Abstract
Plasmids of the incompatibility group IncP-1 can transfer and replicate in many genera of the Proteobacteria. They are composed of backbone genes that encode a variety of essential functions and accessory genes that have implications for human health and environmental remediation. Although it is well understood that the accessory genes are transferred horizontally between plasmids, recent studies have also provided examples of recombination in the backbone genes of IncP-1 plasmids. As a consequence, phylogeny estimation based on backbone genes is expected to produce conflicting gene tree topologies. The main goal of this study was therefore to infer the evolutionary history of IncP-1 plasmids in the presence of both vertical and horizontal gene transfer. This was achieved by quantifying the incongruence among gene trees and attributing it to known causes such as 1) phylogenetic uncertainty, 2) coalescent stochasticity, and 3) horizontal inheritance. Topologies of gene trees exhibited more incongruence than could be attributed to phylogenetic uncertainty alone. Species-tree estimation using a Bayesian framework that takes coalescent stochasticity into account was well supported, but it differed slightly from the maximum-likelihood tree estimated by concatenation of backbone genes. After removal of the gene that demonstrated a signal of intergroup recombination, the concatenated tree was congruent with the species-tree estimate, which itself was robust to inclusion/exclusion of the recombinant gene. Thus, in spite of horizontal gene exchange both within and among IncP-1 subgroups, the backbone genome of these IncP-1 plasmids retains a detectable vertical evolutionary history.
Keywords: plasmid, phylogeny, species tree, genomics, horizontal gene transfer
Introduction
Self-transferable broad-host-range plasmids of Proteobacteria play a crucial role in bacterial adaptation because they can exchange genes among phylogenetically distant bacteria (Adamczyk and Jagura-Burdzy 2003). These extrachromosomal DNA molecules provide bacteria with a variety of novel phenotypic traits and contribute to the alarmingly rapid spread of multidrug resistance in human pathogens. The most promiscuous plasmids (i.e., those with the broadest host range) belong to the incompatibility groups IncP (Adamczyk and Jagura-Burdzy 2003), IncW (Fernandez-Lopez et al. 2006), IncU (Rhodes et al. 2004) and the recently defined group PromA (Gstalder et al. 2003; Van der Auwera et al. 2009). Plasmids belonging to the same group are said to be incompatible because they cannot coexist in the same cell line. IncP, also called IncP-1, plasmids are considered to be among the most promiscuous and carry many kinds of accessory genes (Thomas and Smith 1987; Adamczyk and Jagura-Burdzy 2003; Schlüter et al. 2007). They replicate in different classes within Proteobacteria and can also mobilize nonself-transferable plasmids into Gram-positive bacteria (Mazodier et al. 1989), cyanobacteria (Kreps et al. 1990), and even eukaryotes (Heinemann and Sprague 1990). Not only are they present in diverse environments such as manure (Binh et al. 2008), agricultural soils (Top et al. 1995; Sen et al. 2011), streams (Smalla et al. 2006; Akiyama et al. 2010), and wastewater treatment plants (Schlüter et al. 2007), they are also a cause for concern in the clinic because of the drug resistance they often encode (Ingram et al. 1973; Novais et al. 2006).
Plasmids typically consist of backbone genes that are involved in replication, stable inheritance and control, and conjugative transfer (fig. 1), in addition to accessory genes that confer variable host-beneficial traits. Adamczyk and Jagura-Burdzy (2003) and Schlüter et al. (2007) provide excellent reviews of backbone genes of IncP-1 plasmids. Of the nearly 45 backbone genes found on IncP-1 plasmids, approximately 33 are shared by all plasmids that have been completely sequenced to date. The conservation of this complement of backbone genes across the IncP-1 plasmids suggests that they may share a common phylogenetic history.
Fig. 1.
Genetic map of a typical IncP-1 plasmid showing the different functional modules: region involved in initiating replication, composed of origin of replication (oriV), and replication initiation gene (trfA); trb, involved in mating bridge formation during conjugation; tra, involved in DNA processing for transfer during conjugation; ctl, also called central control region, is composed of regulatory genes and involved in maintaining plasmid stability; and accessory regions are composed of host-beneficial genes. Genes that were included in this study are colored dark gray. Numerals inside the circle indicate tree topologies that were shared by several genes (topologies 1–4) or unique to one gene (U); topologies were inferred in this study by maximum likelihood (fig. 2).
Phylogenies inferred from single or concatenated backbone genes have shown the IncP-1 group to be composed of five diverse subgroups: IncP-1α, -β, -γ, -δ, and -ε (Pansegrau et al. 1994; Thorsted et al. 1998; Vedler et al. 2004; Haines et al. 2006; Bahl et al. 2007). Recently, three new potential IncP-1 subgroups have been identified: ζ (Norberg et al. 2011), η (Sen D, Yano H, Bauer M, Rogers LM, Van der Auwera GA, Brown C, and Top E, unpublished), and an unnamed subgroup (Pachulec and van der Does 2010), thus indicating the tremendous diversity that exists within this plasmid group. The biological significance of these subgroups is unknown, and more work is required to distinguish between them phenotypically. Furthermore, the validity of classifying plasmids into subgroups based on the phylogeny of a single gene or few genes is tenuous because of evidence of horizontal gene transfer (HGT) between plasmid backbones in the group. HGT among plasmid backbones is mediated by conjugation and subsequent recombination between coexisting plasmids, resulting in chimeric plasmid genomes. In such cases, phylogenetic inference based on both recombinant and nonrecombinant regions may lead to conflicting results. The first evidence of recombination between IncP-1 plasmids was clearly shown in the IncP-1β plasmid, pB10 (Schlüter et al. 2003). We recently showed another instance of recombination in the IncP-1δ plasmid pIJB1, which has two sets of the replication and transfer genes trfA–trbE, one of which was acquired from an IncP-1β plasmid and the other was native to its subgroup (Sen et al. 2010). Recently, more IncP-1 plasmids (namely, pB3, pBP136, and pAOVO02) were identified as recombinants through the analysis of concatenated alignments of backbone genes (Norberg et al. 2011). It is thus becoming increasingly apparent that although IncP-1 plasmids are incompatible over long periods of time, they may coexist long enough to allow recombination, which may also be true for other incompatibility groups. Thus, the contribution of recombination to the evolution of plasmids may be greater than previously thought, and a phylogenomic approach is required to elucidate the extent of HGT and how it impacts phylogenetic signal and phylogeny estimation.
It is known that phylogenies inferred from multiple loci often contradict each other (Rokas et al. 2003; Pollard et al. 2006) and that this incongruence among gene phylogenies can have three causes: phylogenetic uncertainty, coalescent stochasticity or random sorting of ancestral polymorphisms (Maddison 1997), and HGT (Maddison 1997). Phylogenetic uncertainty can be attributed to random error, caused by the sample of characters chosen (Graybeal 1998), and/or systematic error (Yang et al. 1994) caused by the introduction of analytical bias during phylogeny estimation. Coalescent stochasticity is caused by the random sorting of polymorphisms in an ancestral population and sometimes occurs in a way that is not in agreement with the species history, such that nonsister taxa or subgroups share the same states. This is often seen in large populations that have undergone recent speciation events (Maddison 1997). In the IncP-1 plasmid, recombination within subgroups (i.e., intragroup HGT) may lead to differential sorting of ancestral polymorphisms at multiple loci in the same way that recombination within sexually reproducing species does. HGT is the transfer of genetic material between different taxa, here plasmids of different subgroups (Maddison 1997). Classically, resolution of incongruence and estimation of species trees were accomplished either with consensus methods (Bryant 2003) or with total evidence methods (Kluge 2004). Consensus methods rely on estimating a summary tree from a collection of gene trees, whereas total evidence involves the analysis of a single-gene tree from a collection of genes that are concatenated into a supermatrix. More recently, however, several methods have been developed to estimate species trees using coalescent models (e.g., BEST, Liu 2008; STEM, Kubatko et al. 2009; *BEAST, Heled and Drummond 2010). These methods assume that incongruence is being generated exclusively by the stochastic sorting of ancestral polymorphisms (Liu et al. 2009), although they appear to be robust to HGT/hybridization to at least some degree (Chung and Anè 2011).
The goal of this study was to determine the evolutionary history of the backbone of IncP-1 plasmids by systematically examining congruence among gene trees estimated from the backbone genes. Results showed extensive incongruence among the trees, as expected. Therefore, we applied a series of phylogenetic analyses to estimate the evolutionary history of these plasmids in the face of this incongruence. This is the first gene-by-gene analysis of backbone genes of IncP-1 plasmids, and in spite of strong incongruence among genes, we derive a strongly supported estimate of the relationships among the five well-known IncP-1 subgroups by using species-tree estimation approaches.
Materials and Methods
Plasmids and Genes
Complete sequences of IncP-1 plasmids were extracted from GenBank or determined by us, a total of 65. Only backbone genes that were common to all plasmids were used for phylogeny estimation (dark-gray open reading frames [ORFs] in fig. 1). For those plasmids that showed identical sets of backbone gene sequences, only one representative plasmid was retained. In addition, plasmids such as pEST4011 that were missing a large section of backbone sequence shared by all other plasmids were not included nor was plasmid pIJB1 because of its duplicated trfA and trb genes of IncP-1δ and IncP-1β descent (Sen et al. 2010). The plasmids from the recently proposed ζ, η, and an unnamed subgroups (see Introduction) were not included because their sequences only became publically available in the middle of this study and because of their wide sequence divergence and lack of evidence that they are physically incompatible with the prototype IncP-1 plasmids. The final set of plasmids used in this study is presented in table 1. All plasmids, including those that were previously published, are referenced only by their Genbank accession numbers or National Center for Biotechnology Information reference numbers.
Table 1.
General Features of the Plasmids Included in This Study Listed by Subgroup.
| Plasmids | IncP-1 Subgroup | Origin/Isolation Method/Hosta | Accession Numberb |
|---|---|---|---|
| pB5 | IncP-1α | Municipal WWTP in Germany, exogenous | CP002151 |
| pBS228 | IncP-1α | Wastewater of antibiotic factory in Russia, host unknown | NC_008357 |
| pG527 | IncP-1α | Pig manure, Germany, exogenous | JX469830 |
| pSP21 | IncP-1α | Municipal WWTP in Germany, exogenous | CP002153 |
| pTB11 | IncP-1α | WWTP in Germany, exogenous | NC_006352 |
| pWEC911 | IncP-1α | Sugar beet rhizosphere in the United Kingdom, exogenous | JX469833 |
| RK2 | IncP-1α | Hospital in the United Kingdom, Pseudomonas aeruginosa and Klebsiella aerogenes | NC_001621 |
| pA1 | IncP-1β | Soil in Japan, Sphingomonas sp. A1 | NC_007353 |
| pA81 | IncP-1β | PCB contaminated soil in Czech Republic, Achromobacter xylosoxidans A8 | AJ515144 |
| pADP-1 | IncP-1β | Soil in the United States, Pseudomonas sp. ADP | NC_004956 |
| pAKD1 | IncP-1β | Agricultural soil in Norway, exogenous | JN106164 |
| pAKD18 | IncP-1β | Agricultural soil in Norway, exogenous | JN106169 |
| pAKD26 | IncP-1β | Agricultural soil in Norway, exogenous | JN106171 |
| pAMMD1 | IncP-1β | Pea rhizosphere in the United States, Burkholderia ambifaria AMMD | NC_008385 |
| pAOVO02 | IncP-1β | Polluted soil in the United States, Acidovorax sp. JS42 | NC_008766 |
| pB1 | IncP-1β | Municipal WWTP in Germany, exogenous | JX469829 |
| pB3 | IncP-1β | Municipal WWTP in Germany, exogenous | NC_006388 |
| pB4 | IncP-1β | Municipal WWTP in Germany, exogenous | AJ431260 |
| pB8 | IncP-1β | Municipal WWTP in Germany, exogenous | NC_007502 |
| pB10 | IncP-1β | Municipal WWTP in Germany, exogenous | NC_004840 |
| pB12 | IncP-1β | Municipal WWTP in Germany, exogenous | JX469826 |
| pBP136 | IncP-1β | Diseased whooping cough patient in Japan, Bordetella pertussis BP136 | NC_008459 |
| pC11 | IncP-1β | Municipal WWTP in Germany, Delftia acidovorans C1 | HQ891317 |
| pCNB1 | IncP-1β | Industrial WWTP in China, Comamonas sp. CNB-1 | NC_010935 |
| pDS3 | IncP-1β | Creek in the United States, exogenous | JX469834 |
| pKS212 | IncP-1β | Hospital WWTP in Belgium, exogenous | JX469831 |
| pKV29 | IncP-1β | Municipal WWTP in Germany, Delftia sp. KV29 | JN648090 |
| pNB1 | IncP-1β | Orchard soil in Belgium Delftia acidovorans LME1 | JF274988 |
| pRSB222 | IncP-1β | Municipal WWTP in Germany, exogenous | JX469824 |
| JX469825 | |||
| pRSB223 | IncP-1β | Municipal WWTP in Germany, exogenous | JX469828 |
| pTB30 | IncP-1β | Agricultural soil in Belgium, Comamonas testosteroni TB30 | JF274987 |
| pTP6 | IncP-1β | Contaminated river sediments in Kazakhstan, exogenous | NC_007680 |
| pUO1 | IncP-1β | Industrial WWTP in Japan, Delftia acidovorans B | NC_005088 |
| pWDL7 | IncP-1β | Orchard soil in Belgium, Comamonas testosteroni WDL7 | GQ495894 |
| pYS1 | IncP-1β | Polluted soil in Japan, Burkholderia cepacia | JX469832 |
| R751 | IncP-1β | Hospital in the United Kingdom, Klebsiella aerogenes | NC_001735 |
| pKS208 | IncP-1γ | Hospital WWTP in Belgium, exogenous | JQ432564 |
| pMBUI1 | IncP-1γ | University of Idaho Arboretum Pond, exogenous | JQ432563 |
| pQKH54 | IncP-1γ | River in the United Kingdom, exogenous | NC_008055 |
| pAKD4 | IncP-1δ | Agricultural soil in Norway, exogenous | GQ983559 |
| pAKD16 | IncP-1ε | Agricultural soil in Norway, exogenous | JN106167 |
| pAKD25 | IncP-1ε | Agricultural soil in Norway, exogenous | JN106170 |
| pEMT3 | IncP-1ε | Agricultural soil in the United States, exogenous | JX469827 |
| pHH128 | IncP-1ε | Manured soil in Germany, exogenous | JQ004406 |
| pHH3414 | IncP-1ε | Manured soil in Germany, exogenous | JQ004408 |
| pKJK5 | IncP-1ε | Manured soil in Denmark, exogenous | NC_008272 |
Note.—WWTP, wastewater treatment plant.
aThe original hosts of plasmids captured by exogenous isolation are not known.
bSee Materials and Methods for references to studies that described the plasmids whose sequences were not previously published. All previously published plasmids are only referred to here by their RefSeq or Genbank/EMBL/DDBJ accession numbers.
Sequencing and Annotation
The following 15 plasmids were sequenced as part of this study (references refer to the studies that first isolated and described the plasmids): pB1 and pB12 (Dröge et al. 2000), pEMT3 (Top et al. 1995), pG527 (Götz et al. 1996), pC11 and pNB1, (Boon et al. 2001), pKV29 (Stolze et al. 2012), pTB30 (Dejonghe et al. 2002), pKS208 and pKS212 (Heuer et al. 2002), pRSB222, pRSB223 (Schlüter and Sczcepanowski, unpublished), pWEC911 (Smalla and Hill, unpublished), pYS1 (Sota, unpublished), and pDS3 and pMBUI1 (Sen, Yano, Bauer, Rogers, Brown, and Top, unpublished). All plasmid sequences were determined at the DOE Joint Genome Institute (Walnut Creek, CA) by either of two methods. Pyrosequencing of plasmids pDS3, pMBUI1, pRSB222, and pRSB223 was performed on a GS FLX with the titanium-sequencing chemistry to approximately 90x coverage (Roche/454 Life Sciences, Branford, CT). Sequence data were assembled using the Newbler software (Roche/454 Life Sciences, Branford, CT). Plasmids pB1 and pB12, pC11, pEMT3, pG527, pKS208, pKS212, pNB1, pTB30, pWEC911, and pYS1 were sequenced using the Sanger method. Approximately 3-kb clone libraries were constructed for DNA sequencing of 384 clones, and sequences were determined for each plasmid in both directions. These sequences were assembled at JGI using PGA a platform for comparative genome assembly based on genetic algorithm optimization (Zhao et al. 2009). Any gap closure and polishing were done in house by primer walking. Automatic annotations were provided by the IGS Annotation Engine at the Institute for Genome Sciences, School of Medicine, University of Maryland (http://ae.igs.umaryland.edu/cgi/index.cgi) for plasmids pDS3, pMBUI1, pRSB222, and pRSB223 and by the J. Craig Venter Institute Annotation Service (http://www.jcvi.org/cms/research/ projects/annotation service) for the rest of the plasmids. These were followed by manual annotation by the authors. GenBank accession numbers are provided in table 1.
Nucleotide Sequence Alignments and Model Selection
The amino acid sequences of each gene were aligned with ClustalX (Thompson et al. 2002). Tranalign (Rice et al. 2000) was used to align the nucleotide sequences of each gene guided by the aligned amino acid sequences. Nexus formatted files were created from aligned nucleotide sequences for analyses in PAUP* (Swofford 2003) and MrBayes v 3.1.2 (Ronquist and Huelsenbeck 2003), and PHYLIP-formatted files were created for analyses in RAxML (Stamatakis 2006). For the concatenated tree, individual genes were aligned and concatenated, and the concatenated alignment was partitioned by codon positions. Model selection for maximum likelihood (ML) and Bayesian estimation were done with the program DT-ModSel (Minin et al. 2003). A list of models selected for each analysis is presented in table 2.
Table 2.
Nucleotide Substitution Modelsa Chosen for the 28 Genes.
| Genes | Models Selected for ML Analyses | Models Selected for Bayesian Analyses |
|---|---|---|
| Concatenated data | 1st codon: GTR + Γ | 1st codon: GTR + I + Γ |
| 2nd codon: GTR + Γ | 2nd codon: GTR + Γ | |
| 3rd codon: GTR + Γ | 3rd codon: GTR + I + Γ | |
| trfA2 | HKY + Γ | HKY + Γ |
| trbA | K81uf + Γ | GTR + Γ |
| trbB | TrN + Γ | GTR + Γ |
| trbC | HKY + Γ | HKY + Γ |
| trbD | HKY + I | HKY + I |
| trbF | TrN + Γ | GTR + Γ |
| trbG | TrN + I | GTR + I |
| trbI | TrN + I + Γ | GTR + I + Γ |
| trbJ | TrN + I + Γ | GTR + I + Γ |
| trbK | TrN + Γ | GTR + Γ |
| traD | TrN + I | GTR + I |
| traE | GTR + Γ | GTR + Γ |
| traF | HKY + Γ | HKY + Γ |
| traG | TIM + I + Γ | GTR + I + Γ |
| traH | GTR + Γ | GTR + Γ |
| traI | TrN + I + Γ | GTR + I + Γ |
| traJ | HKY + Γ | HKY + Γ |
| traK | TVM + Γ | GTR + Γ |
| traL | TVM + Γ | GTR + Γ |
| kfrC | HKY + Γ | HKY + Γ |
| kfrB | HKY + Γ | HKY + Γ |
| kfrA | TrN + Γ | GTR + Γ |
| korB | TrN + I + Γ | GTR + I + Γ |
| incC | HKY + Γ | HKY + Γ |
| korA | HKY + Γ | HKY + Γ |
| kleE | TVM + Γ | GTR + Γ |
| korC | TVM + Γ | GTR + Γ |
| klcA | HKY + I + Γ | HKY + I + Γ |
aHKY, variable base frequencies, different transition, and transversion rates; K81uf, variable base frequencies and three substitution rates; TrN, variable base frequencies, equal transversion rates, and variable transition rates; TVM, variable base frequencies, equal transition rates, and variable transversion rates; TIM, variable base frequencies, variable transition rates, and two transversion rates; GTR, variable base frequencies and six substitution rates; Γ, gamma distributed rate variation among sites; I, proportion of unchanging sites.
ML Analyses
For gene-tree estimation, iterative heuristic searches were performed using PAUP*, and the iterative approach was described by Sullivan et al. (2005). ML searches were carried out with tree bisection and reconnection branch swapping on 20 random starting trees generated by stepwise addition. For the concatenated data, an ML tree was inferred using the program RAxML (Stamatakis 2006) with the GTR + Γ model and parameters estimated separately for the three-codon partitions. Support values were estimated from 100 nonparametric bootstrap replicates (Felsenstein 1985). The tree with the highest likelihood was used as the ML estimate of the concatenated tree, referred to as MLconcat later.
Bayesian Posterior Probability Distributions
The program MrBayes v 3.1.2 (Ronquist and Huelsenbeck 2003) was used for estimating the posterior probability distributions of gene trees for each backbone gene and also for the concatenated data. A Markov Chain Monte Carlo algorithm was used to sample the posterior distribution of trees by running four chains for up to 8 million generations and sampling trees every 100 generations. Convergence of chains was assessed by plotting the standard deviation of split frequencies against the number of generations. A separate partitioned analysis was carried out for the concatenated data using GTR + I + Γ for the first and third codon positions and GTR + Γ for the second position. For both gene and concatenated data sets, trees sampled before convergence were discarded, and the remaining trees were used for Bayesian hypothesis testing.
Congruence Tests
Initial assessment of phylogenetic uncertainty as the cause of incongruence among gene trees was conducted using parametric bootstrap analyses (i.e., SOWH tests; Goldman et al. 2000). ML searches for each gene were conducted in PAUP* (Swofford 2003), as described earlier to provide an ML estimate of the gene tree, MLgene. Model parameters and branch lengths were reoptimized after exclusion of missing and ambiguous characters. ML searches were constrained to fit the topology of the concatenated tree to find the best fit of the individual gene data to the hypothesis that phylogenetic error is the only source of incongruence among the gene trees, MLhyp. The test statistic was the difference in log-likelihood scores of the two trees (δ = ln L[MLhyp] − ln L[MLgene]) the significance of which was evaluated under a frequentist framework by simulation. The constrained tree (MLhyp) was treated as the true tree on which 100 replicate data sets were simulated with SEQ-GEN (Rambaut and Grassly 1997) using the same model parameters that were optimized from the real data. The lengths of the simulated sequences were set to be identical to the length of each gene, and PAUP* was used to find the ML tree and the best tree constrained to fit the topology of the concatenated tree for every replicate. This provided the null distribution against which we compared the test statistic (δ) to evaluate the probability that phylogenetic uncertainty can explain the observed incongruence among the backbone gene trees.
Because the SOWH tests rely on point estimates of model parameters in simulation of the null distribution, we also assessed phylogenetic uncertainty with a Bayesian framework that marginalizes across uncertainty in model parameters. The tree filters option in PAUP* (Swofford 2003) was used to assess the proportion of trees in the posterior distribution of trees for each gene that was congruent with the topology of the concatenated ML tree, MLconcat. We also assessed the reciprocal congruence (i.e., proportion of trees in the posterior distribution of trees for the concatenated data consistent with the topology of each ML gene tree). This yielded the posterior probability that the incongruence is due to phylogenetic uncertainty.
Finally, we used the conservative nonparametric Shimodaira-Hasegawa test (Shimodaira and Hasegawa 1999) to assess the incongruence between each gene’s ML tree and the concatenated tree. To render this test conservative, we included each of the 28 single-gene ML trees and the concatenated tree in the centering step. We then calculated P values for each gene using RELL bootstrap with 1,000 replicates.
Species-Tree Estimation with *BEAST
To account for coalescent stochasticity, we applied the traditional notion of a species to IncP-1 subgroups identified by earlier studies. Thus, intragroup HGT can be treated as analogous to recombination within sexually reproducing species and intergroup HGT treated as analogous to introgressive hybridization. Nexus-formatted files of the 28 genes were used as input for *BEAST (Heled and Drummond 2010). Substitution models were chosen as earlier and were unlinked across genes with parameters estimated separately for each gene. Plasmids were assigned to the five subgroups (a proxy for species) based on previous studies. In BEAST (v 1.6.0), a Markov Chain Monte Carlo algorithm was used to sample the posterior distribution of trees by conducting five independent runs of 100 million generations each using a Yule prior for the species tree, a piecewise linear and constant root prior for population size, and uncorrelated, lognormal, relaxed clocks. Postburnin trees were combined with the program LogCombiner (BEAST v 1.6.0), and chains were assumed to converge when the average standard deviation of split frequencies was found to be <0.011. The maximum clade credibility tree with posterior probability of each node was computed with the program TreeAnnotator (BEAST v 1.6.0).
Detection of Recombinants
To detect recombination among the plasmids in the data set, alignment files of the backbone genes were concatenated in the order and orientation in which they appear on IncP-1 plasmids (fig. 1). The recombination detection programs RDP, GENECONV, BootScan, MaxChi, and Chimaera, which are implemented in RDP3 (Martin et al. 2010), were run with default parameters. Only recombinants that were identified by at least two programs were considered.
Results
Plasmids and Genes
To infer the evolutionary history of plasmids from the incompatibility group IncP-1, a set of 65 completely sequenced IncP-1 plasmid genomes was retrieved from Genbank and our own plasmid sequence collection. They were selected based on previously published assignment to one of the five major IncP-1 plasmid subgroups (α-ε) or our own comparative sequence analysis. A total of 28 backbone genes were found to be common to all plasmids and therefore included in this study (fig. 1, dark gray ORFs). After removing duplicates (plasmids with identical sets of backbone gene sequences), and including our 15 newly sequenced plasmids, a final set of 46 plasmids was obtained (table 1). For example, the backbone gene sequences of pJP4 were identical to those of pB10 and therefore not included. Visual inspection suggested that all alignments were of good quality. Because the 5′-ends of genes trfA1 and kfrC did not appear to be homologous between different subgroups, they were excluded from the analysis.
ML Analyses
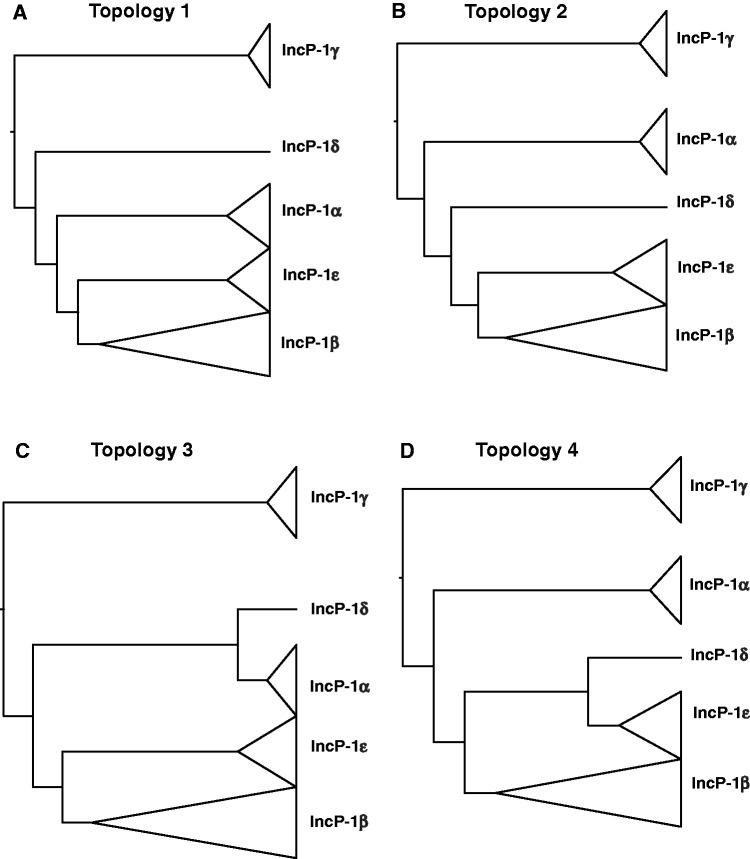
Gene trees were produced by separate ML analyses of the 28 backbone genes using the nucleotide substitution models in table 2 (supplementary fig. S1, Supplementary Material online). Analyses of 21 of those genes produced four topologies that were similar and differed only in the placement of the IncP-1δ and IncP-1α plasmids (fig. 2). The remaining seven gene trees were very different from each other and did not agree with any of the four common topologies (supplementary fig. S1, Supplementary Material online). Topology 1 (fig. 2A) was consistent with trees inferred from almost half of the genes (13 of 28; trfA2, trbA, trbC, trbG, traG, traH, traI, kfrC, kfrB, kfrA, korB, korA, and kleE). Topology 2 (fig. 2B) was found for three genes (trbD, trbK, and traJ) and differed from topology 1 in that it swapped the positions of the IncP-1δ plasmid pAKD4 and the IncP-1α plasmids. Topology 3 (fig. 2C) grouped pAKD4 with the IncP-1α plasmids and was inferred from the three genes trbF, trbI, and traE. Topology 4 (fig. 2D) grouped pAKD4 with the epsilon plasmids and was supported by the klcA and korC gene trees (fig. 2D). The genes corresponding to topologies 1–4 and the unique topologies (U) are also indicated on figure 1 (inside circle). Multiple topologies indicate incongruence among gene phylogenies.
Fig. 2.
Cladograms showing four topologies produced by 21 gene trees. (A) Topology 1: supported by 46% of gene trees, namely, those of trfA2, trbA, trbC, trbG, traG, traH, traI, kfrA, kfrB, kfrC, korB, korA, and kleE. (B) Topology 2: supported by gene trees of trbD, trbK, and traJ. (C) Topology 3: supported by gene trees of trbF, trbI, and traE. (D) Topology 4: supported by gene trees of korC and klcA. Trees were rooted using IncP-1γ as outgroup.
Examination of Congruence
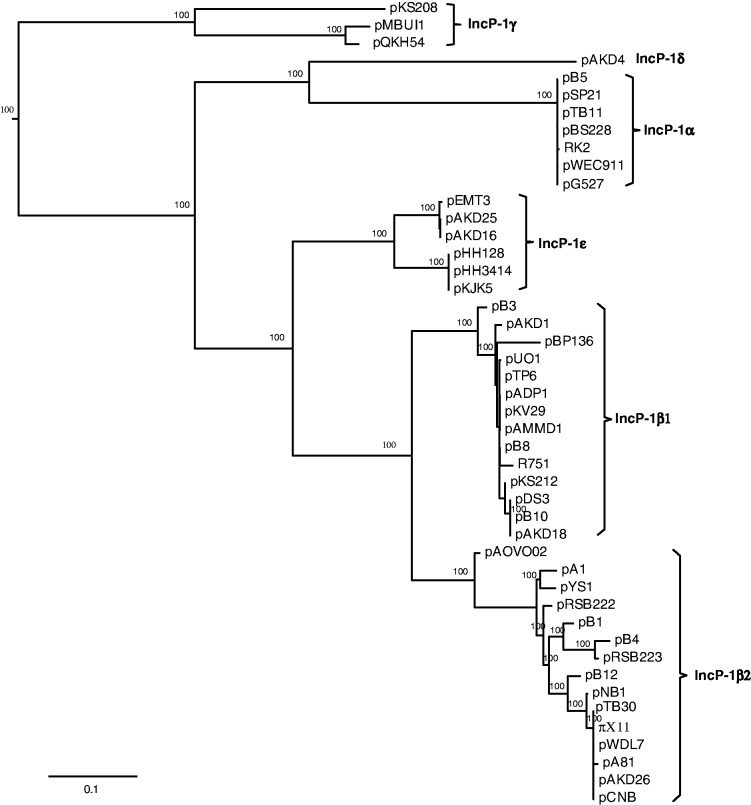
To test the hypothesis that the 28 backbone genes of IncP-1 plasmids have a single evolutionary history and that the incongruence described earlier is only due to phylogenetic uncertainty, each gene tree was compared statistically to the concatenated tree first using the parametric bootstrap. For the concatenated tree (shown in fig. 3), individual genes were aligned and concatenated in the order in which they appear on IncP-1 plasmids: trfA2, trbA, trbB, trbC, trbD, trbF, trbG, trbI, trbJ, trbK, traD, traE, traF, traG, traH, traI, traJ, traK, traL, kfrC, kfrB, kfrA, korB, incC, korA, kleE, korC, and klcA (fig. 1). The concatenated tree had the same topology as topology 3 described (fig. 2C). It represents the null hypothesis that all genes have a single history (i.e., all gene trees are estimates of a single-gene tree) as would be the case in the absence of recombination, either within or among groups. The observed test statistic was evaluated against the distribution of test statistics generated under the null hypothesis. The observed test statistics were significantly larger (P value < 0.01) than the null distributions generated for all 28 genes; in spite of being congruent with the concatenated tree at the deeper nodes, topology 3 still had significant differences at the terminal nodes. Therefore, the incongruence observed between each gene tree and the concatenated tree could not be attributed to phylogenetic uncertainty alone. Thus, the 28 backbone genes have multiple evolutionary histories (i.e., gene trees) because of horizontal transfer and/or coalescent stochasticity.
Fig. 3.
ML tree of concatenated data estimated from a partitioned analysis based on codon position. Nodal support is shown as nonparametric ML bootstrap values. The tree was rooted using IncP-1γ as outgroup.
We use the trbB gene to illustrate incongruence between a gene tree and the concatenated tree because the difference between these two trees was the largest of all, with a test statistic of 651 log-likelihood units (fig. 4A–C). After 14 plasmids were removed from the analysis (several IncP-1β plasmids, the IncP-1-δ plasmid, and all IncP-1ε plasmids, based on recombination detection—see later), the difference in the scores of the ML trees for trbB (MLtrbB) and the concatenated data (MLhyp) decreased to 0 (fig. 4D–F). The new test statistic fell within the null distribution, and the P value was calculated to be 0.49 (fig. 4D–F). Thus, the null hypothesis of discordance due to phylogenetic uncertainty could not be rejected for this analysis, which illustrates that the plasmids we removed were responsible for the incongruence between the concatenated tree and the gene tree observed for trbB.
Fig. 4.
Congruence test for trbB. (A) ML tree for trbB, MLtrbB, (B) ML tree constrained to fit concatenated tree, MLHyp, (C) null distribution and test statistic δ (ln L[MLhyp] − ln L[MLtrbB]) = 651.6. P value of obtaining a test statistic higher than 651.6 is <0.01. (D–F) Re-evaluated difference between MLtrbB and MLhyp after removing 14 plasmids (from top to bottom in panel A: two IncP-1β plasmids pB3 and pAOVO02; all six IncP-1ε plasmids pAKD16, pAKD25, pEMT3, pHH128, pHH3414, and pKJK5; the IncP-1δ plasmid pAKD4; and IncP-1β plasmids pB4, pB12, pA1, pRSB222, and pYS1).
In contrast to the frequentist approach used earlier, the Bayesian approach determines the conditional probability of the hypothesis that incongruence between a gene tree and the concatenated tree is due to phylogenetic uncertainty. MrBayes v 3.1.2. (Ronquist and Huelsenbeck 2003) was used to generate the posterior probability distribution of trees for each gene. The tree filter option in PAUP* (Swofford 2003) was then used to estimate the proportion of trees in the distribution that have the same topology as the concatenated tree, MLconcat. The fraction of trees retained in the filter represents the posterior probability of the hypothesis, and the probabilities of each gene tree being congruent with the concatenated tree can therefore be calculated. No trees were retained by the filtering procedure for any of the genes; therefore, the probability that incongruence between each of the gene trees and the concatenated tree is due to phylogenetic uncertainty approaches zero. Again, elimination of the same 14 plasmids resulted in 6,925 trees out of 7,419 trees in the posterior distribution for the concatenated data set that were consistent with the topology of the trbB tree (P value = 0.93). This agrees with the results from the parametric bootstrap analysis that incongruence was caused by the 14 plasmids. Overall, Bayesian hypothesis testing and parametric bootstrap analyses show that incongruence among gene trees is not due to phylogenetic uncertainty alone.
Not surprisingly, the results of the nonparametric SH tests are not as uniform (table 3). This test suggested that 14 of the 28 gene trees are not significantly different than the concatenated tree, but the other 14 are different. Thus, even our conservative implementation of this relatively low power test detected significant incongruence between the ML estimate of the gene tree and the concatenated tree for half of the backbone genes.
Table 3.
Results of SH Tests.
| Genes | Difference in -lnL | P |
|---|---|---|
| klcA | 409.04842 | 0.000* |
| kfrC | 40.94226 | 0.404 |
| kfrB | 63.47913 | 0.154 |
| kfrA | 127.54953 | 0.179 |
| incc1 | 237.69133 | 0.003* |
| trfA2 | 163.54237 | 0.023* |
| trbK | 39.36975 | 0.346 |
| trbJ | 535.70115 | 0.000* |
| trbI | 408.5485 | 0.002* |
| trbG | 230.63018 | 0.030* |
| trbF | 351.9579 | 0.000* |
| trbD | 37.1931 | 0.318 |
| trbC | 78.0097 | 0.141 |
| trbB | 700.09528 | 0.000* |
| trbA | 133.64327 | 0.014* |
| traL | 132.82945 | 0.051 |
| traK | 210.90414 | 0.006* |
| traJ | 31.30619 | 0.417 |
| traI | 162.64551 | 0.158 |
| traH | 52.51454 | 0.226 |
| traG | 159.0419 | 0.105 |
| traF | 87.27873 | 0.116 |
| traE | 254.17049 | 0.054 |
| traD | 196.59165 | 0.009* |
| korC | 340.93166 | 0.000* |
| korB | 113.80035 | 0.196 |
| kleE | 187.10069 | 0.000* |
| korA | 110.91598 | 0.002* |
Note.—Each gene tree was compared with the concatenated tree. *indicates significant incongruence.
Species Tree Estimation with *BEAST
*BEAST (Heled and Drummond 2010) was used for estimating the phylogenetic history represented by a species tree for IncP-1 plasmids. Figure 5A shows the maximum clade credibility trees estimated by *BEAST. The species trees are consistent with topology 1, which was found for almost half of the ML gene trees (13 genes out of 28, fig. 2A). This suggests that the incongruence detected in the parametric bootstrap and Bayesian tests above may largely be attributable to coalescent stochasticity (i.e., recombination within subgroups).
Fig. 5.

(A) Species tree estimated as maximum clade credibility tree by *BEAST. (B) Species tree estimated as maximum clade credibility tree by *BEAST after removal of recombinant gene traE. Nodal support is shown as posterior probabilities.
Effect of Recombination on Tree Estimation
To detect recombination between plasmids, individual genes were aligned and concatenated in the order and orientation in which they appear in IncP-1 plasmids (fig. 1, all dark gray ORFs, clockwise starting with trfA2). The concatenated alignment was analyzed using RDP, GENECONV, BootScan, MaxChi, and Chimaera, commonly used algorithms for detecting recombination among nucleotide sequences. Extensive recombination was detected within the IncP-1β subgroup, supporting the conclusion that incongruence is attributable to coalescent stochasticity. In contrast, only one instance of recombination between subgroups was detected from positions 7293 to 9902 of the concatenated alignment of the IncP-1α plasmids. This region corresponds to most of the traE and traD genes, specifically from nucleotide 26 of traE to 36 nucleotides from the end of traD (fig. 1; the genes are approximately 2,000 bp and 400 bp long, respectively). The recombination appears to have been between the ancestor of the IncP-1α plasmids and an IncP-1δ plasmid similar to pAKD4. Visual inspection of the aligned trbB genes also clearly showed a recombination event that included pAKD4, several of the IncP-1β plasmids, and the ancestor of the IncP-1ε plasmids, which would help explain the highly incongruent trbB gene tree described earlier. The amount of recombination among the IncP-1β plasmids may have masked this recombination event from the detection programs.
The low nodal support (posterior probability) in the species tree at the node uniting IncP-1α, -β, -δ, and -ε plasmids in figure 5A prompted us to exclude the long putative recombinant gene traE in species-tree estimation. After exclusion of traE, the same topology was obtained but with higher posterior probability at the relevant node (fig. 5B). Similarly, when we excluded traE and kept all other ML parameters constant, an MLconcat tree was obtained that was now congruent with topology 1 and no longer with topology 3 (data not shown). Topology 1 was the topology that was congruent with almost half of the gene trees and the species tree. These results support the conclusion that the traE gene has been involved in intergroup recombination.
Discussion
Our goal was to infer the evolutionary history of IncP-1 plasmids from their backbone genes in the presence of HGT both within and among subgroups. Recent studies have shown that these genes have evolved not only by acquiring mutations during vertical gene transfer but also by recombining with homologs on other IncP-1 plasmids (Schlüter et al. 2003; Sen et al. 2010; Norberg et al. 2011). Our challenge was therefore to infer the phylogeny of these plasmids in the presence of both vertical and horizontal inheritance. Our approach was to examine congruence among the inferred phylogenies of the backbone genes and determine the causes of incongruence, so that they could be accommodated in phylogeny estimation. To rule out phylogenetic uncertainty as one of the possibilities, we compared each gene tree to a tree obtained from concatenating alignments of all 28 backbone genes. Concatenation ignores multiple histories of the underlying data and represents a single history for all 28 backbone genes. The null hypothesis that each gene tree is consistent with that single history was rejected for all 28 genes; the observed incongruence could not be attributed to phylogenetic uncertainty alone, indicating the presence of coalescent stochasticity and/or intergroup HGT.
Assessing the impact of coalescent stochasticity in plasmid phylogeny is somewhat less straightforward, but we have applied species-tree estimation procedures that attempt to model stochastic lineage sorting. In these analyses, we have used existing plasmid “taxonomies” based on usually a single gene to group plasmid backbone genomes into putative taxa (subgroups). Within these subgroups, we have assumed that recombination behaves in a manner analogous to independent assortment in sexually reproducing species. We thus used *BEAST (Heled and Drummond 2010) to estimate the backbone “species” tree and accommodate the stochastic process of sorting ancestral polymorphisms. The output from *BEAST included a maximum clade credibility tree (fig. 5A) with the same topology as topology 1 (fig. 2A) and not topology 3 (fig. 2C) as consistent with the concatenated data set. However, the posterior probability of the node uniting the IncP-1α, IncP-1β, and IncP-1ε plasmids was moderate, 0.89 (fig. 5A). To address indirectly if this relatively low nodal probability was due to intergroup recombination, and therefore violation of the assumption that all incongruence is due to coalescent stochasticity, we identified the long traE gene of IncP-1α plasmids as a putative recombinant with an IncP-1δ pAKD4-like plasmid and excluded it from the species-tree estimation. This is analogous to eliminating putative hybrids from phylogenetic analysis. The topology of the species tree estimated in the absence of traE (fig. 5B) was identical to that produced before removing it but support for the node in question increased (fig. 5A). This suggests that inclusion of this putatively chimeric gene generated by intergroup HGT is the cause of reduced support. Recombination in this gene was also detected previously by Norberg et al. (2011) and Sen et al. (2010).
Plasmids of the IncP-1β subgroup, which can be further divided into IncP-1β1 and IncP-1β2 plasmids based on reciprocal monophyly (fig. 3), have undergone extensive recombination. It is important to note that these plasmids or the corresponding recombinant genes were not excluded from species-tree estimation because they all occurred within a subgroup (treated here as species) and were not expected to interfere with the analysis. Recombinants can be grouped into 1) recombination events within the IncP-1β2 subgroup (pB4, pNB1, pA1, pB1, pRSB222, pRSB223, and pYS1) and 2) recombination events between IncP-1β1 and IncP-1β2 plasmids (pAOVO02, pAKD18, pDS3, and pB10). Recombination in plasmid pAOVO02 and between pB10 and a pB4-like plasmid had been suggested earlier (Schlüter et al. 2003; Heuer et al. 2004; Norberg et al. 2011). Interestingly, there were fewer observations of recombination between members of the different subgroups. Except for recombination between the IncP-1α and IncP-1δ plasmid, few other instances were observed: those in the trbB gene and another between members of the IncP-1β1 subgroup and the IncP-1ε plasmid pEMT3 (data not shown). Recombination crossover points previously detected just upstream and downstream from trbB by Norberg et al. (2011) support our finding that the trbB region is prone to recombination. There are two possible explanations for the limited recombination between members of different subgroups; one is that as similarity between members of different subgroups decreases so does the possibility of recombination. The second is that because the other subgroups do not have as many sequenced plasmids as the IncP-1β subgroup, the genomes of putative recombinants have not yet been sequenced.
Our study provides yet another example of how concatenation may fail to produce accurate estimates of the species tree in complex data sets. Although almost half of the gene trees (13 out of 28) supported topology 1, the concatenated tree supported topology 3. Exclusion of traE from the concatenated data set and keeping all other ML tree estimation parameters constant, yielded a tree that was congruent with topology 1. Thus, the number or configuration of variable sites in the long traE gene may have been enough to dominate the phylogenetic signal in the other genes, a phenomenon called data swamping, such that one or a few partitions provide all the signal in a concatenated analysis (e.g., Edwards 2009).
Although the relationships among the subgroups are largely congruent among gene trees, caution must be exercised in choosing genes for inferring phylogenies. Genes such as trfA2, responsible for plasmid replication and traI, responsible for conjugative transfer are often used for inferring plasmid phylogenies. Their gene trees (supplementary fig. S1, Supplementary Material online) were largely in agreement with topology 1 (fig. 2A) and the species trees estimated by *BEAST (fig. 5), and therefore, these genes are recommended to infer the phylogenetics of IncP-1 plasmids. Other genes that would be suitable because they showed the same topology as the species tree are traG, traH, trbA, trbC, trbG, kfrA, kfrB, kfrC, korB, korA, and kleE. Previous studies have generally used these same genes, establishing in many cases the species tree defined here. Vedler et al. (2004) used individual trfA2, traG, and korA gene trees to clearly establish that the then newly defined IncP-1δ subgroup was separate from the IncP-1α subgroup. Similarly, three of the four genes chosen to infer the phylogeny of the then novel IncP-1ε group, trfA2, korB, and trbA, are part of this set of genes (Bahl et al. 2007). To define the InP-1γ subgroup, Haines et al. (2006) built a tree using five concatenated genes, korA, incC2, korB, korC, and kfrC; three of these (korA, korB, and kfrC) generated a tree with topology 1 in our study, whereas the korC tree had topology 4 and the incC tree a unique topology (fig. 2 and supplementary fig. S1, Supplementary Material online). Interestingly, their tree based on the five concatenated sequences is more similar to topology 4 than to topology 1, with the IncP-1δ and -β plasmids sharing a common ancestor rather than the IncP-1α and -β plasmids. Unfortunately, there were no IncP-1ε plasmids available at that time, so the effect of the five-gene concatenation on the topology with respect to that subgroup could not be evaluated. To infer the phylogeny of two novel IncP-1β2 catabolic plasmids, a tree was recently generated based on 24 concatenated backbone protein sequences, including TraE (Król et al. 2012); interestingly, its topology corresponded to topology 3 of the concatenated tree in this study. On the basis of our findings, topology 1 most closely represents the true evolutionary history of IncP-1 plasmids. Therefore, we recommend using the genes that generated trees of topology 1 (fig. 2).
To summarize, the backbones of IncP-1 plasmids have evolved by a combination of vertical and HGT, with the majority of recombination events being restricted to within the IncP-1β subgroup. Why recombination is seen so frequently in IncP-1β plasmids and why the traE gene of IncP-1α and IncP-1δ plasmids underwent recombination remains unknown. These recombination events may either be neutral or selectively advantageous to their hosts. In fact, recombination may be a tool that adds further flexibility to plasmids by allowing them to rapidly adapt to changing bacterial hosts and environmental conditions.
Supplementary Material
Supplementary figure S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank the following people for providing us with some of the plasmids that we sequenced and used in this study: N. Boon (pNB1 and pC11); W. Dejonghe (pTB30 and pWDL7); A. Schlüter and R. Szczepanowski (pB1, pB12, pKV29, pRSB222, and pRSB223); K. Smalla and H. Heuer (pG527, pKS208, pKS212, and pWEC911); and M. Sota (pYS1). We also appreciate the help of J. Król and H. Yano for finalizing, annotating, and submitting a few plasmid genome sequences (pNB1, pC11, pTB30, and pMBUI1, pKS208). New plasmid genome sequence data were generated with financial support from the National Science Foundation Grant EF-0627988. We are grateful to Kerrie Barry, Brian Foster, and Alla Lapidus at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI) for providing draft genome sequences, supported by the DOE Office of Science under Contract No. DE-AC02-05CH11231. We also acknowledge the National Institutes of Health NCRR COBRE grant P20RR16448 for an IBEST fellowship to D.S. and for support for the IBEST Computational Resources Core at the University of Idaho.
References
- Adamczyk M, Jagura-Burdzy G. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim Pol. 2003;50:425–453. [PubMed] [Google Scholar]
- Akiyama T, Asfahl KL, Savin MC. Broad-host-range plasmids in treated wastewater effluent and receiving streams. J Environ Qual. 2010;39:2211–2215. doi: 10.2134/jeq2010.0228. [DOI] [PubMed] [Google Scholar]
- Bahl MI, Hansen LH, Goesmann A, Sørensen SJ. The multiple antibiotic resistance IncP-1 plasmid pKJK5 isolated from a soil environment is phylogenetically divergent from members of the previously established α, β and δ sub-groups. Plasmid. 2007;58:31–43. doi: 10.1016/j.plasmid.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Binh CT, Heuer H, Kaupenjohann M, Smalla K. Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol Ecol. 2008;66:25–37. doi: 10.1111/j.1574-6941.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- Boon N, Goris J, De Vos P, Verstraete W, Top EM. Genetic diversity among 3-chloroaniline- and aniline-degrading strains of the Comamonadaceae. Appl Environ Microbiol. 2001;67:1107–1115. doi: 10.1128/AEM.67.3.1107-1115.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D. A classification of consensus methods for phylogenetics. BioConsensus. 2003:163–183. [Google Scholar]
- Chung Y, Ané C. Comparing two Bayesian methods of gene tree/species tree reconstructions: simulations with incomplete lineage sorting and horizontal gene transfer. Syst Biol. 2011;60:261–275. doi: 10.1093/sysbio/syr003. [DOI] [PubMed] [Google Scholar]
- Dejonghe W, Goris J, Dierickx A, De Dobbeleer V, Crul K, De Vos P, Verstraete W, Top E. Diversity of 3-chloroaniline and 3,4-dichloroaniline degrading bacteria isolated from three different soils and involvement of their plasmids in chloroaniline degradation. FEMS Microbiol Ecol. 2002;42:315–325. doi: 10.1111/j.1574-6941.2002.tb01021.x. [DOI] [PubMed] [Google Scholar]
- Dröge M, Pühler A, Selbitschka W. Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge. Mol Gen Genet. 2000;263:471–482. doi: 10.1007/s004380051191. [DOI] [PubMed] [Google Scholar]
- Edwards SV. Is a new and general theory of molecular systematics emerging? Evolution. 2009;63:1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fernandez-Lopez R, Garcillan-Barcia MP, Revilla C, Lazaro M, Vielva L, de la Cruz F. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol Rev. 2006;30:942–966. doi: 10.1111/j.1574-6976.2006.00042.x. [DOI] [PubMed] [Google Scholar]
- Goldman N, Anderson JP, Rodrigo AG. Likelihood-based tests of topologies in phylogenetics. Syst Biol. 2000;49:652–670. doi: 10.1080/106351500750049752. [DOI] [PubMed] [Google Scholar]
- Götz A, Pukall R, Smit E, Tietze E, Prager R, Tschäpe H, van Elsasa JD, Smalla K. Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl Environ Microbiol. 1996;62:2621–2628. doi: 10.1128/aem.62.7.2621-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybeal A. Is it better to add taxa or characters to a difficult phylogenetic problem? Syst Biol. 47. 1998:9–17. doi: 10.1080/106351598260996. [DOI] [PubMed] [Google Scholar]
- Gstalder ME, Faelen M, Mine N, Top EM, Mergeay M, Couturier M. Replication functions of new broad host range plasmids isolated from polluted soils. Res Microbiol. 2003;154:499–509. doi: 10.1016/S0923-2508(03)00143-8. [DOI] [PubMed] [Google Scholar]
- Haines AS, Akhtar P, Stephens ER, Jones K, Thomas CM, Perkins CD, Williams JR, Day MJ, Fry JC. Plasmids from freshwater environments capable of IncQ retrotransfer are diverse and include pQKH54, a new IncP-1 subgroup archetype. Microbiology. 2006;152:2689–2701. doi: 10.1099/mic.0.28941-0. [DOI] [PubMed] [Google Scholar]
- Heinemann JS, Sprague GFJ. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1990;340:205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer H, Krögerrecklenfort E, Wellington EM, et al. (11 co-authors) Gentamicin resistance genes in environmental bacteria: prevalence and transfer. FEMS Microbiol Ecol. 2002;42:289–302. doi: 10.1111/j.1574-6941.2002.tb01019.x. [DOI] [PubMed] [Google Scholar]
- Heuer H, Szczepanowski R, Schneiker S, Pühler A, Top EM, Schlüter A. The complete sequences of plasmids pB2 and pB3 provide evidence for a recent ancestor of the IncP-1ß group without any accessory genes. Microbiology. 2004;150:3591–3599. doi: 10.1099/mic.0.27304-0. [DOI] [PubMed] [Google Scholar]
- Ingram LC, Richmond MH, Sykes RB. Molecular characterization of the R factors implicated in the carbenicillin resistance of a sequence of Pseudomonas aeruginosa strains isolated from burns. Antimicrob Agents Chemother. 1973;3:279–288. doi: 10.1128/aac.3.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge AG. On total evidence: for the record. Cladistics. 2004;20:205–207. doi: 10.1111/j.1096-0031.2004.00020.x. [DOI] [PubMed] [Google Scholar]
- Kreps S, Ferino F, Mosrin C, Gerits J, Mergeay M, Thuriaux P. Conjugative transfer and autonomous replication of a promiscuous IncQ plasmid in the cyanobacterium Synechocystis PCC 6803. Mol Biol Genet. 1990;221:129–133. [Google Scholar]
- Król JE, Penrod JT, McCaslin H, Rogers LM, Yano H, Dejonghe W, Brown CJ, Parales RE, Wuertz S, Top EM. Genomic and functional analysis of the IncP-1β plasmids pNB8c and pWDL7::rfp explains their role in 3-chloroaniline catabolism. Appl Environ Microbiol. 2012;78:828–838. doi: 10.1128/AEM.07480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubatko LS, Carstens BC, Knowles LL. STEM: species tree estimation using maximum likelihood for gene trees under coalescence. Bioinformatics. 2009;25:971–973. doi: 10.1093/bioinformatics/btp079. [DOI] [PubMed] [Google Scholar]
- Liu L. BEST: Bayesian estimation of species trees under the coalescent model. Bioinformatics. 2008;24:2542–2543. doi: 10.1093/bioinformatics/btn484. [DOI] [PubMed] [Google Scholar]
- Liu L, Yu L, Kubatko L, Pearl DK, Edwards SV. Coalescent methods for estimating phylogenetic trees. Mol Phylogenet Evol. 2009;53:320–328. doi: 10.1016/j.ympev.2009.05.033. [DOI] [PubMed] [Google Scholar]
- Maddison W. Gene trees in species trees. Syst Biol. 1997;46:523–536. [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazodier P, Petter R, Thompson C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J Bacteriol. 1989;171:3583–3585. doi: 10.1128/jb.171.6.3583-3585.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin V, Abdo Z, Joyce P, Sullivan J. Performance-based selection of likelihood models for phylogeny estimation. Syst Biol. 2003;52:674–683. doi: 10.1080/10635150390235494. [DOI] [PubMed] [Google Scholar]
- Norberg P, Bergstrom M, Jethava V, Dubhashi D, Hermansson M. The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat Commun. 2011;2:268. doi: 10.1038/ncomms1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais A, Canton R, Valverde A, Machado E, Galan JC, Peixe L, Carattoli A, Baquero F, Coque TM. Dissemination and persistence of blaCTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-α, and IncFI groups. Antimicrob Agents Chemother. 2006;50:2741–2750. doi: 10.1128/AAC.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachulec E, van der Does C. Conjugative plasmids of Neisseria gonorrhoeae. PLoS One. 2010;5:e9962. doi: 10.1371/journal.pone.0009962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W, Lanka E, Barth PT, Figurski DH, Guiney DG, Haas D, Helinski DR, Schwab H, Stanisich VA, Thomas CM. Complete nucleotide sequence of Birmingham IncPα plasmids: compilation and comparative analysis. J Mol Biol. 1994;239:623–663. doi: 10.1006/jmbi.1994.1404. [DOI] [PubMed] [Google Scholar]
- Pollard DA, Iyer VN, Moses AM, Eisen MB. Widespread discordance of gene trees with species tree in Drosophila: evidence for incomplete lineage sorting. PLoS Genet. 2006;2:e173. doi: 10.1371/journal.pgen.0020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Grassly NC. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput Appl Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- Rhodes G, Parkhill J, Bird C, Ambrose K, Jones MC, Huys G, Swings J, Pickup RW. Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity. Appl Environ Microbiol. 2004;70:7497–7510. doi: 10.1128/AEM.70.12.7497-7510.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schlüter A, Heuer H, Szczepanowski R, Forney LJ, Thomas CM, Pühler A, Top EM. The 64,508 bp IncP-1β antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1β group. Microbiology. 2003;149:3139–3153. doi: 10.1099/mic.0.26570-0. [DOI] [PubMed] [Google Scholar]
- Schlüter A, Szczepanowski R, Pühler A, Top EM. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol Rev. 2007;31:449–477. doi: 10.1111/j.1574-6976.2007.00074.x. [DOI] [PubMed] [Google Scholar]
- Sen D, Van Der Auwera G, Rogers L, Thomas CM, Brown CJ, Top EM. Broad-host-range plasmids from agricultural soils have IncP-1 backbones with diverse accessory genes. Appl Environ Microbiol. 2011;77:7975–7983. doi: 10.1128/AEM.05439-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D, Yano H, Suzuki H, Krol JE, Rogers L, Brown CJ, Top EM. Comparative genomics of pAKD4, the prototype IncP-1δ plasmid with a complete backbone. Plasmid. 2010;63:98–107. doi: 10.1016/j.plasmid.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with application to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- Smalla K, Haines AS, Jones K, Krögerrecklenfort E, Heuer H, Schloter M, Thomas CM. Increased abundance of IncP-1β plasmids and mercury resistance genes in mercury-polluted river sediments: first discovery of IncP-1β plasmids with a complex mer transposon as the sole accessory element. Appl Environ Microbiol. 2006;72:7253–7259. doi: 10.1128/AEM.00922-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stolze Y, Eikmeyer F, Wibberg D, et al. (13 co-authors) IncP-1β plasmids of Comamonas sp. and Delftia sp. strains isolated from a wastewater treatment plant mediate resistance to and decolorization of the triphenylmethane dye crystal violet. Microbiology. 2012;158:2060–2072. doi: 10.1099/mic.0.059220-0. [DOI] [PubMed] [Google Scholar]
- Sullivan J, Abdo Z, Joyce P, Swofford DL. Evaluating the performance of a successive-approximations approach to parameter optimization in maximum-likelihood phylogeny estimation. Mol Biol Evol. 2005;22:1386–1392. doi: 10.1093/molbev/msi129. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Sunderland (MA): Sinauer Associates; 2003. PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4. [Google Scholar]
- Thomas CM, Smith CA. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Ann Rev Microbiol. 1987;41:77–101. doi: 10.1146/annurev.mi.41.100187.000453. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002 doi: 10.1002/0471250953.bi0203s00. Chapter 2:Unit 2.3. [DOI] [PubMed] [Google Scholar]
- Thorsted PB, Macartney DP, Akhtar P, et al. (12 co-authors) Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J Mol Biol. 1998;282:969–990. doi: 10.1006/jmbi.1998.2060. [DOI] [PubMed] [Google Scholar]
- Top EM, Holben WE, Forney LJ. Characterization of diverse 2,4-dichlorophenoxyacetic acid-degradative plasmids isolated from soil by complementation. Appl Environ Microbiol. 1995;61:1691–1698. doi: 10.1128/aem.61.5.1691-1698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Krol JE, Suzuki H, Foster B, Van Houdt R, Brown CJ, Mergeay M, Top EM. Plasmids captured in C. metallidurans CH34: defining the PromA family of broad-host-range plasmids. Antonie van Leeuwenhoek. 2009;96:193–204. doi: 10.1007/s10482-009-9316-9. [DOI] [PubMed] [Google Scholar]
- Vedler E, Vahter M, Heinaru A. The completely sequenced plasmid pEST4011 contains a novel IncP1 backbone and a catabolic transposon harboring tfd genes for 2,4-dichlorophenoxyacetic acid degradation. J Bacteriol. 2004;186:7161–7174. doi: 10.1128/JB.186.21.7161-7174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Goldman N, Friday A. Comparison of models for nucleotide substitution used in maximum-likelihood phylogenetic estimation. Mol Biol Evol. 1994;11:316–324. doi: 10.1093/oxfordjournals.molbev.a040112. [DOI] [PubMed] [Google Scholar]
- Zhao F, Hou H, Bao Q, Wu J. PGA4 genomics for comparative genome assembly based on genetic algorithm optimization. Genomics. 2009;94:284–286. doi: 10.1016/j.ygeno.2009.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.