to incomplete degradation by PK and demonstrated the exposure of the C-terminal core domain of mARC1 to the cytosol. Furthermore, following trypsin application a strong reduction of the mARC1 signal was detected. In analogy to the signal intensity differences between full-length and truncated mARC1 upon PK application, a weak band of the full-length protein also remained after trypsin treatment, whereas the intermembrane space control remained unaffected (Fig. 2D). This finding in aggregate suggests the C-terminal core domain of mARC1 to be tightly folded and thus to be partially stable to PK- and trypsin-mediated degradation. Taken together, both protease treatments of mitochondria revealed a consistent localization of mARC1 in the outer mitochondrial membrane with an N(in)-C(out) orientation.
mARC1 Is Targeted to Mitochondria by Its N-terminal Domain
Upon determination of its localization and membrane orientation, we next asked how mARC1 is directed to the outer mitochondrial membrane. The predictions of Fig. 1B suggested the presence of a weak N-terminal mitochondrial targeting signal and a downstream transmembrane domain. To determine the function of those motifs in mitochondrial translocation of mARC1, both elements were fused to GFP and localized in HEK-293 cells. First, we fused the putative N-terminal mitochondrial targeting signal consisting of residues 1–20 to GFP. Expression in HEK-293 cells revealed a heterogeneous subcellular distribution with some cells revealing a complete mitochondrial localization and others showing a weaker mitochondrial targeting, accompanied by a diffuse distribution throughout the cell (Fig. 3A). Statistical analyses revealed approximately 50% of the cells to display a complete and efficient mitochondrial targeting, although full-length mARC1-GFP was targeted exclusively to mitochondria in nearly all cells (Fig. 3B). These distributions suggested residues 1–20 to constitute a weak mitochondrial targeting signal being in line with the predictions of MITOPROT II and the low amphipathic character with only two basic residues within this region.
FIGURE 3.
Mitochondrial targeting motifs of mARC1. A and D–F, mARC1(Δ21–337)-GFP (A), mARC1(Δ41–337)-GFP (D), mARC1(Δ1–20,Δ41–337)-GFP (green) (E), and mARC1(Δ1–40)-GFP (F) were expressed in HEK-293 cells for 48 h and visualized by confocal laser-scanning microscopy. Mitochondria were stained with MitoTracker Red CMXRos (mitoTR). Scale bar, 10 μm. Sketch illustrates the expression constructs with the predicted mitochondrial targeting signal (light gray), the transmembrane domain (dark gray), the catalytic core (white), and GFP (green). B, mARC1-GFP and mARC1(Δ41–337)-GFP were expressed in HEK-293 cells for 48 h. Cells revealing a complete mitochondrial localization of the respective construct were counted and are depicted as percent of wild type. In each experiment 50 cells were analyzed; error bars represent S.D. of the average of three independent experiments (n = 3). C, mARC1(Δ21–337)-GFP was expressed in HEK-293 cells for 48 h. Mitochondria were enriched and treated with and without 1% Triton X-100 for 10 min prior to the addition of 100 μg/ml PK for 10 min at 4 ºC. After protease inactivation and TCA precipitation, mitochondrial proteins were loaded on a 12% SDS-PAGE followed by Western blotting.
Because classical N-terminal targeting signals are usually not found in outer mitochondrial membrane proteins (18), the principal ability of the N-terminal 20 residues of mARC1 to target a heterologous protein to the mitochondrial matrix was investigated next. mARC1Δ21–337-GFP expressed in HEK-293 cells accumulated in purified mitochondria and was protected against externally added protease, although protease treatment after solubilization of mitochondrial membranes resulted in efficient degradation (Fig. 3C). Thus, residues 1–20 of mARC1 were confirmed to target GFP to the mitochondrial matrix. However, full-length mARC1 was clearly shown not to be targeted to the mitochondrial matrix, and the general transport efficiency of the protein was demonstrated to exceed the targeting capability of the isolated N terminus. This suggested the presence of an additional targeting motif within the mARC1 protein, which would on one hand ensure a complete mitochondrial localization and on the other hand induce sorting of mARC1 to the outer mitochondrial membrane.
To analyze the combined impact of N-terminal targeting signal and the downstream transmembrane domain for mitochondrial targeting, residues 1–40 of mARC1 were fused to GFP and expressed in HEK-293 cells. In contrast to the isolated N-terminal targeting signal, fusion with the transmembrane domain triggered GFP translocation to mitochondria as efficiently as was seen with the full-length protein (Fig. 3D). The reconstitution of the entire mitochondrial translocation upon the fusion of the hydrophobic domain indicated a function of the latter in mitochondrial transport of mARC1. This was confirmed by fusion of the isolated hydrophobic domain, predicted to be a transmembrane helix, with GFP and expression in HEK-293 cells. Residues 21–40 were sufficient to mediate a complete mitochondrial localization of the fused GFP, demonstrating the hydrophobic domain to constitute a mitochondrial targeting signal on its own (Fig. 3E). Thus, the mARC1 N terminus contains two autonomous motifs triggering mitochondrial sorting of mARC1. Finally, the catalytic core of mARC1 was fused to GFP and expressed in HEK-293 cells. No mitochondrial localization was observed, and instead, the construct was diffusely distributed within the entire cell (Fig. 3F). In conclusion, residues 41–337 of mARC1 do not contain any further mitochondrial targeting signals, suggesting that the mitochondrial localization of mARC1 is mediated solely by its two N-terminal motifs.
The Subcellular Localization of mARC1 Is Independent of Moco
In a previous study, we have uncovered the essential role of Moco for the targeting of sulfite oxidase, a second mitochondrial Mo enzyme (19). In absence of Moco, sulfite oxidase accumulated in the cytosol despite its functional N-terminal targeting signal. As this finding and the underlying mechanisms were novel as well as unexpected and mARC1 belongs to the same family of Moco enzymes, we next analyzed a possible impact of Moco on mitochondrial targeting of mARC1. For this purpose, we expressed mARC1 containing a C-terminal myc tag in human Moco-deficient fibroblasts derived from a patient with a mutation in the MOCS1 gene. After demonstrating the loss of Moco in those fibroblasts (20) (Fig. 4A), we examined the subcellular distribution of mARC1-myc by immunocytochemistry and confocal laser-scanning microscopy (Fig. 4B). In contrast to sulfite oxidase, mARC1 revealed an efficient mitochondrial targeting even in absence of Moco. Therefore, the N-terminal domain is sufficient for accurate localization, although Moco is not involved directly in cellular routing of mARC1.
FIGURE 4.
Moco-independent mitochondrial targeting of mARC1. A, Moco content of WT and MOCS1-deficient fibroblasts was determined by the nit-1 assay. Different amounts of cell extract were applied in the assay, and protein-specific activities were determined. Error bars represent S.D. (n = 3). B, mARC1-myc was expressed in MOCS1-deficient fibroblasts for 48 h and visualized by anti-myc immunostaining (green) using confocal laser-scanning microscopy. Mitochondria were stained with MitoTracker Red CMXRos (mitoTR). Scale bar, 10 μm. Sketch illustrates the expression construct with the predicted mitochondrial targeting signal (light gray), the transmembrane domain (dark gray), the catalytic core (white), and the C-terminal myc tag (black).
The Mitochondrial Import of mARC1 Is Not Strictly Dependent on the Membrane Potential, Requires ATP, and Results in the Formation of High Oligomeric Complexes in the Outer Membrane
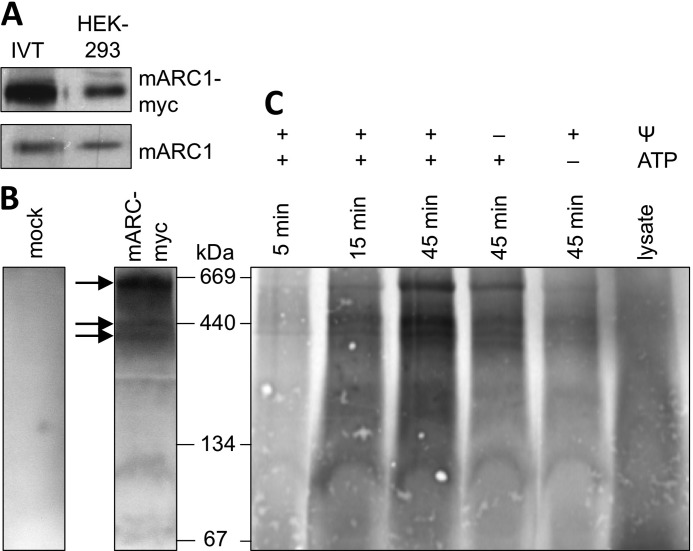
After identification and dissection of the internal motifs responsible for mitochondrial localization of mARC1, we characterized the import process of mARC1 precursor protein into isolated mitochondria. The N-terminal 20 residues of mARC1 have properties similar to those of typical mitochondrial targeting signals (amphipathic helix/positively charged). These motifs are usually cleaved after import by mitochondrial peptidases, resulting in N-terminal truncation of the respective mature proteins. In addition, predictions executed by MITOPROT II suggested the presence of a potential peptidase cleavage site at position 44 (13). Therefore, we first probed mARC1 for mitochondrial processing. Whereas the unprocessed precursor was obtained by in vitro translation using a reticulocyte lysate system, mARC1 was simultaneously expressed in HEK-293 cells to compare the size of in vitro and in vivo synthesized proteins, which would highlight proteolytic processing. Both untagged and myc-tagged mARC1 expressed in HEK-293 cells revealed similar sizes compared with the respective unprocessed precursors, demonstrating that mARC1 is not processed in mitochondria and retains both previously identified and described targeting motifs during maturation (Fig. 5A).
FIGURE 5.
In vitro characterization of mARC1 mitochondrial import. A, mARC1 and mARC1-myc were translated in vitro (IVT) and in parallel expressed in HEK-293 cells for 48 h. Each 4-μl lysate and 30 μg of cell extract (HEK-293), respectively, were loaded on a 12% SDS-gel followed by Western blotting. In vitro translated mARC1 was detected by x-ray film exposure, while mARC1 expressed in HEK-293 cells was subsequently detected by anti-mARC1 antibody staining. Images of both detections were merged for final size comparisons. B, mitochondria were enriched from HEK-293 cells expressing mARC1-myc or from untransfected cells, and mitochondrial proteins were extracted from enriched mitochondria by detergent application. Proteins were loaded on a 4–16% Bis-Tris acrylamide gradient BN-PAGE followed by Western blotting. mARC1-myc-containing complexes were detected by anti-myc antibody detection. Arrows point to high molecular mass bands that were also identified upon in vitro import of mARC1-myc in C. C, import of in vitro translated mARC1-myc occurred into mitochondria purified from HEK-293 cells. Import reactions were performed at 37 ºC and stopped after 5, 15, or 45 min. Membrane potential-deficient mitochondria (−Ψ) were obtained by the addition of 20 μg/ml valinomycin prior to the import reaction. The presence (+) or absence (−) of ATP is indicated. After import, mitochondrial proteins were extracted by detergent application and subsequently loaded on a 4–16% Bis-Tris acrylamide gradient BN-PAGE, followed by Western blotting. Radioactive signals were detected by 2 weeks exposure to an x-ray film.
To describe the kinetics and character of the mitochondrial import process of mARC1, we next incubated in vitro translated and myc-tagged precursors with mitochondria purified from HEK-293 cells. Detection of successful in vitro import into mitochondria is usually performed based on protection to external protease application or based on processing and truncation of the imported precursor. However, because mARC1 is exposed to the cytosol and not processed following successful import, we aimed to monitor mitochondrial translocation based on the proposed ability of mARC1 to form a complex with its electron donors cytochrome b5 reductase and cytochrome b5.
First, we determined mARC1 complex formation by expression of the protein in HEK-293 cells. Mitochondria were purified, and extracts were loaded on BN-PAGE to monitor mARC1 oligomerization in the outer membrane and thus to define the native mARC1 protein pattern to be expected upon successful in vitro import. Western blotting against the C-terminal myc tag revealed mARC1 to reside in three main high oligomeric protein complexes of more than 350 kDa in size, although mitochondria purified from nontransfected cells did not deliver any signals, thus ensuring antibody specificity and successful expression of mARC1 (Fig. 5B).
Successful in vitro import of mARC1 was expected to reproduce the complexes seen after in vivo expression of the protein in HEK-293 cells. The incubation of in vitro synthesized precursors with purified mitochondria indeed resulted in a time-dependent formation of similar high oligomeric complexes, suggesting successful import and integration of mARC1 (Fig. 5C). These complexes were also detected upon depletion of the membrane potential by valinomycin, demonstrating that, as known for proteins residing in the outer membrane (18), an intact membrane potential seems not to be required for mitochondrial transport of mARC1. Because the general function of N-terminal targeting signals is based on an intact membrane potential, the putative membrane potential-independent integration of mARC1 into the outer mitochondrial membrane further indicates that the N-terminal targeting signal does not traverse the mitochondrial inner membrane as conventional matrix targeting signals. Depletion of external ATP significantly decreased the efficiency of complex formation, illustrating that extramitochondrial ATP is required for mARC1 assembly into high oligomeric structures. The specificity of complex formations was ensured by application of precursors in the absence of mitochondria, which did not result in any detectable oligomerizations (Fig. 5C).
In summary, mARC1 is targeted to the outer mitochondrial membrane by a membrane potential-independent mechanism. This results in ATP- and time-dependent formation of high oligomeric structures, demonstrating that mARC1 is assembled into multiprotein complexes upon its integration into the outer membrane.
DISCUSSION
In a screen for the missing third component of the pro-drug-activating N-reductive system, mARC2 was identified as the fifth eukaryotic Moco-containing enzyme (5). Different views on the subcellular localization of mARC1 have been reported, localizing mARC1 to the outer or inner membrane of mitochondria (9, 10).
In this study, we have resolved the subcellular localization of mARC1 and demonstrate its integration into the outer mitochondrial membrane. Assuming a similar localization of mARC1 and mARC2, our results are in line with the previous studies detecting mARC2 in purified outer membrane fractions (5, 9). In addition, for the first time we were also able to clarify the orientation of one of both mARC proteins in the outer membrane, demonstrating the exposure of the C-terminal catalytic domain toward the cytosol. The enzymatic activity of mARC requires its integration into a three-component enzyme system, in which electrons are transferred from NADH cytochrome b5 reductase via cytochrome b5 to mARC (8). Both, NADH cytochrome b5 reductase (21, 22) as well as cytochrome b5 (23, 24) are well known components of the outer mitochondrial membrane exposing their catalytic domains to the cytosol. Our findings of an N(in)-C(out) orientation of mARC1 in the outer mitochondrial membrane are thus in well agreement with the catalytic interplay of the three components, which all expose their functional domains to the cytosol to build an efficient intermolecular electron transport chain.
The outer membrane localization of mARC1 is triggered by its first 40 N-terminal residues, which form a weak N-terminal mitochondrial targeting signal of the first 20 residues, followed by a hydrophobic domain. Based on our findings in respect to structure and localization, mARC1 can be classified as a novel signal-anchored protein of the outer mitochondrial membrane. This class of proteins shares α-helical membrane-spanning segments in their N termini, which are required for sorting as well as membrane anchoring (25). In most cases, the transmembrane domain is sufficient for mitochondrial targeting (26–28), and consistently, a fusion of GFP with the mARC1 transmembrane domain resulted in efficient mitochondrial localization. Therefore, the transmembrane domain of mARC1 simultaneously depicts a targeting signal and membrane anchor, confirming the classification of mARC1 as a novel signal-anchored protein.
The N terminus of mARC1 contains a weak N-terminal mitochondrial targeting signal, as illustrated by the inefficient mitochondrial sorting of a fusion of residues 1–20 to GFP and the presence of only two basic residues with a much less amphipathic character compared with other classical N-terminal targeting signals (29). In previous studies, the soluble N-terminal domain upstream of the transmembrane segment of signal-anchored proteins was shown to have supportive functions by increasing the efficiency of the targeting, which is primarily mediated by the transmembrane domain (26, 28). This supporting function was based on the presence of basic residues within the N-terminal domain, which following their depletion, was abolished. In contrast to our findings, the isolated soluble N termini of other signal-anchored proteins did not, however, constitute classical mitochondrial targeting signals and were not sufficient for mitochondrial localization (27). In either case, we suggest that the N terminus of mARC1 exerts similar supportive functions for mitochondrial sorting. We propose its identity as a weak matrix-targeting signal to be suppressed by the downstream and transmembrane domain, resulting in an insertion of the protein into the outer mitochondrial membrane. Consequently, the mARC1 N terminus may thus function as a primary receptor for the outer mitochondrial membrane, as described for the analogous segments of other signal-anchored proteins such as TOM70 and TOM20. The N termini of the latter resemble the mARC1 N terminus in the presence of one or two basic residues, suggesting a smooth transition between outer membrane receptor and weak matrix-targeting signal of signal-anchored proteins. Therefore, the transmembrane domain seems to ensure correct localization and to overwrite potential matrix signals within the upstream segment. Yeast Mcr1 contains a similar N-terminal targeting signal and a downstream transmembrane domain, which were interestingly shown to mediate dual localization of the protein to the outer membrane and to the intermembrane space (30). According to MITOPROT II (13) and the increased number of basic residues compared with the N terminus of mARC1, the N-terminal targeting signal of Mcr1 appears to be stronger, although the hydrophobicity of the transmembrane domain is weaker compared with mARC1 (31). The intensities of N-terminal targeting signals and downstream transmembrane domains may therefore regulate insertion of mARC1 and Mcr1 into the outer or inner mitochondrial membrane.
Our analysis of the mitochondrial import of mARC1 revealed characteristics typical for proteins residing in the outer mitochondrial membrane. Therefore, mARC1 was not processed during import and seemed not to strictly require a membrane potential across the inner mitochondrial membrane, given that the addition of valinomycin resulted in only a slight reduction of complex intensities. The depletion of external ATP from the import buffer, however, significantly interfered with successful targeting of mARC1. We suggest that ATP is required for the release of mARC1 from cytosolic chaperons and thereby to be essential for an efficient integration into the outer membrane. A similar role of external ATP during mitochondrial import has been described earlier for other membrane-anchored proteins (32).
Interestingly, membrane integration of mARC1 resulted in the formation of high oligomeric complexes as depicted by BN-PAGE following in vitro import and expression of mARC1 in HEK-293 cells, suggesting that mARC1 is part of these complexes in its final and active form. Whereas mARC was suggested to interact with cytochrome b5 and its reductase (total ∼90 kDa), both being required for mARC1 activity, the complex sizes of more than 350 kDa suggest mARC to be part of a yet unknown membrane-bound multienzyme complex probably hosting other still to be identified proteins. Isolation and characterization of these complexes will thus constitute an exciting future prospect.
In addition to its mitochondrial targeting, a peroxisomal localization has been described for mARC2 (33). Considering the similarities of both mARC proteins, an additional peroxisomal localization of mARC1 might possible, although our data did not reveal any significant extramitochondrial localization of mARC1.
In summary, we have identified mARC1 as a novel signal-anchored protein of the outer mitochondrial membrane. Localization and N(in)-C(out) membrane orientation of mARC are both triggered by the N-terminal domain, in which the transmembrane-helix motif constitutes an essential targeting signal as well as anchor, although the N-terminal targeting signal serves as supportive receptor for the outer mitochondrial membrane. The integration of mARC1 into the outer mitochondrial membrane is membrane potential-independent but requires external ATP, finally resulting in mARC1 assembly into high oligomeric protein complexes.
Acknowledgments
We thank Simona Jansen and Joana Fischer (University of Cologne, Germany) for technical assistance.
This work was supported by a grant from the Studienstiftung des Deutschen Volkes (to J. M. K.), the German Science Foundation (to G. S.), and the Fonds der Chemischen Industrie (to G. S.).
- Mo
- molybdenum
- Moco
- molybdenum cofactor
- Bis-Tris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- BN-PAGE
- Blue NativePAGE
- mARC
- amidoxime-reducing component
- PK
- proteinase K
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Schwarz G., Mendel R. R. (2006) Molybdenum cofactor biosynthesis and molybdenum enzymes. Annu. Rev. Plant Biol. 57, 623–647 [DOI] [PubMed] [Google Scholar]
- 2. Schwarz G., Mendel R. R., Ribbe M. W. (2009) Molybdenum cofactors, enzymes and pathways. Nature 460, 839–847 [DOI] [PubMed] [Google Scholar]
- 3. Mendel R. R., Bittner F. (2006) Cell biology of molybdenum. Biochim. Biophys. Acta 1763, 621–635 [DOI] [PubMed] [Google Scholar]
- 4. Cohen H. J., Betcher-Lange S., Kessler D. L., Rajagopalan K. V. (1972) Hepatic sulfite oxidase: congruency in mitochondria of prosthetic groups and activity. J. Biol. Chem. 247, 7759–7766 [PubMed] [Google Scholar]
- 5. Havemeyer A., Bittner F., Wollers S., Mendel R., Kunze T., Clement B. (2006) Identification of the missing component in the mitochondrial benzamidoxime prodrug-converting system as a novel molybdenum enzyme. J. Biol. Chem. 281, 34796–34802 [DOI] [PubMed] [Google Scholar]
- 6. Kotthaus J., Wahl B., Havemeyer A., Kotthaus J., Schade D., Garbe-Schönberg D., Mendel R., Bittner F., Clement B. (2011) Reduction of Nω-hydroxy-l-arginine by the mitochondrial amidoxime reducing component (mARC). Biochem. J. 433, 383–391 [DOI] [PubMed] [Google Scholar]
- 7. Gruenewald S., Wahl B., Bittner F., Hungeling H., Kanzow S., Kotthaus J., Schwering U., Mendel R. R., Clement B. (2008) The fourth molybdenum containing enzyme mARC: cloning and involvement in the activation of N-hydroxylated prodrugs. J. Med. Chem. 51, 8173–8177 [DOI] [PubMed] [Google Scholar]
- 8. Wahl B., Reichmann D., Niks D., Krompholz N., Havemeyer A., Clement B., Messerschmidt T., Rothkegel M., Biester H., Hille R., Mendel R. R., Bittner F. (2010) Biochemical and spectroscopic characterization of the human mitochondrial amidoxime reducing components hmARC-1 and hmARC-2 suggests the existence of a new molybdenum enzyme family in eukaryotes. J. Biol. Chem. 285, 37847–37859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neve E. P., Nordling A., Andersson T. B., Hellman U., Diczfalusy U., Johansson I., Ingelman-Sundberg M. (2012) Amidoxime reductase system containing cytochrome b5 type B (CYB5B) and MOSC2 is of importance for lipid synthesis in adipocyte mitochondria. J. Biol. Chem. 287, 6307–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Da Cruz S., Xenarios I., Langridge J., Vilbois F., Parone P. A., Martinou J. C. (2003) Proteomic analysis of the mouse liver mitochondrial inner membrane. J. Biol. Chem. 278, 41566–41571 [DOI] [PubMed] [Google Scholar]
- 11. Reiss J., Gross-Hardt S., Christensen E., Schmidt P., Mendel R. R., Schwarz G. (2001) A mutation in the gene for the neurotransmitter receptor-clustering protein gephyrin causes a novel form of molybdenum cofactor deficiency. Am. J. Hum. Genet. 68, 208–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schägger H., von Jagow G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 13. Claros M. G., Vincens P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 14. Käll L., Krogh A., Sonnhammer E. L. (2004) A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036 [DOI] [PubMed] [Google Scholar]
- 15. Gakh O., Cavadini P., Isaya G. (2002) Mitochondrial processing peptidases. Biochim. Biophys. Acta 1592, 63–77 [DOI] [PubMed] [Google Scholar]
- 16. Fujiki Y., Hubbard A. L., Fowler S., Lazarow P. B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. John G. B., Shang Y., Li L., Renken C., Mannella C. A., Selker J. M., Rangell L., Bennett M. J., Zha J. (2005) The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell 16, 1543–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walther D. M., Rapaport D. (2009) Biogenesis of mitochondrial outer membrane proteins. Biochim. Biophys. Acta 1793, 42–51 [DOI] [PubMed] [Google Scholar]
- 19. Klein J. M., Schwarz G. (August 1, 2012) Cofactor-dependent maturation of mammalian sulfite oxidase links two mitochondrial import pathways. J. Cell Sci., doi 10.1242/jcs.110114 [DOI] [PubMed] [Google Scholar]
- 20. Nason A., Lee K. Y., Pan S. S., Ketchum P. A., Lamberti A., DeVries J. (1971) In vitro formation of assimilatory reduced nicotinamide adenine dinucleotide phosphate: nitrate reductase from a Neurospora mutant and a component of molybdenum enzymes. Proc. Natl. Acad. Sci. U.S.A. 68, 3242–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Borgese N., Aggujaro D., Carrera P., Pietrini G., Bassetti M. (1996) A role for N-myristoylation in protein targeting: NADH-cytochrome b5 reductase requires myristic acid for association with outer mitochondrial but not ER membranes. J. Cell Biol. 135, 1501–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Borgese N., Pietrini G. (1986) Distribution of the integral membrane protein NADH-cytochrome b5 reductase in rat liver cells, studied with a quantitative radioimmunoblotting assay. Biochem. J. 239, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fukushima K., Ito A., Omura T., Sato R. (1972) Occurrence of different types of cytochrome b5-like hemoprotein in liver mitochondria and their intramitochondrial localization. J. Biochem. 71, 447–461 [PubMed] [Google Scholar]
- 24. Ito A. (1980) Cytochrome b5-like hemoprotein of outer mitochondrial membrane: OM cytochrome b. I. Purification of OM cytochrome b from rat liver mitochondria and comparison of its molecular properties with those of cytochrome b5. J. Biochem. 87, 63–71 [DOI] [PubMed] [Google Scholar]
- 25. Shore G. C., McBride H. M., Millar D. G., Steenaart N. A., Nguyen M. (1995) Import and insertion of proteins into the mitochondrial outer membrane. Eur. J. Biochem. 227, 9–18 [DOI] [PubMed] [Google Scholar]
- 26. Kanaji S., Iwahashi J., Kida Y., Sakaguchi M., Mihara K. (2000) Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 151, 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McBride H. M., Millar D. G., Li J. M., Shore G. C. (1992) A signal-anchor sequence selective for the mitochondrial outer membrane. J. Cell Biol. 119, 1451–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suzuki H., Maeda M., Mihara K. (2002) Characterization of rat TOM70 as a receptor of the preprotein translocase of the mitochondrial outer membrane. J. Cell Sci. 115, 1895–1905 [DOI] [PubMed] [Google Scholar]
- 29. Neupert W., Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 30. Hahne K., Haucke V., Ramage L., Schatz G. (1994) Incomplete arrest in the outer membrane sorts NADH-cytochrome b5 reductase to two different submitochondrial compartments. Cell 79, 829–839 [DOI] [PubMed] [Google Scholar]
- 31. Kyte J., Doolittle R. F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 32. Wachter C., Schatz G., Glick B. S. (1994) Protein import into mitochondria: the requirement for external ATP is precursor-specific whereas intramitochondrial ATP is universally needed for translocation into the matrix. Mol. Biol. Cell 5, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wiese S., Gronemeyer T., Ofman R., Kunze M., Grou C. P., Almeida J. A., Eisenacher M., Stephan C., Hayen H., Schollenberger L., Korosec T., Waterham H. R., Schliebs W., Erdmann R., Berger J., Meyer H. E., Just W., Azevedo J. E., Wanders R. J., Warscheid B. (2007) Proteomics characterization of mouse kidney peroxisomes by tandem mass spectrometry and protein correlation profiling. Mol. Cell Proteomics 6, 2045–2057 [DOI] [PubMed] [Google Scholar]