Abstract
Plant extracts and compounds are applied to a wide variety of diseases in which traditional drugs have proven ineffective. A quickly developing trend in biomedicine is the therapeutic use of siRNA (short interfering RNA) structures. The focus of this study was on evaluating the gene expression involved in the modulation of apoptosis, in cases of combinatorial treatment (−)-epigallocatechin-3-gallate (EGCG) and/or p53siRNA. EGCG in combination with p53siRNA exerts synergic pro-apoptotic effects that are greater than those of each agent taken individually. There is a cumulative antiproliferative effect, induced by EGCG and p53siRNA treatment, and it is mediated through the activation of a large number of pro-apoptotic genes and the inhibition of anti-apoptotic protein expression levels. p53siRNA promotes the convergence of the extrinsic and intrinsic pathways in a synergic manner with EGCG. The chemotherapeutic effects of EGCG in combination with p53siRNA therapy induced a synergic pro-apoptotic effect, indicating the potential for development of promising new anticancer therapies.
Keywords: p53siRNA, apoptosis, HeLa cells
Introduction
As cancer is a multifactorial disease, it may require treatment with compounds able to target multiple intracellular components. Phytochemicals, including catechins, are able to modulate many components of intercellular and intracellular signaling pathways.1 Catechins belong to the polyphenol class of compounds and are in the flavan-3-ol subclass, that includes (−)-epigallocatechin-3-gallate (EGCG), (−)-epigallocatechin (EGC), (−)-epicatechin (EC), and (−)-catechin-3-gallate (ECG). EGCG is the major component of green tea. Flavan-3-ols can modulate multiple metabolic pathways and may have different biological effects, including dose-dependent pro- or antioxidant properties,3 antiproliferative effects through blockades of the cell cycle, apoptotic effects, anti-angiogenic effects through the inhibition of pro-angiogenic factors and their receptors, anti-inflammatory effects through the suppression of cytokines and cell adhesion molecules, suppression of tumor growth and inhibition of metastasis.2–7 Thus, the multiple actions of flavan-3-ols suggest that they may have a role as gene co-activators, that have direct implications in cancer therapy.6,8
A developing trend in biomedicine is the therapeutic use of siRNA (short interfering RNA), an approach that has been combined with nanotechnology for the treatment of a variety of diseases, with promising results. RNAi is a post-transcriptional method of inhibiting gene expression, and it is a promising approach to treat cancer at a molecular level.9,10 Recent studies suggest that new therapeutic approaches based on RNAi therapy selectively target genes at various molecular pathways. RNAi technology can be directed against cancer using different strategies, including modulation of the immune response, inhibition of overexpressed oncogenes, promotion of apoptosis, regulation of the cell cycle, anti-angiogenesis, and enhancement of the efficacy of chemo- and radiotherapy.6 It has also been observed that blocking the expression of the p53 tumor suppressor gene or anti-apoptotic genes, such as Bcl2, survivin, XIAP, and RhoGDI, increases the susceptibility of tumor cells to chemotherapy-induced apoptosis. Based on this hypothesis, the efficacy of p53siRNA is being evaluated in the treatment of acute renal failure, in which apoptosis plays a key role.9,11 The restoration of tumor suppressor genes is the most intuitive application of gene therapy for the treatment of cancer. Several approaches have been designed in order to restore the tumor suppressing function of p53, including rescuing the mutated p53 gene or delivering exogenous p53 as a therapeutic strategy taking into account that the p53 protein is probably the best known of all tumor suppressors. p53 plays a fundamental role in protecting cells against malignant transformation by causing cell-cycle arrest and apoptosis, and it also delays cellular senescence. Mutant p53 that is detected in over 50% of cancer cases not only loses its tumor suppressing capacity, but it also gains oncogenic function.12
An improved understanding of the patho-physiological basis of the disease will bring more sophisticated diagnostic opportunities and yield enhanced therapies by inducing apoptosis. For decades, the standard approach to cancer treatment has typically been a combination of surgery, chemotherapy, or radiation therapy. Chemotherapeutic agents routinely produce unwanted side effects, because, in addition to the cancer cells, healthy cells are killed as well. Additionally, many tumor cells eventually develop multi-drug resistance after being exposed to one or more anticancer agent. Thus, single-agent chemotherapy is curative in only a very limited number of cancers. Most chemotherapeutic drugs act as antiproliferative or pro-apoptotic agents, acting through different cellular and molecular mechanisms.1,2
The current treatment involves the use of natural phytochemical compounds, associated with siRNA gene therapy for targeting apoptosis in a synergic way. Apoptosis is a complex process in which p53 plays an important role, but it is insufficient for a general approach; a complex investigation using polymerase chain reaction (PCR)-array technology at gene expression level is needed. Preliminary tests for the optimization of p53siRNA transfection were performed. One of the major purposes of the current study was to assess the impact of the post-transcriptional inhibition of p53 expression at the RNA level using RNAi in parallel with EGCG treatment, to evaluate the expression profile of genes involved in cellular survival and apoptosis using the HeLa cell line as a model system. EGCG, either alone or in combination with p53siRNA, can be a useful tool in cancer treatment.
Materials and methods
Cell cultures
The HeLa human cervical carcinoma cell line was grown at 37°C in a humidified atmosphere with 5% CO2 in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, St Louis, MO) supplemented with 10% fetal bovine serum (Sigma-Aldrich), 2 mM glutamine, 100 U/mL penicillin, and 100 mg/mL streptomycin (Sigma-Aldrich).
Cell treatment
HeLa cells were transfected with siRNA using neofection protocol after preliminary optimization steps in order to have optimal cell density, concentration of siPORT™ NeoFX™ and p53siRNA with no side effects. After the transfection step, EGCG (Sigma-Aldrich) was added in order to achieve final concentration of 10 μM, with/without p53siRNA treatment. Each experiment was performed in triplicate.
p53siRNA transfection
For this experiment, 5 × 105 HeLa cells per well were reverse-transfected in 6-well plates with the siPort NeoFx transfection agent (Ambion) according to the manufacturer’s recommended protocol and p53siRNA, in the presence or absence of 10 μM EGCG. The p53siRNA was chosen as a pool of three to five target-specific 19–25 nt siRNAs designed to knock-down gene expression (sc-29435, Santa Cruz). p53siRNA was used at a final concentration of 50 nM in serum-free OptiMem medium (Gibco BRL, no 31985-070, Invitrogen, Romania), according to the manufacturer’s instructions. All experiments were conducted under conditions of serum starvation, in order to induce cell stress and stimulate the cells to produce growth factors. Reverse transfection was performed using the siPort NeoFx transfection reagent, at a volume of 5 μL, to induce the maximum gene inhibition with acceptable levels of cell viability and no alteration of gene expression, as observed in previous experiments.
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay
HeLa cells (2 × 104) were treated in a 96-well plate, as described above, and were incubated for 24, 48, and 72 hours. The MTT assay was performed by removing the culture medium, washing the cells with phosphate buffered saline (Sigma-Aldrich) and adding 150 μL Hanks salt (Life Technologies, Carlsbad, CA) containing MTT to each well at a final concentration of 455 μg of MTT per mL of Hanks salt. After 2 hours of incubation under standard conditions, the MTT solution was removed, and 200 μL of dimethyl sulfoxide (Merck Millipore Romania) were added to each well. The absorbance was measured at 490 nm using a BioTek Synergy HT Microplate Plate Reader (BioTek Instruments, Inc, Winooski, VT).
RNA extraction and quality control
The total RNA of treated and untreated HeLa cells was isolated using TriReagent at 24 hours post-treatment (Sigma-Aldrich, Bucuresti, Romania), according to the manufacturer’s instructions. The extracted RNA was subsequently treated with DNAse (Qiagen, Bucharest, Romania cat no 79254) and purified using the RNeasy Mini kit (Qiagen, cat no 74106). RNA concentrations were measured using a NanoDrop-1100 spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The quality of the RNA was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), and all samples had an RNA integrity number in the 8–10 range.
RT2 profiler™ PCR array technology
For this experiment, 84 genes involved in apoptosis were simultaneously evaluated using The Human Apoptosis RT2 Profiler™ PCR Array (SABiosciences, Bucharest, Romania, cat no PAHS-012Z). Also, 500 ng of total RNA from each sample were reverse transcribed using the RT2 First Strand Kit (SABiosciences). The resulting cDNA was then amplified with ready-to-use RT2 SYBR Green qPCR Master Mix (SABiosciences) in a 98-well plate (The Human Apoptosis RT2 Profiler™ PCR Array) using a LightCycler 480 instrument Roche (Bucharest, Romania) as follows: 10 minutes at 95°C for enzyme activation, followed by 45 cycles at 95°C for 15 seconds, and 1 minute at 60°C for the amplification step.
PCR array data analysis
The data were analyzed using the software package for the PCR Array system according to the ΔΔCt method. A statistical analysis was performed to compare the gene expression values for the treated group versus the untreated group. P-values < 0.05 were considered to be statistically significant.
Results
p53siRNA and EGCG increase cytotoxicity in a synergic manner
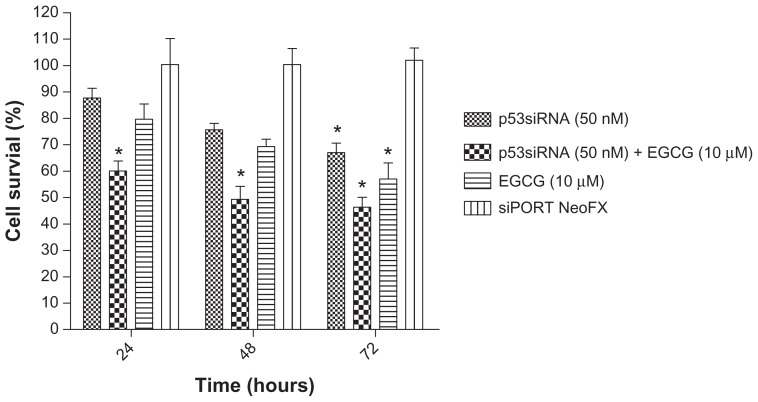
The MTT values for HeLa cells after 24, 48, and 72 hours of incubation with p53siRNA, in the presence or absence of 10 μM EGCG, are represented as the percent of control (Figure 1). The reduction of cell proliferation was obtained by reporting all the data to the control, which is considered 100%.
Figure 1.
Cytotoxic effects as measured by MTT assay after 24 hours incubation with 50 nM p53siRNA, in the presence or absence of 10 μM EGCG, and siPORT™ NeoFX™ transfection agent on HeLa cell line (mean ± standard deviation, n = 6; P ≤ 0.05 compared with the control group).
Abbreviations: EGCG, (−)-epigallocatechin-3-gallate; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; siRNA, short interfering RNA.
The cytotoxic effect of the cell treatment and the impact of siRNA-mediated p53 inhibition on cell proliferation were analyzed using the MTT assay. The loss of cell viability was time-dependent, with the highest antiproliferative effect observed at 72 hours. The results show that p53siRNA+EGCG induces a decrease in the number of live cells by 60% after 24 hours of treatment, without any noticeable cytotoxicity associated with siRNA delivery (untransfected cells compared with siPORT NeoFX-treated cells) (Figure 1). An unexpected finding was that p53siRNA and EGCG had a strong inhibitory effect on the proliferation of HeLa cells even at 24 hours. HeLa cells treated only with EGCG or p53siRNA did not show a similar antiproliferative response at 24 and 48 hours, while at 72 hours a statistically significant reduction of cell proliferation was observed (Figure 1).
p53siRNA and EGCG modulate apoptotic genes in a synergic manner
The activation of apoptosis is a key mechanism for most antitumor therapies; previous data proved that p53siRNA and EGCG act in a synergic manner using Annexin V-FITC/PI flow cytometry (unpublished data). RNA from cells treated with p53siRNA (50 nM), p53siRNA+EGCG (10 μM), EGCG, or siPORT NeoFX (control group) was used to analyze the expression profile of 84 key genes involved in modulating apoptosis, using quantitative real-time PCR. The array includes tumor necrosis factor (TNF) ligands and their receptors, members of the bcl-2 family, caspases, inhibitor of apoptosis proteins, TNF receptor-associated factor (TRAF), caspase recruitment domain (CARD), death effector domain and CIDE families, and genes involved in the p53 DNA damage pathway. Three housekeeping genes (B2M, HPRT1, and GAPDH) were used to normalize gene expression. The ΔΔCt method was used to calculate the fold change in gene expression. The genes with a fold change ≥ 1.25 were considered to be of interest.
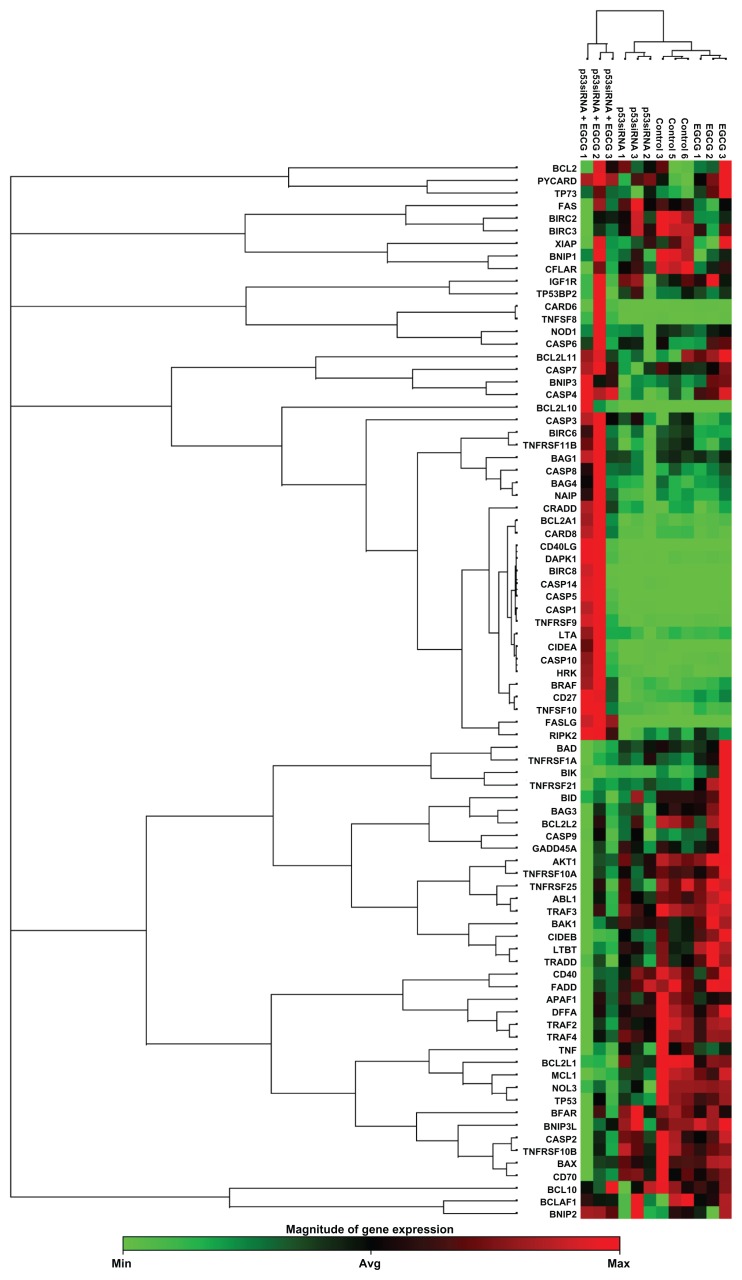
The 84 genes were used to generate clustergrams between treatment groups (p53siRNA, p53siRNA+EGCG, EGCG) and the control group (Figure 2). Genes of interest were separated into functional gene groups based on their involvement in modulating apoptosis (Table 1). In the p53siRNA treatment group, there were two genes that were overexpressed (indicated in red) and 10 genes that were downregulated at mRNA level. In contrast, there were nine genes upregulated and 27 genes that were downregulated in the p53siRNA+EGCG group. The results show that there are significant differences in gene expression for eleven genes, six of which were upregulated and five downregulated, resulting from co-treatment with EGCG. Furthermore, p53siRNA treatment in the presence of EGCG results in increased cytotoxic activity, due to the co-activation of apoptotic pathways.
Figure 2.
The clustergram generated for differentially expressed genes (P < 0.05, cut-off fold change 1.25) in treated versus control samples.
Table 1.
Apoptosis-related genes with altered expression levels following 24 hours of incubation with p53siRNA, p53siRNA+EGCG, or EGCG
| Gene family | p53siRNA | p53siRNA+EGCG | EGCG |
|---|---|---|---|
| TNF ligand family | TNFSF10 (+1.47) | FASLG (+1.29), TNFSF10 (+9.05), TNF (−1.54), TNFRSF10A (−2.31), TNFRSF10B (−2.56), TNFRSF25 (−2.80), CD70 (−1.48) | TNFRSF10 (+2.3), TNFRSF11B (−1.27) |
| TNF receptor family | TNFRSF11B (−1,43), CD27 (−1.72) | CD27 (+3.87), CD40 (−1.67), LTBR (−1.68) | LTBR (+1.307), TNFRSF21 (+1.5), CD27 (+1.54) |
| Bcl-2 family | BCL2L1 (−1.47), BCL2L2 (−1.27), MCL1 (−1.46), NOL3 (−1.48) | BCL2A1 (+4.08), BAD (−1.61), BAX (−2.06), BCL2L1 (−2.06), BCL2L2 (−2.53), MCL1 (−1.89), NOL3 (−1.89) | BAK1 (+1.27), BNIP1 (−1.25) |
| Caspase family | CASP1 (+2.66) | CASP4 (+1.28), BIRC2 (−1–33), BIRC3 (−1.34), CASP2 (−1.56) | BIRC2 (−1.33) |
| TRAF family | TRAF2 (−1.5) | TRAF2 (−2.98), TRAF3 (−1.52), TRAF4 (−1.96) | |
| CARD family | CARD8 (−1.41) | CRADD (+1.55), PYCARD (+1.29), RIPK2 (+1.25), APAF1 (−1.31), CASP2 (−1.56) | CARD8 (−1.39) |
| Death domain family | FADD (−1.73), TNFRSF10A (−2.31), TNFRSF10A (−2.31), TNFRSF10B (−2.56), TNFRSF25 (−2.80) | TNFRSF21 (+1.5), TNFRSF11B (−1.27) | |
| Death effector domain family | CFLAR (−1.29), TNFRSF11B (−1.43) | CFLAR (−1.5) | CFLAR (−1.34) |
| CIDE domain family | CIDEB (2.94), DFFA (1.54) | ||
| p53 and DNA damage response | TP53 (−1.73), BCL2L1 (−1.47) | TP53 (−2.06), ABL1 (−1.53), | TP73 (+1.81) |
| Anti-apoptosis | BCL2L2 (−1.27) | BRAF (+2.11), AKT1 (−1.31) |
Abbreviations: CARD, caspase recruitment domain; EGCG, (−)-epigallocatechin-3-gallate; siRNA, short interfering RNA; TNF, tumor necrosis factor; TRAF, TNF receptor-associated factor.
Functional pathway analysis was performed using Ingenuity Systems (Redwood City, CA) and several canonical pathways were identified as a response to p53siRNA, p53siRNA+EGCG and EGCG treatment. Several genes involved in apoptosis signaling and death receptor pathway are shown to have a high importance in this study, being presented in Figures S1 and S2. The networks generated by Ingenuity and all the available data from literature contributed to the canonical network presented in Figure 1. There is a connection between the treatment and the major signal transduction pathway that regulates apoptosis and cell death pathways, proving a higher efficiency for p53+EGCG than for the compounds administered separately.
Discussion
The objective of this study was to use natural phytochemicals to enhance the efficacy of siRNA therapy. Catechins are a class of biologically active natural compounds that have beneficial effects on human health and have recently gained attention due to their low toxicity, low cost, and high availability.1,2 EGCG is the major compound found in green tea and seems to have the most powerful bioactive properties.2 Although earlier studies have presented a correlative role of p53siRNA+EGCG in the reduction of cell proliferation,4,8,13–15 the current study has unequivocally established the higher efficiency of p53siRNA in the presence of EGCG in cell proliferation reduction. This effect is demonstrated by the MTT data and is confirmed at the molecular level using a PCR-based expression array.
EGCG may have possible interactions either with the transfection agent or with p53siRNA. The transfection agent may increase the stability of EGCG. There have been limited data available concerning EGCG drug interactions.16 EGCG may have synergic effects with p53siRNA; meanwhile, Golden et al have found that EGCG antagonizes bortezomib on in-vitro and in-vivo model systems.17
The analysis of mRNA gene expression provides evidence of a distinct expression profile of genes involved in apoptosis.13 EGCG in combination with p53siRNA has synergic pro-apoptotic effects, which are greater than each agent taken individually and induce an immediate expression of anti-apoptotic genes (mRNA), as shown in Figure S1. In contrast, the expressions of BAD, BAX, BCL2L1, BCL2L2, MCL1, and NOL3 were reduced. The specific inhibition of pro-apoptotic genes is also observed, including ligands and receptors of the TNF family,15,18TRAF, CARD, the death effector domain family (Figure S2), and members of the p53 gene family.19
The two apoptotic pathways, of which one is activated by death receptors and the other regulated by Bcl-2 family members, do not function in isolation.20,23 The Bcl-2 family consists of pro- and anti-apoptotic genes. It is the balance in expression between these two gene lineages that may determine the death or survival of a cell. The present study reveals the inhibition of anti-apoptotic genes, including the unexpected inhibition of anti-apoptotic BCL-2 family members BAX and BAD that may be associated with the resistance to apoptosis in the case of the combined treatment with EGCG and p53siRNA.
The intrinsic apoptotic pathway is coordinated by a complex network of anti- and pro-apoptotic factors, including Bcl-2, Bax, and the p53 tumor suppressor gene (Figure S2). Treatment with EGCG leads to an increased Bax/Bcl-2 ratio and results in the induction of apoptosis and stimulation of caspases.21,22
Caspases play a central role as executioners in most types of apoptosis, including activation-induced cell death. CASP1 was downregulated at mRNA level after treatment with p53siRNA, while its adaptor PYCARD was upregulated in the p53siRNA+EGCG combination treatment group; EGCG may have an inhibitory effect on CASP1 activation. CASP1 and CASP4 might play important roles in cell homeostasis and proliferation. In the case of p53siRNA+EGCG treatment, BIRC2 and BIRC3 were downregulated at mRNA level in the present study. These results are partially confirmed by a similar study.26
The extrinsic apoptotic pathway is also regulated by p53, although the overall contribution of this regulation to p53-mediated cell death is poorly understood. The most intuitive link between p53-mediated transactivation and apoptosis comes from its ability to control the transcription of pro-apoptotic members of the Bcl-2 family, including the “multidomain” Bcl-2 family member, Bax.23 Also, TRAIL and the gene encoding Fas ligand are, each, direct targets of p53.8,14,27 Moreover, the ability of p53 to transactivate Bcl-2 family gene members may facilitate crosstalk between the extrinsic and intrinsic pathways. Consequently, p53 may sensitize cells to death receptor ligands (Figure S2), either inducing apoptosis directly or enhancing cell death in ligand-rich environments.27
TRAIL has been shown to induce apoptosis independently of p53 function, but may be dependent on p73, as we can see from the treatment with EGCG and other natural compounds.23,26 Also, p73 is a member of the p53 family, which can also promote apoptosis either in parallel or in concert with p53.22,27 Moreover, they revealed a number of targets, like death receptors and ligands for EGCG-induced apoptosis that were expressed in the absence of functional p53. These death receptors were also reported to induce anti-angiogenic signals, as was shown in a previous study.8 At the same time, a crucial role for p73 in EGCG-induced apoptosis was identified along with a number of previously unidentified p73 target genes.8
EGCG activates the death receptor pathway in the presence of p53siRNA by inducing changes in the expression of TNF receptors (TNFSF10, FASLG), which contain an intracellular region, called a death domain, that is responsible for transmitting apoptotic signals,16,24,25 especially the intrinsic apoptosis machinery.26
Changes in mRNA expression levels of the TNFRSF10A and TNFRSF10B, TRAIL receptors, which are also known as DR4 (death receptor 4) and DR5 (death receptor 5), were observed. The upregulation of these receptors suggests that p53siRNA in the presence of EGCG could promote TRAIL-induced apoptosis in a synergic manner, as opposed to the individual treatments, clearly observed in Figure S2. Wiley et al27 hypothesized that TRAIL can induce apoptosis in tumorigenic or transformed cells, but not in normal cells, and later proved that a variety of tumor cells, but not normal tissues, express both DR4 and DR5.17,27 This can be exploited as a potential cancer therapy, as stimulation of the DR4 and DR5 pro-apoptotic receptors has shown promising safety and efficacy profiles in several preclinical cancer models.28,29 However, not all tumor cells are sensitive to TRAIL therapy.17 Studies on the intracellular mechanism of TRAIL resistance identified anti-apoptotic or pro-survival molecules as being responsible for treatment failure.18 In this present study, the apoptotic effects of p53siRNA+EGCG are demonstrated to be mediated by death domain regulation, specifically by the TNF family of receptors and ligands. The TNFSF10 ligand is upregulated (p53siRNA, +1.47; p53siRNA+EGCG, +9.05; EGCG, +2.3) and has been found to have a profound apoptotic effect in neoplastic cells but not in normal ones.30TRAIL can also bind to decoy receptors (TNSF10A/DR4 or TNFSFR10A/DR5) that cannot induce apoptosis.12 The differential expression of TRAIL, its receptors and/or regulatory proteins, such as CFLAR, have been associated with resistance to TRAIL-mediated apoptosis in certain cancers. The TNFR binding receptor CD27 (TNFRSF7) and its related receptors activate inflammatory and antigenic signals. Therefore, these receptors typically act in the context of an immune response and not only in the induction of apoptosis. Many of the receptors are also induced upon immune activation, including CD27.31
TNFRSF21 (death receptor-6) has been involved in the regulation of the immune response; it may also be involved in tumor-cell survival and immune evasion.18,24,25 The TNFRSF21 mRNA level is upregulated (1.5, P = 0.005) only in the presence of EGCG. The protein encoded by the TNFRSF25 gene is an adaptor molecule that interacts with various cell surface receptors and mediates apoptotic cell signals. TNFRSF25 and TNFSF10 participate in cell death signaling initiated by these receptors in the case of p53siRNA+EGCG.
TNFR family members bind to TRAF family adaptors (TRAF2, −2.98; TRAF3, −1.52; TRAF4, −1.96), which stimulate the nuclear factor kappa B (NFκB) and mitogen activated protein kinase (MAPK) signaling pathways.18,22,25 The data in this present study show changes in the expression of numerous genes associated with apoptosis that are involved in the NFκB and MAPK signaling pathways. The NFκB pathway is well known for its anti-apoptotic effect, which proceeds through the transcriptional upregulation of Bcl-2 family members and inhibitor of apoptosis proteins. BRAF expression occurs through a common mechanism involving the MEK-ERK signaling cascade. However, the specific role of oncogenic BRAF in cancer has not been fully elucidated. Recent studies reveal that there was no correlation in vivo between the activation status of BRAF and the proportion of phospho-ERK-positive cells or the clinical course of the disease.8,27
Conclusion
The development and testing of novel nanoparticle delivery systems is an important task for cancer therapy research. RNAi nanotechnology has emerged as a novel and exciting area for the therapy and diagnosis of multiple diseases. The selectivity and ability to target molecular factors therapeutically renders siRNA applications encouraging. The present study provides evidence that p53siRNA has synergic effects when combined with EGCG in the induction of apoptosis, as shown by MTT and PCR-array data. This is evidence for the fact that the p53 network is complex. Although challenging, a full understanding of p53 biology is likely to facilitate the development of new cancer therapies. The advantage of combining EGCG with p53-targeted RNAi therapy is that it leads to the activation of alternative apoptosis-inducing pathways and suggests promising new therapies for cancer inhibition. This study also provides key information on apoptosis signaling pathways modulated by EGCG in the presence of p53siRNA. This novel therapy is based on the concept of synergy between EGCG and p53-targeted RNAi, which may have limited side effects.
The present study reveals an increase in the efficacy of p53 gene therapy using siPORT NeoFX as a transfection agent, in the presence of EGCG on the HeLa cervical tumor cell line. These two agents specifically induce apoptosis at a cellular level. The co-activation of pro-apoptotic processes is mediated through specific receptor–ligand interactions of cell death pathways, which are present on the cell membrane surface. The EGCG modulation of multiple genes involved in apoptosis suggests a role as a co-activator of gene expression with direct implications for cancer therapy. These data explain the synergic action of p53siRNA and EGCG, but still remain to be validated on animal models.
Supplementary figures
The canonical network associated with the genes involved in cancer apoptosis mediated by (A) EGCG, (B) p53siRNA, and (C) p53+EGCG.
The canonical network associated with the genes involved in death receptor signalling mediated by (A) EGCG, (B) p53siRNA, and (C) p53+EGCG.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Teiten MH, Eifes S, Dicato M, Diederich M. Curcumin – the paradigm of a multi-target natural compound with applications in cancer prevention and treatment. Toxins (Basel) 2010;2(1):128–162. doi: 10.3390/toxins2010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tachibana H. Molecular basis for cancer chemoprevention by green tea polyphenol. EGCG Forum Nutr. 2009;61:156–169. doi: 10.1159/000212748. [DOI] [PubMed] [Google Scholar]
- 3.Chedea V, Braicu C, Socaciu C. Antioxidant/prooxidant activity of a polyphenolic grape seed extract. Food Chem. 2010;121(1):132–139. [Google Scholar]
- 4.Nakazato T, Ito K, Ikeda Y, Kizaki M. Green tea component, catechin, induces apoptosis of human malignant B cells via production of reactive oxygen species. Clin Cancer Res. 2005;11(16):6040–6049. doi: 10.1158/1078-0432.CCR-04-2273. [DOI] [PubMed] [Google Scholar]
- 5.Kanadzu M, Yuquan L, Morimoto K. Dual function of (−)-epigallocatechin gallate (EGCG) in healthy human lymphocytes. Cancer Lett. 2006;241(2):250–255. doi: 10.1016/j.canlet.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 6.Khan N, Afaq F, Saleem M, Ahmad N, Mukhtar H. Targeting multiple signaling pathways by green tea polyphenol (−)-epigallocatechin-3-gallate. Cancer Res. 2006;66(5):2500–2505. doi: 10.1158/0008-5472.CAN-05-3636. [DOI] [PubMed] [Google Scholar]
- 7.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457(7228):405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 8.Tudoran O, Soritau O, Balacescu O, et al. Early transcriptional pattern of angiogenesis induced by EGCG treatment in cervical tumor cells. J Cel Mol Med. 2012;16(3):520–530. doi: 10.1111/j.1582-4934.2011.01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castanotto D, Rossi J. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457(7228):426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fire AZ, Mello CC. The Nobel Prize in Physiology or Medicine 2006, “for their discovery of RNA interference-gene silencing by double-stranded RNA”. [Accessed July 15, 2012];2006 Available from: http://www.nobelprize.org/nobel_prizes/medicine/laureates/2006/ [Google Scholar]
- 11.Storvold GL, Andersen TI, Perou CM, Frengen E. siRNA: a potential tool for future breast cancer therapy? Crit Rev Oncog. 2006;12(1–2):127–150. doi: 10.1615/critrevoncog.v12.i1-2.70. [DOI] [PubMed] [Google Scholar]
- 12.Sigal A, Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 2000;60(24):6788–6793. [PubMed] [Google Scholar]
- 13.Zhang D, Al-Hendy M, Richard-Davis G, Montgomery-Rice V, Rajaratnam V, Al-Hendy A. Antiproliferative and proapoptotic effects of epigallocatechin gallate on human leiomyoma cells. Fertil Steril. 2010;94(5):1887–1893. doi: 10.1016/j.fertnstert.2009.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinreb O, Mandel S, Youdim M. cDNA gene expression profile homology of antioxidants and their antiapoptotic and proapoptotic activities in human neuroblastoma cells. FASEB J. 2003;17(8):935–937. doi: 10.1096/fj.02-0712fje. [DOI] [PubMed] [Google Scholar]
- 15.Qin J, Xie LP, Zheng XY, et al. A component of green tea, (−)-epigallocatechin-3-gallate, promotes apoptosis in T24 human bladder cancer cells via modulation of the PI3K/Akt pathway and Bcl-2 family proteins. Biochem Biophys Res Commun. 2007;354(4):852–857. doi: 10.1016/j.bbrc.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Ahmed S. Green tea polyphenol epigallocatechin 3-gallate in arthritis: progress and promise. Arthritis Res Ther. 2010;12(2):208. doi: 10.1186/ar2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golden EB, Lam PY, Kardosh A, et al. Green tea polyphenols block the anticancer effects of bortezomib and other boronic acid-based proteasome inhibitors. Blood. 2009;113(23):5927–5937. doi: 10.1182/blood-2008-07-171389. [DOI] [PubMed] [Google Scholar]
- 18.Gasco M, Shami S, Crook T. The p53 pathway in breast cancer. Breast Cancer Res. 2002;4(2):70–76. doi: 10.1186/bcr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacroix M, Toillo RA, Leclercq G. p53 and breast cancer, an update. Endocr Relat Cancer. 2006;13(2):293–325. doi: 10.1677/erc.1.01172. [DOI] [PubMed] [Google Scholar]
- 20.Wang M, Windgassen D, Papoutsakis E. A global transcriptional view of apoptosis in human T-cell activation. BMC Med Genomics. 2008;1:53. doi: 10.1186/1755-8794-1-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nandakumar V, Singh T, Katiyar S. Multi-targeted prevention and therapy of cancer by proanthocyanidins. Cancer Lett. 2008;269(2):378–387. doi: 10.1016/j.canlet.2008.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa S, Utan A, Cervellati R, Speroni E, Guerra MC. Catechins: natural free-radical scavengers against ochratoxin A-induced cell damage in a pig kidney cell line (LLC-PK1) Food Chem Toxicol. 2007;45(10):1910–1917. doi: 10.1016/j.fct.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 23.Fridman J, Lowe S. Control of apoptosis by p53. Oncogene. 2003;22(56):9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- 24.Dyer MJ, MacFarlane M, Cohen GM. Barriers to effective TRAIL-targeted therapy of malignancy. J Clin Oncol. 2007;25(28):4505–4506. doi: 10.1200/JCO.2007.13.1011. [DOI] [PubMed] [Google Scholar]
- 25.Mahalingam D, Szegezdi E, Keane M, de Jong S, Samali A. TRAIL receptor signaling and modulation: are we on the right TRAIL? Cancer Treat Rev. 2009;35(3):280–288. doi: 10.1016/j.ctrv.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 26.Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. 1998;281(5381):1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 27.Wiley SR, Schooley K, Smolak PJ, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3(6):673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 28.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3(9):745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 29.Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer. 2002;2(6):420–430. doi: 10.1038/nrc821. [DOI] [PubMed] [Google Scholar]
- 30.Peperzak V, Veraar EA, Keller AM, Xiao Y, Borst J. The Pim kinase pathway contributes to survival signaling in primed CD8+ T cells upon CD27 costimulation. J Immunol. 2010;185(11):6670–6678. doi: 10.4049/jimmunol.1000159. [DOI] [PubMed] [Google Scholar]
- 31.Denoeud J, Moser M. Role of CD27/CD70 pathway of activation in immunity and tolerance. J Leukoc Biol. 2011;89(2):195–203. doi: 10.1189/jlb.0610351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The canonical network associated with the genes involved in cancer apoptosis mediated by (A) EGCG, (B) p53siRNA, and (C) p53+EGCG.
The canonical network associated with the genes involved in death receptor signalling mediated by (A) EGCG, (B) p53siRNA, and (C) p53+EGCG.