Abstract
Why is agreement on one particular name for each gene important? As one genome after another becomes sequenced, it is imperative to consider the complexity of genes, genetic architecture, gene expression, gene-gene and gene-product interactions and evolutionary relatedness across species. To agree on a particular gene name not only makes one's own research easier, it aids automated text-mining algorithms and search engines, which are increasingly employed to find relationships in the millions of abstracts in the medical research literature and sequence databases. A common nomenclature system will also be helpful to the present generation, as well as future generations, of graduate students and postdoctoral fellows who are about to enter genomics research. In this paper, the authors present some problems that arose when two separate research communities decided to choose the same root, CYP, for naming their gene families. They then offer a logical solution, by renaming the cyclophilin genes with a common root, such a cyn- in Caenorhabditis and CYN- in mammals (Cyn in mouse), and using evolutionary divergence to cluster genes of the highest level of relatedness.
Keywords: human genome, mouse genome, Caenorhabditis elegans genome, cytochrome P450 (CYP) gene superfamily, cyclophilin gene family, immunophilins, peptidylprolyl cis-trans isomerases, FK506-binding proteins, tacrolimus, parvulin
Introduction
A previous paper in this series [1] summarised the steps that one is strongly encouraged to follow in order to ensure proper nomenclature of any gene. Three examples were given to illustrate how and why one should strive for a standardised gene nomenclature system. In these examples, the focus of the paper was on using the gene names as search terms, rather than comparing a DNA or protein sequence that has just been determined by searching via BLAST [2]. The three examples included: PTGS1 and PTGS2 as the correct gene names for prostaglandin G/H synthase-1 and -2, also known as cyclooxygenase-1 and -2 and commonly erroneously nicknamed 'COX-1' and 'COX-2' in many journals; the short- and long-chain fatty acid synthase gene families, for which there is currently no official agreed-upon nomenclature (although FASN on human chromosome 17q25 is the official symbol for the fatty acid synthase gene); an POR as the correct name for the NADPH-P450 oxidoreductase gene [1]. Before deciding upon a new gene symbol, the reader is encouraged to visit the website describing this topic [3].
This theme is extended in the current paper, which shows how two completely separate research communities adopted the same gene root name, while not realising that the other group had done the same thing.
Cyclophilins as 'cyp-' in a Caenorhabditis elegans database
As Head of the Cytochrome P450 (CYP) Superfamily Gene Nomenclature Committee, David Nelson maintains a website dedicated to cytochrome P450 gene nomenclature [4]. The C. elegans genome has 76 full-length P450 genes and nine pseudogenes, which have been assembled by Nelson during the past several years and were nearly completed after the genome sequence had been published. Recently, Dan Lawson of WormPep [5] asked Nelson to review assemblies of these genes, following the recent revision of the worm's genome. While carrying out this request, Nelson discovered that Dan Lawson had referred to several P450 genes as 'ccp-xx'. Nelson then explored WormPep further and confirmed that ccp was being used as the root for cytochrome P450 genes. Although the usual root for P450 gene names is CYP, this term (cyp) was being used in the C. elegans protein database for the cyclophilins (Table 1). Can this be a problem -- i.e. the same gene root being used for different gene families by colleagues in two separate, very distant research fields?
Table 1.
List of cyclophilin and P450 genes in C. elegans.
| Cyclophilin genes | Alternative name | WormPep accession # | |
|---|---|---|---|
| cyp-1Y49A3A.5 | CGC approved | CE22213 | |
| cyp-2ZK520.5 | CGC approved | CE16730 | |
| cyp-3Y75B12B.5 | CGC approved | CE20374 | |
| cyp-4F59E10.2 | mog-6 | CGC approved | CE01596 |
| cyp-5F31C3.1 | CGC approved | CE17730 | |
| cyp-6F42G9.2 | CGC approved | CE01301 | |
| cyp-7Y75B12B.2 | CGC approved | CE20371 | |
| cyp-8D1009.2a | D1009.2b | CGC approved | CE04286 |
| cyp-9T27D1.1 | CGC approved | CE03745 | |
| cyp-10B0252.4a | B0252.4b | CGC approved | CE02420 |
| cyp-11T01B7.4 | CGC approved | CE03588 | |
| cyp-12C34D4.12 | CGC approved | CE17506 | |
| cyp-13Y116A8C.34 | CGC approved | CE24152 | |
| cyp-14F39H2.2a | F39H2.2b | CGC approved | CE32410 |
| cyp-15Y87G2A.6 | CGC approved | CE24686 | |
| cyp-16Y17G7B.9 | CGC approved | CE19042 | |
| cyp-17ZC250.1 | CGC approved | CE28157 | |
| P450 genes | |||
| ccp-13A7T10B9.10 | CGC approved | CE01655 | |
| ccp-14A5F08F3.7 | CGC approved | CE09262 | |
| ccp-31A1 | C01F6.3 | CGC approved | unavailable |
| ccp-44ZK177.5 | cyp-44 | CGC approved | CE25682 |
Data taken from Jonathan Hodgkin, CGC Genetic Map and Nomenclature Curator (Caenorhabditis Genetics Center), Genetics Unit, Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, UK.
CYP for cytochrome P450 genes in all species
The mammalian cytochrome P450 (CYP) superfamily encodes enzymes involved in: the metabolism of pharmaceuticals, foreign chemicals and pollutants; arachidonic acid metabolism and eicosanoid biosynthesis; cholesterol, sterol and bile acid biosynthesis; steroid synthesis and catabolism; vitamin D3 synthesis and catabolism; retinoic acid hydroxylation; biogenic amine and neuroamine metabolism; and several orphan CYP genes still of unknown function [6]. There are 102 and 57 putatively functiona CYP genes in the mouse and human, respectively [7]. To date, more than 3,400 P450 sequences have been named with the three-letter root of CYP. This nomenclature has been in place [8,9] since 1987, and is growing every day [4]. The official root names for mouse and human P450s are Cyp and CYP, respectively. The Drosophila nomenclature [10] also use Cyp. There are now 727 genes in rice an Arabidopsis that have been named CYP [4]. It is anticipated that the number of named P450 genes will exceed 4,000 by the end of 2004.
Whereas continuing to use the CYP root for cyclophilin genes will be a nightmare for cyclophilin researchers, P450 researchers might find this an annoyance but not really much of a problem. To prevent conflicts over nomenclature, it becomes increasingly urgent to rename the cyclophilin genes. What is the best root name for these genes?
Finding the best root for the cyclophilin genes
The three families of immunophilins, known as peptidylprolyl cis-trans isomerases (PPIases), include the cyclophilins, the FK506-binding proteins (FKBPs) and parvulin [11-13]. All three gene families are found in animals, plants and eubacteria. While two cyclophilins and two types of FKBPs exist in archaebacteria, no parvulin homologue has been found. Parvulin is unique among the immunophilins. A search of the LocusLink [14], HUGO Gene Nomenclature Committee [15], and the National Center for Biotechnology Information (NCBI) UniGene [16] websites using 'parvulin', shows a single gene; Pin4 and PIN4 are the approved mouse and human gene names, respectively. 'PIN' is an abbreviation for peptidylprolyl cis-trans isomerase NIMA-interacting-4. 'NIMA' stands for 'never-in-mitosis-gene-a', which was first isolated as a series of conditional cell cycle mutants that failed to enter mitosis in Aspergillus nidulans [17,18]. There are 11 genes (NEK1, NEK2, ... NEK11) in the human genome that encode NIMA-related mitotic kinases and are involved in DNA replication and genotoxic stress responses [19,20]. Although parvulin has peptidylprolyl cis-trans isomerase activity, it shares no evolutionary homology with the FKBPs or cyclophilins.
Immunophilins are defined as receptors for immuno-suppressive drugs including cyclosporin-A, FK506 and rapamycin. FK506 is also called tacrolimus, a macrolide of fungal origin (produced by Streptomyces tsukubaensis) and having strong immunosuppressive actions. FK506- and rapamycin-binding proteins are abbreviated as FKBPs and share no evolutionary homology with the cyclophilins or parvulin. A search of the LocusLink, HUGO Gene Nomenclature Committee and the NCBI UniGene websites using 'fkbp', shows more than 80 FKBP genes in the human and mouse (FKBP1, FKBP2, ... FKBP82). These gene products have many unique features, such as targeting BCL2 to the mitochondria and inhibiting apoptosis [21].
Cyclophilins, the third and last class of the PPIases, comprise cyclosporin-A-binding proteins [22] ranging in size from 17 kDa to 324 kDa [12]. This class of immunophilins carries out a wide range of functions -- including acting as a chaperone to facilitate the nuclear transport of the somatolactogenic hormones [23], facilitating the calcium-regulated mitochondrial permeability transition pore which precedes apoptosis [24] and participating in the pre-mRNA splicing machinery [25]. Cyclophilin-binding drugs are emerging as potential leads to novel targets for interference with interleukin-12 production [26] and, therefore, to the possibility of treating conditions such as multiple sclerosis and rheumatoid arthritis. Cyclosporin-A also has activity against helminth and protozoan parasites [27].
A search of the LocusLink, HUGO Gene Nomenclature Committee and the NCBI UniGene websites using 'cyclophilin', shows 15 putatively functional genes and 22 pseudogenes. The 15 putatively functional gene names (Table 2) include PPIA through to PPIH (for peptidylprolyl isomerase-A, -B, ... -H; cyclophilin A-, B-, ... H-related), one PPIA-like (PPIAL3), and six cyclophilin-like (PPIL1, PPIL2, ... PPIL6).
Table 2.
List of putatively functional human cyclophilin genes.
| Approved gene symbol | Approved gene name | Chromosomal location |
|---|---|---|
| PPIA | Peptidylprolyl isomerase A (cyclophilin A) | 7p13-p11.2 |
| PPIAL3 | Peptidylprolyl isomerase A (cyclophilin A)-like-3 | 21 |
| PPIB | Peptidylprolyl isomerase B (cyclophilin B) | 15 |
| PPIC | Peptidylprolyl isomerase C (cyclophilin C) | [reserved] |
| PPID | Peptidylprolyl isomerase D (cyclophilin D) | 4 |
| PPIE | Peptidylprolyl isomerase E (cyclophilin E) | 1p32 |
| PPIF | Peptidylprolyl isomerase F (cyclophilin F) | 10q22-q23 |
| PPIG | Peptidylprolyl isomerase G (cyclophilin G) | 2q31.1 |
| PPIH | Peptidylprolyl isomerase H (cyclophilin H) | 1p34.1 |
| PPILI | Peptidylprolyl isomerase (cyclophilin)-like 1 | 6p21.1 |
| PPIL2 | Peptidylprolyl isomerase (cyclophilin)-like 2 | 22 |
| PPIL3 | Peptidylprolyl isomerase (cyclophilin)-like 3 | 2 |
| PPIL4 | Peptidylprolyl isomerase (cyclophilin)-like 4 | 6q24-25 |
| PPIL5 | Peptidylprolyl isomerase (cyclophilin)-like 5 | 14q21.3 |
| PPIL6 | Peptidylprolyl isomerase (cyclophilin)-like 6 | 6q21 |
Not included here are PPIAL, PPIAL2, PPIAP, PPI AP2, PPIAP3, PPIAP4, PPIAP5, PPIAP6, PPIHP1, PPIHP2, PPIL1P1, PPIP1, PPIP2, PPIP3, PPIP4, PPIP5, PPIP6, PPIP7, PPIP8, PPIP9, PPIP10 and PPIP11, which represent the 22 cyclophilin pseudogenes in the Human Genome Project (HGP) database.
PPID has the synonym 'CYP-40', but this is no longer the official name. Unfortunately, the mouse RIKEN full-length cDNAs that match this sequence are being called CYP40, not PPID, so the name is propagating itself in the literature and into the databases in an uncontrollable way. The cloning and naming of 11 cyclophilin genes from C. elegans (Cyp-1 to Cyp-11)[28] was reported in 1996. A search of GenBank for CYP20 finds AY568517, a Arabidopsis thylakoid lumen cyclophilin [29], named CYP20-2. (CYP20A1 is a chordate cytochrome P450 of unknown function, possibly involved in development.)[4] The date on this Arabidopsis CYP20 GenBank entry is 15th April, 2004, showing that the problem is not going away. In fact, the PubMed link from the GenBank entry leads to a publication [30] in which a nomenclature system for the 29 cyclophilin genes in the Arabidopsis thaliana genome is presented using CYP as the root.
What is the solution?
Solutions -- like politics -- are local. We have contacted the C. elegans community and alerted them to this nomenclature conflict. They are responding and will select a new root for cyclophilins and change their P450 gene names to cyp-, from the current ccp- root. This will go into the official WormPep and WormBase nomenclature and will eventually prevent use of the cyp- root in C. elegans (and, hopefully, C. briggsae) for cyclophilins. Additional effort will be needed for the Arabidopsis community, as well as for the human and mouse gene databases.
What might be the best root for the cyclophilin gene family? Cyn has been used for cyclone, a mouse gene in LocusLink; CPN1 and CPN2 are being used for carboxypeptidase N-1 and -2; Cph was considered, but CPH1 has been used to refer to a cryptochrome or phytochrome (light-sensing protein) [31]. Because of the sharing of this paper (while still being written) with Lois Maltais of Mouse Genome Informatics (MGI), she consulted with the authors of the mouse cyclone gene paper and they have now agreed to use Cycn, in order to free up Cyn and CYN for the mouse and human cyclophilin, respectively. After searching databases and search engines for conflicts, the present authors suggest that Cyn- might be the most suitable root for C. elegans cyclophilins, but this needs to be decided among members of the worm community. It is unfortunate that some databases (eg worm, yeast and bacteria) are mandating that gene names be limited to three letters. The authors suspect that three-letter root names for the ~19,000 C. elegans genes may not be enough. For example, 10,000 families will require the same number of roots. 26 cubed is only 17,576; this will require the use of odd letter combinations that have no symbolic meaning, such a xyz1, cxq, rzx, etc. Also, the nature of language is to use some letters more often than others, which will put great pressure on naming the genes that begin with the most often-used letters. CYN has now been officially approved as the root to unify all mammalian cyclophilins.
Using evolutionary trees to assign names to genes in the P450 superfamily [4,8,9], in the authors' experiences, has been very positive. This can work in general for any other homologous group of genes and, in fact, has been used for at least 124 families and/or superfamilies to date [32]. To illustrate this point, a simple sequence alignment (Figure 1) and tree (Figure 2) are presented for the C. elegans cyclophilins.
Figure 1.
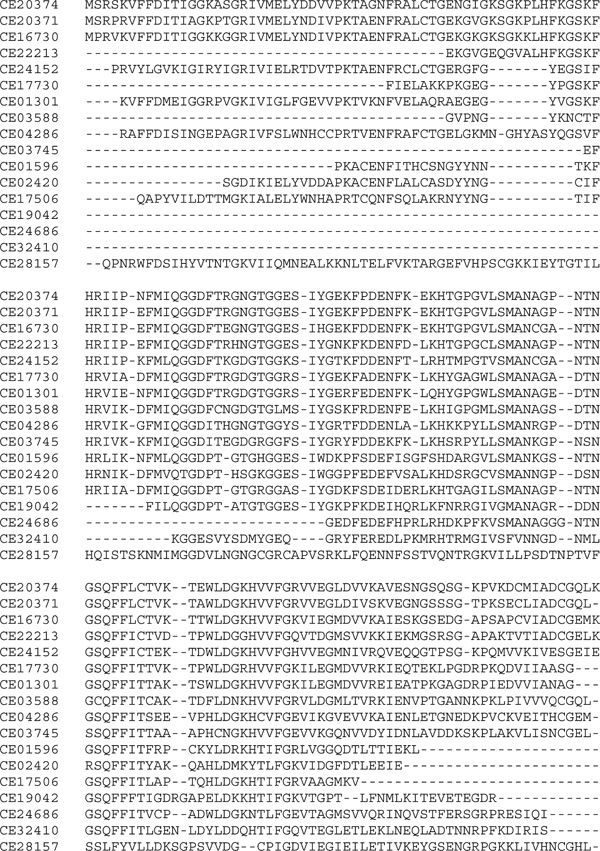
Sequence alignment of the conserved regions of 17 Caenorhabditis elegans cyclophilin proteins.
Figure 2.
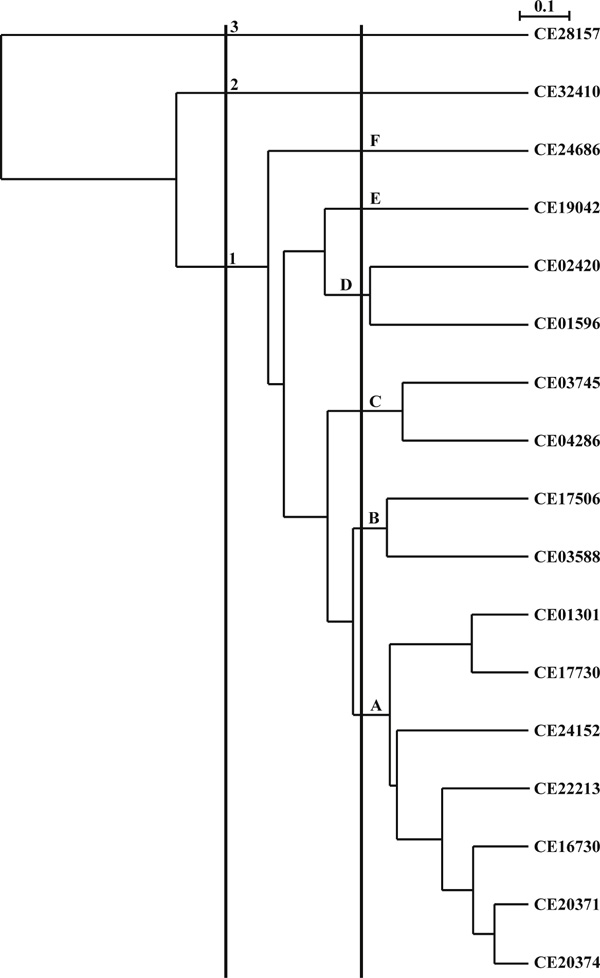
UPGMA tree and possible family and subfamily divisions of the Caenorhabditis elegans cyclophilin gene family, based on evolutionary divergence. The root of cyn-, is acceptable because it has not yet been used by any other gene database, except 'cyclone' in mouse. Families are designated by Arabic numerals and represent amino acid identity of 40 per cent or greater. Subfamilies are designated by letters and represent amino acid identity of 54 per cent or greater. Individual genes within subfamilies are then given Arabic numbers. Identifiers are the WormPep accession numbers.
The vertical lines in Figure 2 are suggested break-points for family and subfamily designations. Branches on the tree intersected by the lines would define family and subfamily clusters. The lines could be moved to modify the number of families and subfamilies. As drawn, there are six subfamilies in family 1, and one each in families 2 and 3. Moving the subfamily line to the left could reduce the number of subfamilies in family 1 from six to three. If cyn were used, CE28157 (at the top of Figure 2) would be named cyn3a1 and CE20374 (at the bottom of Figure 2) would be named cyn1a1, and so on.
A method for creating a network of 'gene co-occurrences' from the literature, and portioning it into communities of related genes, has recently been presented [33]. In that paper, a program is described (but not named) which searches all Medline titles and abstracts and OMIM entries for occurrences and co-occurrences of gene symbols, gene names and diseases; the databases contain more than 12 million abstracts. Relationships are identified by automated bioinformatics methods between genes, and between genes and diseases, that might not be detected by less computationally intense methods. Such methods must rely on consistent names, or they have to deal with a list of synonyms.
Conclusions
The cyclophilin gene nomenclature has several problems. First, many in the cyclophilin field continue to use CYP, which has been the gene root for the large cytochrome P450 gene superfamily since 1987. Secondly, the gene root chosen by the HUGO Human/Mouse Gene Nomenclature Committees had been PPI for peptidylprolyl cis-trans isomerase, although -- as detailed above -- the cyclophilins represent just one of three classes of the PPIases that are perhaps functionally related but evolutionarily unrelated. Thirdly, the authors suggest the root Cyn for the C. elegans cyclophilin genes. Fourthly, eight of the 15 putatively functional human cyclophilin genes end in the letters 'A' through to 'H', while the others end in two groups of numbers (one PPIA-like and six PPI-like). It is strongly recommended that these genes be named by families and subfamilies, according to evolutionary divergence, as shown in Figure 3. Because of discussions related to the writing of this paper, the CYN root has now been officially approved for mammals. It would be desirable to incorporate as many species as possible into the naming scheme. One additional source of nomenclature friction is the strict use of three letter roots for gene names in C. elegans, yeast and bacteria; this automatically creates conflicts when human and mouse root names can be much longer than three letters, as in PPIAL3 or NIPSNAP1; however, that is a battle for another day.
Figure 3.
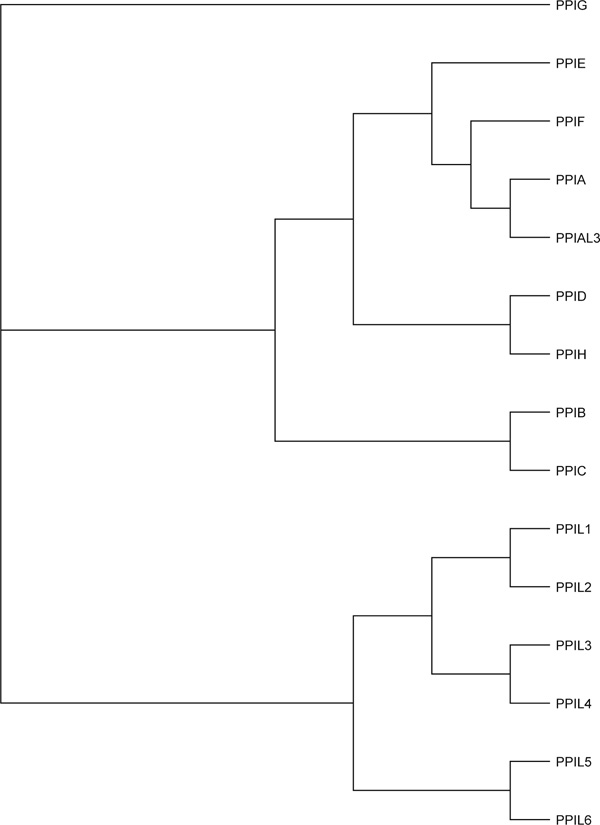
Rectangular cladogram of the 15 human cyclophilin proteins aligned. If one were to drop vertical lines, as in Figure 2, one might name: PPIL1 through to PPIL6 as CYNIA1 through to CYNIA4, and CYNIB1 and CYNIB2, respectively; PPIB and PPIC as CYN2A1 and CYN2A2, respectively; PPIE, PPIF, PPIA and PPIAL3 as CYN2C1, CYN2C2, CYN2C3 and CYN2C4, respectively; PPIG as CYN3; and so on.
Acknowledgements
The writing of this article was funded, in part, by NIH grant P30 ES06096 (D.W.N.).
References
- Nebert DW, Wain HM. 'Update on human genome completion and annotations: Gene nomenclature'. Hum Genomics. 2003;1:66–71. doi: 10.1186/1479-7364-1-1-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.ncbi.nlm.nih.gov/BLAST/
- http://www.gene.ucl.ac.uk/nomenclature/information/check.shtml
- http://drnelson.utmem.edu/cytochromeP450.html
- The C. elegans protein database. http://www.sanger.ac.uk/Projects/C_elegans/wormpep/
- Nebert DW, Russell DW. 'Clinical importance of the cytochromes P450'. Lancet. 2002;360:1155–1162. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Zeldin D, Hoffman S. et al. 'Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes including nomenclature recommendations for genes, pseudogenes, and alternative-splice variants'. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Adesnik M, Coon MJ. et al. 'The P450 gene superfamily. Recommended nomenclature'. DNA. 1987;6:1–11. doi: 10.1089/dna.1987.6.1. [DOI] [PubMed] [Google Scholar]
- Nelson DR, Koymans L, Kamataki T. et al. 'Cytochrome P450 superfamily: Update on new sequences, gene mapping, accession numbers, and nomenclature'. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- http://flybase.bio.indiana.edu/
- Maruyama T, Furutani M. 'Archaeal peptidyl prolyl cis-trans isomerases (PPIases)'. Front Biosci. 2000;5:D821–D836. doi: 10.2741/maruyama. [DOI] [PubMed] [Google Scholar]
- Galat A. 'Peptidylprolyl cis/trans isomerases (immunophilins): Biological diversity -- targets -- functions'. Curr T op Med Chem. 2003;3:1315–1347. doi: 10.2174/1568026033451862. [DOI] [PubMed] [Google Scholar]
- He Z, Li L, Luan S. 'Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis'. Plant Physiol. 2004;134:1248–1267. doi: 10.1104/pp.103.031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.ncbi.nlm.nih.gov/LocusLink/
- http://www.gene.ucl.ac.uk/nomenclature/
- http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = unigene
- Osmani SA, May GS, Morris NR. 'Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans'. J Cell Biol. 1987;104:1495–1504. doi: 10.1083/jcb.104.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani SA, Pu RT, Morris NR. 'Mitotic induction and maintenance by over-expression of a G2-specific gene that encodes a potential protein kinase'. Cell. 1988;53:237–244. doi: 10.1016/0092-8674(88)90385-6. [DOI] [PubMed] [Google Scholar]
- Krien MJ, West RR, John UP. et al. 'The fission yeast NIMA kinase Fin1p is required for spindle function and nuclear envelope integrity'. EMBO J. 2002;21:1713–1722. doi: 10.1093/emboj/21.7.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi K, Fukazawa H, Murakami Y. et al. 'Nek11, a new member of the NIMA family of kinases, involved in DNA replication and genotoxic stress responses'. J Biol Chem. 2002;277:39655–39665. doi: 10.1074/jbc.M204599200. [DOI] [PubMed] [Google Scholar]
- Shirane M, Nakayama KI. 'Immunophilin FKBP38, an inherent inhibitor of calcineurin, targets BCL2 to mitochondria and inhibits apoptosis'. Nippon Rinsho. 2004;62:405–412. [PubMed] [Google Scholar]
- Jin L, Harrison SC. 'Crystal structure of human calcineurin complexed with cyclosporine-A and human cyclophilin'. Proc Natl Acad Sci USA. 2002;99:13522–13526. doi: 10.1073/pnas.212504399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rycyzyn MA, Clevenger CV. 'Role of cyclophilins in somatolactogenic action'. Ann NY Acad Sci. 2000;917:514–521. doi: 10.1111/j.1749-6632.2000.tb05416.x. [DOI] [PubMed] [Google Scholar]
- Halestrap AP, McStay GP, Clarke SJ. 'The permeability transition pore complex: Another view'. Biochimie. 2002;84:153–166. doi: 10.1016/S0300-9084(02)01375-5. [DOI] [PubMed] [Google Scholar]
- Pemberton TJ, Rulten SL, Kay JE. 'Identification and characterization of Schizosaccharomyces pombe cyclophilin-3, a cyclosporin A-insensitive orthologue of human USA-CyP'. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;786:81–91. doi: 10.1016/S1570-0232(02)00738-9. [DOI] [PubMed] [Google Scholar]
- Vandenbroeck K, Alloza I, Gadina M. 'Inhibiting cytokines of the interleukin-12 family: Recent advances and novel challenges'. J Pharm Pharmacol. 2004;56:145–160. doi: 10.1211/0022357022962. [DOI] [PubMed] [Google Scholar]
- Chappell LH, Wastling JM. 'Cyclosporin A: Antiparasite drug, modulator of the host-parasite relationship, and immunosuppressant'. Parasitology. 1992;105:S25–S40. doi: 10.1017/S0031182000075338. [DOI] [PubMed] [Google Scholar]
- Page AP, MacNiven K, Hengartner MO. 'Cloning and biochemical characterization of the cyclophilin homologues from the free-living nematode Caenorhabditis elegans'. Biochem J. 1996;317:179–185. doi: 10.1042/bj3170179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano PGN, Edvardsson A, Ruban AV. et al. 'Arabidopsis AtCYP20-2 is a light-regulated cyclophilin-type peptidyl-prolyl cis-trans isomerase associated with the photosynthetic membranes'. Plant Physiol. 2004;134:1244–1247. doi: 10.1104/pp.104.041186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano PG, Horton P, Gray JE. 'The Arabidopsis cyclo-philin gene family'. Plant Physiol. 2004;134:1268–1282. doi: 10.1104/pp.103.022160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisdorph NA, Small GD. 'The CPH gene of Chlamy-domonas reinhardtii encodes two forms of cryptochrome whose levels are controlled by light-induced proteolysis'. Plant Physiol. 2004;134:1546–1554. doi: 10.1104/pp.103.031930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.gene.ucl.ac.uk/nomenclature/genefamily.shtml
- Wilkinson DM, Huberman BA. 'A method for finding communities of related genes'. Proc Natl Acad Sci USA. 2004;101:S5241–S5248. doi: 10.1073/pnas.0307740100. [DOI] [PMC free article] [PubMed] [Google Scholar]


