Abstract
Association studies hold great promise for the elucidation of the genetic basis of diseases. Studies based on functional single nucleotide polymorphisms (SNPs) or on linkage disequilibrium (LD) represent two main types of designs. LD-based association studies can be comprehensive for common causative variants, but they perform poorly for rare alleles. Conversely, functional SNP-based studies are efficient because they focus on the SNPs with the highest a priori chance of being associated. Our poor ability to predict the functional effect of SNPs, however, hampers attempts to make these studies comprehensive. Recent progress in comparative genomics, and evidence that functional elements tend to lie in conserved regions, promises to change the landscape, permitting functional SNP association studies to be carried out that comprehensively assess common and rare alleles. SNP genotyping technologies are already sufficient for such studies, but studies will require continued genomic sequencing of multiple species, research on the functional role of conserved sequences and additional SNP discovery and validation efforts (including targeted SNP discovery to identify the rare alleles in functional regions). With these resources, we expect that comprehensive functional SNP association studies will soon be possible.
Keywords: functional SNPs, association studies, human disease
Introduction
Association studies of common, complexly inherited human diseases have the potential to provide us with insights into causes of enormous human suffering [1]. While thousands of such studies have been published (typically using single nucleotide polymorphisms [SNPs]), only a handful of these finding have been clearly and consistently replicated. While some findings are doubtless real, [2] debate continues over most. There are only a small number of genetic variants that have been clearly and consistently associated with a common disease, many of which are listed in Table 1.
Table 1.
Some clear, consistent common disease associations.
| Gene | Disease | Presumed causative variant |
Functional effect | Approximate frequency (in ethnic population of first positive study) |
Frequency information For other populationsa |
|---|---|---|---|---|---|
| PTPN22 | Rheumatoid arthritis [3-13] |
R620W | nsSNP | 9-10% (Caucasian)[3-13] | 0% in n = 1,600 Japanese; 0% in n = 60 Africans [3,14] |
| - | Type 1 diabetes [7,9,15-20] |
- | - | - | - |
|
CFH (factorH) |
Macular degeneration [21-26] |
Y402H | nsSNP | 30 40% (Caucasians)[21-26] |
Unknown |
|
FV (factor5) |
Deep venous thrombosis [27-34] |
R506W | nsSNP | 3 7% (Caucasians)[35] | 0% in n = 800 from Africa, South-East Asia, Australasia and the Americas (Native) [35,36] |
|
F2 (prothrombin) |
Deep venous thrombosis [34,37-40] |
G20210A | 3' utr mRNA cleavage site [41] |
1 3% (Caucasians)[42] | 0% in Asians;0% in Africans [36,40,42] |
|
CARD15 (NOD2) |
Crohn's disease [43-46] |
1007fs | Frame shift causing truncated protein |
~2% (Caucasians) | 0% in [Q3] n = 888 Asians; 0% in n = 640 Gambians [47-49] |
| - | - | R702W | nsSNP | ~4% (Caucasians) | < 0.1% in 888 Asians; 0% in 640 Gambians [47-49] |
| - | - | G908R | nsSNP | ~1% (Caucasians) | < 0.1% in n = 888 Asians; 0% in n = 640 Gambians [47-49] |
| - | - | Several very rare variants |
nsSNPs | < 1% (Caucasians) | Unknown |
| CHEK2 | Breast cancer [50-56] | 1100delC | Frame shift causing truncated protein |
0.5-1.5% (Caucasians)[50-56] | Unknown |
| APOE | Alzheimer's disease [57-60] |
C112R | nsSNP | ~15% (Caucasians)[57-61] | 25 40% in Africans; 8% in Asians [62-65] |
| KCNJ11 | Type 2 diabetes [66,67] | E23K | nsSNP | ~40% (Caucasians)[68] | Unknown |
| CCR5 | HIV infection [69-73] | Delta32 | Frame shift causing truncated protein | 8 10% (Caucasians)[69-73] | Absent in Africans and Asians; 2 5% in the Middle East, India, Europe [74] |
|
HLA- various genes |
Many autoimmune diseases |
Varied | Largely nsSNPs/haplotypes |
Varied | Varied; many show striking population frequency differences |
Abbreviations: nsSNP = non-synonymous single nucleotide polymorphisms; utr = untranslated regions.
a With the exception of HLA and APOE, none of the presumed causative variants have been shown to be present above 1 per cent in multiple major ethnic populations. While the 112R allele of APOE (which defines APOE*4 from the major allele, APOE*3) is seen in African, Asians and Caucasians, this variant is not associated with Alzheimer's disease in African populations [63].
Types of association studies
Researchers, typically, carefully weigh comprehensiveness and efficiency in designing an association study. A highly comprehensive study would assess every variant in the region(s) under study, regardless of type, location and allele frequency. A highly efficient study would be designed to reduce costs, including genotyping and/or multiple testing costs. Genotyping costs can be saved by determining which SNPs are in linkage disequilibrium (LD). For example, if you knew that two SNPs were in complete LD in the specific population of interest, you would only need to genotype one to assess them both. Multiple testing costs can be reduced by only looking at SNPs with a high a priori chance of being associated. Note that as multiple testing correction should account for the effective number of independent tests performed, genotyping only one of two SNPs in complete LD does not reduce multiple testing costs; if the SNPs are in complete LD, only one effective independent test is being performed, regardless of whether one or two SNPs are genotyped (Bonferroni correction is overly conservative). As 'per SNP' genotyping costs continue to fall, it seems likely that multiple testing costs will become the predominant concern in efficiency. Therefore, we discuss efficiency in terms of the a priori likelihood for an SNP to be associated with the phenotype studied.
Different types of large-scale association studies and the balance they strike are shown in Figure 1, although, obviously, many studies are hybrids of these types. These approaches, which have been applied to candidate genes, regions and recently to the whole genome, [21,76] are discussed in detail below, along with another technique (re-sequencing), which can currently only be applied on a small scale. Additional techniques that may be useful in 'special' populations, such as isolated founder and admixed populations, are discussed elsewhere [77-79].
Figure 1.
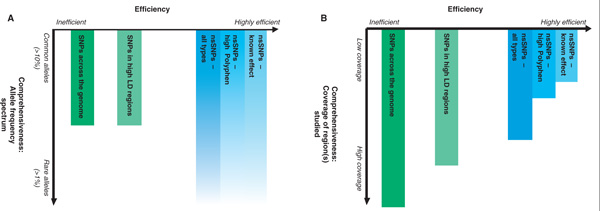
Association study approaches: Efficiency versus comprehensiveness. Studies vary in their efficiency (the a priori likelihood of a tested single nucleotide polymorphism [SNP] being associated with a disease), which has an impact on genotyping and multiple testing costs. Highly efficient designs (as defined by multiple testing costs) are shown on the right, with less efficient designs on the left. Studies also vary in comprehensiveness, both in terms of the allele frequency spectrum assessed (A) and the extent the region under study is assessed (B). Highly comprehensive studies extend from top to bottom. The efficiency (or comprehensiveness) for a specific study type relative to another in this figure is certainly not meant to be quantitative but merely indicative of the direction (bigger or smaller). This figure is applicable to large-scale studies of candidate genes, regions or the whole genome. Different functional SNP approaches are represented in blue, while non-functional approaches are represented in green. Re-sequencing is currently only feasible for examining one or a few candidate genes and is therefore not depicted. (A) Using linkage disequilibrium (LD) approaches, rare alleles are less likely to be tagged and hence the rare allele region is not covered. Since non-synonymous SNPs (nsSNPs) are assessed directly, association with rare alleles can be readily detected; however, this is limited by the availability of these SNPs. The light colour in the rare allele region is to indicate that coverage is dependent on SNP discovery. In this figure, we consider the most obvious functional SNPs, the nsSNPs. We presume the efficiency of the other functional categories may be significantly lower. (B) Typically, there is a trade-off between efficiency and comprehensiveness. One may limit the study to nsSNPs in order to have high efficiency at the cost of comprehensiveness. Further increase in the efficiency (and decrease in comprehensiveness) can be achieved by focusing only on nsSNPs predicted or known to have a functional consequence. Similarly, it has been proposed that a study utilising SNPs that tag the highest number of other SNPs (ie SNPs in high LD regions) would be more efficient (but less comprehensive) than a study aiming at LD coverage of the full genome [75].
Re-sequencing
When there is strong a priori evidence that a gene may be involved in a disease, it is possible to sequence that gene in cases and controls [43,80,81]. This requires no prior knowledge of variants in the region and allows researchers comprehensively to evaluate all variants in a gene, regardless of their allele frequency. Usually, it is necessary to group the very rare variants (< 1 per cent) for power considerations [43,80,81]. While this approach is now possible for one or a few candidate genes, it is by no means comprehensive across the genome and dramatic reductions in sequencing costs are necessary for its implementation on a large scale [82-84].
LD
Given the high rate of LD in the genome, many variants do not need to be directly genotyped in order to be assessed. They may instead be assessed by genotyping another SNP in high LD. The goal of LD-based ('tagging') approaches is to test a sufficient number of common SNPs so that SNPs that are not directly tested are assessed through their high correlation with the genotyped SNPs. This can create efficiency in genotyping but does not reduce multiple testing costs (as discussed previously, multiple testing corrections should account for the effective number of independent tests, rather than the number of SNPs genotyped). Additionally, the efficiency of the approach is modest, since there is a low a priori chance that a specific assessed SNP is associated with disease. By focusing only on regions with high LD (in which a single SNP is likely to tag several other SNPs), one improves the efficiency because there is an increased likelihood for any assessed SNP (ie for one test) to be tagging a functional SNP that is associated with the phenotype of interest [75].
Tagging allows most common SNPs to be comprehensively assessed in linkage regions, [85] candidate genes [86] or the whole genome [87]. Tagging, however, is not comprehensive in terms of allele frequencies because it tends to work poorly on rare polymorphisms [88-92]. Given the clear importance of rare polymorphisms (Box 1), this presents a substantial drawback. While some analytical work suggests that long haplotypes may be used to achieve a degree of 'tagging' of the rare allele, this comes with a dramatic multiple testing cost [106]. The adequate assessment of rare alleles requires direct interrogation.
Functional SNPs
Functional variants are the most likely to be associated with diseases (in fact, non-functional variants should only be associated secondary to LD); therefore, genotyping studies using only functional SNPs are relatively efficient. Since these variants are directly assessed, these studies are comprehensive in terms of allele frequency, covering rare and common variants present in the databases or discovered during focused SNP discovery. Our poor ability to predict functional SNPs, however, means that this approach is generally far from comprehensive in terms of coverage of the region under study. Nevertheless, by focusing on the most obvious classes of potentially functional SNPs, such as those causing non-synonymous changes in proteins, researchers have had notable successes with association studies in candidate genes [107] or linkage regions [3,22]. It is now possible to apply this method on a genome-wide scale, [75,108] which increases comprehensiveness with some reduction in efficiency.
Extending the (potentially) functional SNP approach
There are many attractive features of the functional SNP approach, including its efficiency and ability to assess rare and common alleles. Additionally, a positive association automatically provides a candidate causative polymorphism.
A major criticism of the functional approach is its lack of comprehensiveness, [96] and extending the coverage has been difficult, given our poor ability to predict functional SNPs. We can, however, broadly define functional SNPs as SNPs in any class predicted to have an above-average chance of having a functional effect. Recent progress in comparative genomics is likely to dramatically increase the comprehensiveness of this approach.
Below, we address some traditional functional elements (non-synonymous, splicing and promoter SNPs), as well as functional sequences emerging from the study of genome conservation.
Non-synonymous
The most obvious class of potentially functional SNPs is those causing non-synonymous changes in proteins (nsSNPs). Over 60 per cent of known Mendelian disease mutations and almost all the consistent, common disease mutations in Table 1 involve nsSNPs [109]. While there is a clear ascertainment bias for studying and confirming associations with nsSNPs, they are inarguably important in disease.
Additional evidence that many nsSNPs are functional and subject to selection comes from candidate gene sequencing studies, which find that 60 per cent of the expected number of nsSNPs are missing [110,111]. Furthermore, nsSNPs have lower minor allele frequencies than do synonymous SNPs [110,111]. When we examined all coding SNPs currently in the SNP database (dbSNP), we also found a dearth of nsSNPs; these are expected to comprise two-thirds of coding SNPs [111] but instead comprised less than one-half (20,463 nsSNP out of 42,387 coding SNPs). The deficiency of nsSNPs was even more notable when the analysis was limited to conserved coding regions in which only one-third of SNPs were non-synonymous (8,828 of 23,397). (SNP definitions were derived from the Ensembl database, and conserved regions were as defined previously [112].)
Large-scale studies of nsSNPs maintain high efficiency while allowing reasonable coverage [75]. One could choose to further increase efficiency (and decrease comprehensiveness) by limiting a study only to nsSNPs with a high predicted likelihood of being damaging. A substantial proportion of such SNPs have already been implicated in human disease [103,113].
Splicing
Perhaps the next most obvious class of potentially functional variants is SNPs around splice junctions. Mutations that affect splicing underlie 15 per cent of mutations in Mendelian diseases and hence are likely to play some role in common diseases [114].
Splicing is catalysed by weakly conserved 5' and 3' splice sites and a branch site, as well as exonic and intronic enhancers and silencers. Sites far from splice junctions can affect splicing, and a few mutations in these distant sites have been shown to cause human disease [115-120]. It appears, however, that most control of splicing lies in the 20 base pairs (bp) flanking each side of exon - intron boundaries [120]. These regions contain a high density of splicing enhancers (SEs), [120] have fewer SNPs than sequences further from splice junctions [120] and contain most of the known splicing mutations [114]. We find that these sequences are significantly conserved and have a relative dearth of SNPs (Table 2).
Table 2.
Conservation and relative single nucleotide polymorphism (SNP) density in different types of functional regions.
| Odds ratio ± standard error | Fold conservation | |
|---|---|---|
| Transcriptsa | 0.895 ± 0.003 | 12.0× |
| Transcripts: coding regions | 0.762 ± 0.004 | 16.4× |
| Transcripts: non-coding | 1.072 ± 0.004 | 6.2× |
| Conserved elementsb | 0.748 ± 0.002 | 23.5× |
| Promoterc | 0.995 ± 0.005 | 3.5× |
| Splice junctionsd | 0.780 ± 0.007 | 10.3× |
For each functional region, we report the odds ratio that a nucleotide in that region will be a variant by comparison with the rest of the genome (essentially, the relative SNP density) and standard error. The expected number is obtained using the validated SNP in the genome (4.9 M) and the total number of base pairs of the genome within a particular class of functional elements. A number less than 1 indicates a deficiency in SNP number. We also report the fold conservation (as defined previously [112]) compared with the genome average.
a Includes coding regions and untranslated regions (including RNA genes). All SNPs and the definitions of gene elements were obtained from the Ensembl database http://www.ensembl.org/.
b Defined previously [112] and obtained from the University of California, Santa Cruz website http://genome.ucsc.edu/.
c Within 500 base pairs (bp) upstream of the transcription start site.
d Within 20 bp of splice junctions.
Rather than testing all SNPs within the vicinity of a splice junction, one could increase efficiency by limiting the analysis to SNPs specifically predicted by computational models to affect splicing [121,122]. Conversely, one can increase comprehensiveness by assessing SEs beyond 20 bp of splice junctions. SEs are most prevalent in exons [123,124]. Some synonymous SNPs have also been shown to alter splicing [122]. Several programs are now available to predict SEs [125,126]. In addition to SNPs within 20 bp of the junction, the interrogation of synonymous SNPs predicted to disrupt SE activity [126] increases study comprehensiveness.
Promoters
Promoters are cis-elements that lie upstream of transcription start sites and are responsible for transcription initiation [127]. The existence of regulatory variants affecting transcription has long been established [128,129] and that have been shown to play a role in human disease [130,131].
Even though the exact promoter sequence may not be easily discerned, recent work has shown that the 500 bp upstream from the transcription start site is almost always able to function as a promoter [132]. Defining the promoter, however, requires determining the 5' end of transcripts, which is typically done experimentally and hence is laborious [133-135]. As shown in Table 2, conservation in the promoter sequences is threefold higher than expected.
In addition to promoters, numerous other cis-acting elements (for example enhancers) contribute to gene regulation. These elements have been more difficult to identify because they can lie within coding sequences, introns or as far as 1 megabase away [120,136,137]. Defining these elements is a main goal of the ENCODE project [138]. Genomic work aimed at identifying transcription factor binding sites and other regulatory sequences experimentally and informatically is ongoing, [87,139,140] and study of conserved sequences holds promise for the identification of these regions.
Conserved sequences
Computational efforts have consistently found that approximately 5 per cent of the human genome shows conservation with other species [112,141-148]. Although some regions may be conserved due to low mutation rates, clearly many, and perhaps most, of these regions are functionally important [149]. Indeed, most coding exons and many untranslated regions show interspecies conservation, although these only account for a minority of conserved regions. Conserved elements have been show to affect gene transcription levels, [150-156] RNA editing [112] and genome stability [157]. Additionally, conserved regions are enriched in intronic stretches surrounding alternatively spliced exons and have an excess of predicted secondary structure [112,143,158] and matrix-scaffold attachment regions [159]. Furthermore, they are enriched in stable gene deserts, which have been postulated to contain long range cis-regulatory regions [112]. Two lines of evidence suggest that many SNPs in conserved regions are subject to selection and, hence, are presumably functional: these regions contain a relative dearth of SNPs (Table 2), and the SNPs present there show a shift in allele frequency distribution towards rarer alleles [160,161].
The identification of conserved non-coding elements has generated a paradigm shift for the definition of functional elements. Without knowing the exact function of each element, sequences conserved across species define a map of likely functional regions in the genome and SNPs in the regions are candidates for functional SNP association studies.
The study of conserved regions is a vibrant field, with diverse methods of defining conservation and views on the correct number and types of species to compare. Some groups have focused on very large regions while others have examined conservation of regions as small as 4 bp [112,143,144]. Analyses can be performed using very closely related species (such as primates) or very distant species (such as a range of eukaryotes) [112,143,144]. The study of species that are moderately distant (< 75 million years) has yielded many of the conserved elements, [162] while study of primates has provided insight on primate-specific regulatory elements [146]. In addition to identifying conserved elements subject to purifying selection, comparative genomics has identified genes with evidence of positive selection [163,164]. Similar analyses may eventually be able to identify non-coding elements subjected to positive selection.
The proportion of functional elements that can be identified by comparative genomics is not yet clear. In a study using sequences from multiple yeast species, essentially all the known non-coding regulatory regions were identified as conserved [157]. Another study in yeast could identify conserved elements at the resolution of 6 bp transcription factor binding sites [165]. In mammals, using the currently available genomic sequences, most of the coding sequences and known regulatory sequences are conserved [166]. The analysis of more mammalian genome sequences will undoubtedly refine the current picture of conserved elements, although it is not clear that it will reach the same resolution achieved in yeast [162]. Nevertheless, it is likely that some functional sequences may not be identified through comparative genomics. If these SNPs do not fall into another obvious class of functional elements (like promoter regions), they may be missed by function-based association studies.
Generating a whole genome set of functional SNPs
The current feasibility of genome-wide function association studies depends upon the total number of functional SNPs and the extent to which such SNPs are represented in the databases. In the following discussion, we define functional SNPs as SNPs that fall into any of the above classes (ie non-synonymous, splicing, promoter, conserved [112]). Ongoing improvements in the definition of conserved regions may slightly change these estimates.
To estimate the total number of functional SNPs, we have utilised publicly available data from ENCODE regions. Ten regions (500 kilobases each) were re-sequenced in 48 unrelated individuals (16 Yoruba, 16 Centre D'Etude Du Polymorphisme Humain [CEPH], eight Han Chinese and eight Japanese). The SNPs in these regions, including those already present in the dbSNP and those newly discovered in sequencing, were then genotyped in the full 270 HapMap samples.
We first determined the total number of functional SNPs currently in dbSNP (using the above definitions). We then used the ENCODE regions to determine the allele frequency distribution (ie percentage rare and common) of conserved-region SNPs already in the dbSNP (ignoring those newly discovered by the ENCODE re-sequencing effort). We subsequently used information on the newly discovered ENCODE SNPs and our internal SNP discovery efforts to infer the percentage of SNPs missing from the dbSNP. This allowed us finally to estimate the total number of such SNPs. Implicit in this estimation is that the distribution of the allele frequency of functional SNPs is the same as the distribution of the subset of these SNPs that are in conserved elements (which account for over 75 per cent of the functional SNPs).
There are approximately 380,000 functional SNPs in dbSNP build 124. We infer from the ENCODE data that approximately 190,000 of these are common and 85,000 are rare (the remaining SNPs are very rare or database errors). Results were similar using data from both the CEPH and Yoruban samples. These results differ markedly from the expectations under the standard neutral model that there should be similar numbers of rare and common SNPs, suggesting that rare SNPs are missing in the dbSNP database [167]. Of the conserved region SNPs detected in the ENCODE Yoruban samples, the dbSNP database contained 23 per cent of the rare and 55 per cent of the common SNPs. Coverage was higher for conserved-region SNPs detected in the ENCODE CEPH samples, as the dbSNP database contained 35 per cent of the rare as well as 71 per cent of the common SNPs. Given that limited numbers of chromosomes typically are used for SNP discovery, both the dbSNP database and ENCODE are biased to miss rare SNPsa. The extent of this bias estimated using our internal SNP discovery efforts suggests that dbSNP coverage of rare SNPs is between approximately 25 per cent (in Caucasian) and approximately 15 per cent (in African).
From the above data, we estimate that there are approximately 350,000 common and 570,000 rare functional SNPs in the Yoruban samples and 270,000 common and 340,000 rare functional SNPs in the CEPH samples. Hence, a study that assayed only common functional SNPs would require a similar number of SNPs as an LD tagging study [161,168]. Even greater genotyping efficiency could be found by combining the approaches. Additionally, the number of rare functional SNPs is within the ability of new genotyping technologies [98,99,169].
Discussion
Association studies based on functional SNPs are highly efficient as they study the set of SNPs most likely to cause disease. In the past, these studies have been criticised as not being comprehensive due to our incomplete knowledge of the functional elements of the human genome. Research into conserved sequences and the continuing influx of genomic sequences into the public domain promises to delineate many of these elements and increase the comprehensiveness of functional SNP association studies. The use of functionalbased association studies can, in principle, adequately assess rare alleles, poor coverage of which is a major drawback for LD-based association studies.
It may be possible to improve the balance between the comprehensiveness and efficiency (defined in terms of multiple testing costs) of a functional SNP-based study by incorporating the a priori probability that an SNP is functional into the statistical tests used for analysis. For instance, one might set a less stringent p-value threshold for a nonsense SNP than for one in a putative promoter. Additionally, one might set a lower p-value threshold for an SNP that was in two functional categories rather than in a single functional category. For example, Table 3 indicates that SNP density (which over the whole genome probably reflects selection and, hence, functionality) is particularly low in coding regions that are also conserved or flank splice junctions.
Table 3.
SNP density per kilobase (kb) and counts in different types of functional regions.
| Transcriptsa | Coding regions | Conserved elementsb | Promoterc | Splice junctiond | |
|---|---|---|---|---|---|
| Transcripts | 1.46 ± 0.005e (87065) | ||||
| Coding regions | 1.24 ± 0.006 (42387) | 1.24 ± 0.006 (42387) | |||
| Conserved elements | 1.03 ± 0.006 (31339) | 0.98 ± 0.006 (23397) | 1.22 ± 0.003 (170256) | ||
| Promoter | 1.65 ± 0.038 (1854) | 1.38 ± 0.06 (533) | 1.03 ± 0.02 (2732) | 1.62 ± 0.01 (28463) | |
| Splice junctions | 1.11 ± 0.012 (8728) | 1.06 ± 0.012 (7519) | 1.07 ± 0.013 (7149) | 1.46 ± 0.086 (292) | 1.27 ± 0.009 (19225) |
The diagonal provides single nucleotide polymorphism (SNP) density for each region type and the off-diagonal provides density for regions of two types, either because one type is a subtype (coding is a subtype of transcript) or because of overlapping transcript definitions (a region may be in the promoter of one transcript, yet coding in another).
a Includes coding regions and untranslated regions (including RNA genes). All SNPs and the definitions of gene elements were obtained from the Ensembl database http://www.ensembl.org/.
b Defined previously [112] and obtained from the University of California, Santa Cruz website http://genome.ucsc.edu/.
c Within 500 base pairs (bp) upstream of the transcription start site.
d Within 20 bp of splice junctions.
e SNPs per kb ± standard error of the mean (total number of SNPs).
For comprehensive functional-based association studies to become practical, several goals need to be accomplished. First, the definition of functional elements needs to be refined through the availability of more genomic sequences. Secondly, SNP discovery efforts must be continued and expanded. Targeted re-sequencing in the functional regions may be necessary in order to compensate for bias against rare alleles in the databases, especially those that are population-specific and hence more likely to be functional [105]. The availability of extra sequencing capacity and efficient SNP discovery technologies can help to achieve this goal [170]. Thirdly, SNPs must be genotyped in the major ethnic populations to determine allele frequencies. HapMap now includes millions of SNPs, although these are biased to common SNPs [161]. Given the high-throughput genotyping technologies available, testing additional candidate functional SNPs to identify the common and rare SNPs can be readily performed. Indeed, we have recently undertaken the task of genotyping approximately 30,000 nsSNPs from the public databases to identify a set of approximately 20,000 that are polymorphic in at least one population [105].
With the availability of the functional elements and the SNPs, only approximately 270,000 - 350,000 SNPs must be genotyped to assess common functional SNPs in the genome. Furthermore, the genotyping of 300,000 - 500,000 additional SNPs will allow assessment of rare functional SNPs which have been implicated in many common diseases and are inadequately assessed by other approaches.
Box 1. Common variant/common disease versus rare variant/common disease
For the purposes of this review, we use the standard definition of a polymorphism as a variant whose minor allele frequency (MAF) is above 1 per cent, and define common alleles/polymorphisms as those with MAF > 10 per cent, rare alleles/polymorphisms as those with MAF 1 - 10 per cent and very rare alleles/variants as those with MAF < 1 per cent. In the past decade, there has been substantial debate over the importance of common alleles versus rare alleles (or even very rare variants) in common, complex human diseases. Theoretical work has been used to argue all points of view: that causative common disease alleles are most likely common alleles, or rare alleles, or very rare alleles [93-95].
One key argument for common alleles relies on the perceived greater practical difficulties in studying rare alleles rather than common alleles. First, analysis methods are particularly sensitive to genotyping errors of rare alleles and rare alleles have been particularly prone to genotyping errors [96,97]. Recent improvements in genotyping technologies, however, dramatically lessen these concerns [98,99]. Secondly, rare alleles are more likely to be population specific and therefore are more likely to generate spurious associations due to population substructure. Again, improvements, this time to analytical methods, allow us to detect and adjust for these artifacts [100,101]. Thirdly, it has been argued that the power to detect associations with rare alleles appears low when compared with that to detect common alleles. While this is certainly true if one assumes the same genotypic relative risk, this assumption is arbitrary, and if one instead uses another arbitrary assumption of equal population attributable risk, then the power to detect rare alleles would be significantly better than that for common alleles. Probably, a more reasonable approach is to consider a specific genetic effect size (eg defined by likelihood of the odds (LOD) score in sibling-pair analysis) of a locus and assume that causative alleles generate this specific effect size [102]. Given this assumption, the power to detect common and rare alleles is fairly similar (data not shown). Finally, rare alleles are difficult to 'tag' and therefore need to be assessed directly, creating two problems: alleles must be in databases in order to be assessed and genotyping all of the rare alleles in the genome would be at least an order of magnitude larger than contemplated for the linkage disequilibrium (LD)-based approach for common alleles. These concerns, while substantial, may be addressed by single nucleotide polymorphism (SNP) discovery and focusing genotyping efforts on rare SNPs that are also potentially functional.
One theoretical argument for rare alleles is that purifying selection should keep the frequency of deleterious functional alleles low. Indeed, in a study of approximately 30,000 non-synonymous SNPs, we confirmed previous observations that SNPs predicted by PolyPhen [103,104] to be damaging have significantly lower allele frequencies than SNPs predicted to be benign. This effect is largely due to an enrichment of damaging SNPs in the MAF < 10 per cent category [105].
Perhaps the strongest argument comes from an examination of Table 1, which indicates that both common and rare alleles are important. In light of these data, it is clearly essential for common disease association studies to investigate rare, as well as common, alleles.
End notes
a SNP discovery efforts interrogate a limited number of individuals and hence are more likely to find a common minor allele than a rare minor allele. For example, a study using only one individual (two chromosomes) has a 50 per cent chance of including both alleles of a 50 per cent allele frequency SNP, but only a 2 per cent chance of finding both alleles of a 1 per cent frequency SNP. Hence 1 per cent alleles are more likely to be missed in both dbSNP and the targeted re-sequencing than 10 per cent alleles. In addition, SNPs in dbSNP and those identified in this targeted re-sequencing effort are more biased to be more common in a different ethnic population where they may have been discovered. Indeed when studying alleles that are rare in the Caucasian population, we found the frequency in other populations to be higher for SNPs already in dbSNP than for SNPs identified through SNP discovery in the Caucasian population (MF unpublished results).
References
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Lohmueller KE, Pearce CL, Pike M. et al. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- Begovich AB, Carlton VEH, Honigberg LA. et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75:330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oene M, Wintle RF, Liu X. et al. Association of the lymphoid tyrosine phosphatase R620W variant with rheumatoid arthritis, but not Crohn's disease, in Canadian populations. Arthritis Rheum. 2005;52:1993–1998. doi: 10.1002/art.21123. [DOI] [PubMed] [Google Scholar]
- Simkins HM, Merriman ME, Highton J. et al. Association of the PTPN22 locus with rheumatoid arthritis in a New Zealand Caucasian cohort. Arthritis Rheum. 2005;52:2222–2225. doi: 10.1002/art.21126. [DOI] [PubMed] [Google Scholar]
- Hinks A, Barton A, John S. et al. Association between the PTPN22 gene and rheumatoid arthritis and juvenile idiopathic arthritis in a UK population: Further support that PTPN22 is an autoimmunity gene. Arthritis Rheum. 2005;52:1694–1699. doi: 10.1002/art.21049. [DOI] [PubMed] [Google Scholar]
- Zhernakova A, Eerligh P, Wijmenga C. et al. Differential association of the PTPN22 coding variant with autoimmune diseases in a Dutch population. Genes Immun. 2005;6:459–461. doi: 10.1038/sj.gene.6364220. [DOI] [PubMed] [Google Scholar]
- Viken MK, Amundsen SS, Kvien TK. et al. Association analysis of the 1858C > T polymorphism in the PTPN22 gene in juvenile idiopathic arthritis and other autoimmune diseases. Genes Immun. 2005;6:271–273. doi: 10.1038/sj.gene.6364178. [DOI] [PubMed] [Google Scholar]
- Criswell LA, Pfeiffer KA, Lum RF. et al. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: The PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76:561–571. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AT, Li W, Liew A. et al. The PTPN22 R620W polymorphism associates with RF positive rheumatoid arthritis in a dose-dependent manner but not with HLA-SE status. Genes Immun. 2005;6:129–133. doi: 10.1038/sj.gene.6364159. [DOI] [PubMed] [Google Scholar]
- Orozco G, Sanchez E, Gonzalez-Gay MA. et al. Association of a functional single-nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheum. 2005;52:219–224. doi: 10.1002/art.20771. [DOI] [PubMed] [Google Scholar]
- Steer S, Lad B, Grumley JA. et al. Association of R602W in a protein tyrosine phosphatase gene with a high risk of rheumatoid arthritis in a British population: Evidence for an early onset/disease severity effect. Arthritis Rheum. 2005;52:358–360. doi: 10.1002/art.20737. [DOI] [PubMed] [Google Scholar]
- Seldin MF, Shigeta R, Laiho K. et al. Finnish case-control and family studies support PTPN22 R620W polymorphism as a risk factor in rheumatoid arthritis, but suggest only minimal or no effect in juvenile idiopathic arthritis. Genes Immun. 2005;6:720–722. doi: 10.1038/sj.gene.6364255. [DOI] [PubMed] [Google Scholar]
- Mori M, Yamada R, Kobayashi K. et al. Ethnic differences in allele frequency of autoimmune-disease-associated SNPs. J Hum Genet. 2005;50:264–266. doi: 10.1007/s10038-005-0246-8. [DOI] [PubMed] [Google Scholar]
- Qu H, Tessier MC, Hudson TJ, Polychronakos C. Confirmation of the association of the R620W polymorphism in the protein tyrosine phosphatase PTPN22 with type 1 diabetes in a family based study. J Med Genet. 2005;42:266–270. doi: 10.1136/jmg.2004.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, She JX. Genetic association between a lymphoid tyrosine phosphatase (PTPN22) and type 1 diabetes. Diabetes. 2005;54:906–908. doi: 10.2337/diabetes.54.3.906. [DOI] [PubMed] [Google Scholar]
- Ladner MB, Bottini N, Valdes AM, Noble JA. Association of the single nucleotide polymorphism C1858T of the PTPN22 gene with type 1 diabetes. Hum Immunol. 2005;66:60–64. doi: 10.1016/j.humimm.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Onengut-Gumuscu S, Ewens KG, Spielman RS, Concannon P. A functional polymorphism (1858C/T) in the PTPN22 gene is linked and associated with type I diabetes in multiplex families. Genes Immun. 2004;5:678–680. doi: 10.1038/sj.gene.6364138. [DOI] [PubMed] [Google Scholar]
- Smyth D, Cooper JD, Collins JE. et al. Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes. 2004;53:3020–3023. doi: 10.2337/diabetes.53.11.3020. [DOI] [PubMed] [Google Scholar]
- Bottini N, Musumeci L, Alonso A. et al. A functional variant of lymphoid tyrosine phosphatase is associated with type 1 diabetes. Nat Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY. et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AO, Ritter III, Abel KJ. et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- Conley YP, Thalamuthu A, Jakobsdottir J. et al. Candidate gene analysis suggests a role for fatty acid biosynthesis and regulation of the complement system in the etiology of age-related maculopathy. Hum Mol Genet. 2005;14:1991–2002. doi: 10.1093/hmg/ddi204. [DOI] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV. et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S. et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- Zareparsi S, Branham KEH, Li M. et al. Strong association of the Y402H variant in complement factor H at 1q32 with susceptibility to age-related macular degeneration. Am J Hum Genet. 2005;77:149–153. doi: 10.1086/431426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertina RM, Koeleman BPC, Koster T. et al. Mutation in blood coagulation factor V associated with resistance to activated protein C. Nature. 1994;369:64–67. doi: 10.1038/369064a0. [DOI] [PubMed] [Google Scholar]
- Ridker PM, Hennekens CH, Lindpaintner K. et al. Mutation in the gene coding for coagulation factor V and the risk of myocardial infarction, stroke, and venous thrombosis in apparently healthy men. N Engl J Med. 1995;332:912–917. doi: 10.1056/NEJM199504063321403. [DOI] [PubMed] [Google Scholar]
- Zoller B, Dahlback B. Linkage between inherited resistance to activated protein C and factor V gene mutation in venous thrombosis. Lancet. 1994;343:1536–1538. doi: 10.1016/S0140-6736(94)92940-8. [DOI] [PubMed] [Google Scholar]
- Zoller B, Svensson PJ, He X, Dahlback B. Identification of the same factor V gene mutation in 47 out of 50 thrombosis-prone families with inherited resistance to activated protein C. J Clin Invest. 1994;94:2521–2524. doi: 10.1172/JCI117623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DD, Aboud MR, Williams BG, Isbister JP. Activated protein c resistance (APC) and inherited factor V (FV) mis-sense mutation in patients with venous and arterial thrombosis in a haematology clinic. Aust N Z J Med. 1995;25:151–154. doi: 10.1111/j.1445-5994.1995.tb02828.x. [DOI] [PubMed] [Google Scholar]
- Ridker PM, Miletich JP, Stampfer MJ. et al. Factor V Leiden and risks of recurrent idiopathic venous thromboembolism. Circulation. 1995;92:2800–2802. doi: 10.1161/01.CIR.92.10.2800. [DOI] [PubMed] [Google Scholar]
- Arruda VR, Annichino-Bizzacchi JM, Costa FF, Reitsma PH. Factor V Leiden (FVQ 506) is common in a Brazilian population. Am J Hematol. 1995;49:242–243. doi: 10.1002/ajh.2830490312. [DOI] [PubMed] [Google Scholar]
- Schobess R, Junker R, Auberger K. et al. Factor V G1691A and prothrombin G20210A in childhood spontaneous venous thrombosis -- Evidence of an age-dependent thrombotic onset in carriers of factor V G1691A and prothrombin G20210A mutation. Eur J Pediatr. 1999;158(Suppl 3):S105–S108. doi: 10.1007/pl00014335. [DOI] [PubMed] [Google Scholar]
- Rees DC, Cox M, Clegg JB. World distribution of factor V Leiden. Lancet. 1995;346:1133–1134. doi: 10.1016/S0140-6736(95)91803-5. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawasaki T, Fujimura H. et al. The prothrombin gene G20210A mutation is not found among Japanese patients with deep vein thrombosis and healthy individuals. Blood Coagul Fibrinolysis. 1998;9:451–452. doi: 10.1097/00001721-199807000-00011. [DOI] [PubMed] [Google Scholar]
- Cumming AM, Keeney S, Salden A. et al. The prothrombin gene G20210A variant: Prevalence in a UK anticoagulant clinic population. Br J Haematol. 1997;98:353–355. doi: 10.1046/j.1365-2141.1997.2353052.x. [DOI] [PubMed] [Google Scholar]
- Cattaneo M, Chantarangkul V, Taioli E. et al. The G20210A mutation of the prothrombin gene in patients with previous first episodes of deep-vein thrombosis: Prevalence and association with factor V G1691A, methylenetetrahydrofolate reductase C677T and plasma prothrombin levels. Thromb Res. 1999;93:1–8. doi: 10.1016/S0049-3848(98)00136-4. [DOI] [PubMed] [Google Scholar]
- Margaglione M, Brancaccio V, Giuliani N. et al. Increased risk for venous thrombosis in carriers of the prothrombin G - > A20210 gene variant. Ann Intern Med. 1998;129:89–93. doi: 10.7326/0003-4819-129-2-199807150-00003. [DOI] [PubMed] [Google Scholar]
- Poort SR, Rosendaal FR, Reitsma PH, Bertina RM. A common genetic variation in the 3'-untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood. 1996;88:3698–3703. [PubMed] [Google Scholar]
- Sachchithananthan M, Stasinopoulos SJ, Wilusz J, Medcalf RL. The relationship between the prothrombin upstream sequence element and the G20210A polymorphism: The influence of a competitive environment for mRNA 3'-end formation. Nucleic Acids Res. 2005;33:1010–1020. doi: 10.1093/nar/gki245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees DC, Chapman NH, Webster MT. et al. Born to clot: The European burden. Br J Haematol. 1999;105:564–566. doi: 10.1111/j.1365-2141.1999.01361.x. [DOI] [PubMed] [Google Scholar]
- Lesage S, Zouali H, Cezard JP. et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70:845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J, Cuthbert A, Croucher PJ. et al. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357:1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- Hugot JP, Chamaillard M, Zouali H. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- Kim TH, Rahman P, Jun JB. et al. Analysis of CARD15 polymorphisms in Korean patients with ankylosing spondylitis reveals absence of common variants seen in western populations. J Rheumatol. 2004;31:1959–1961. [PubMed] [Google Scholar]
- Yamazaki K, Takazoe M, Tanaka T. et al. Absence of mutation in the NOD2/CARD15 gene among 483 Japanese patients with Crohn's disease. J Hum Genet. 2002;47:469–472. doi: 10.1007/s100380200067. [DOI] [PubMed] [Google Scholar]
- Stockton JC, Howson JM, Awomoyi AA. et al. Polymorphism in NOD2, Crohn's disease, and susceptibility to pulmonary tuberculosis. FEMS Immunol Med Microbiol. 2004;41:157–160. doi: 10.1016/j.femsim.2004.02.004. [DOI] [PubMed] [Google Scholar]
- CHEK2 Breast Cancer Case-Control Consortium. CHEK2*1100delC and susceptibility to breast cancer: A collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004;74:1175–1182. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeks A, de Witte L, Nooijen A. et al. Excess risk for contralateral breast cancer in CHEK2*1100delC germline mutation carriers. Breast Cancer Res Treat. 2004;83:91–93. doi: 10.1023/B:BREA.0000010697.49896.03. [DOI] [PubMed] [Google Scholar]
- Cybulski C, Gorski B, Huzarski T. et al. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet. 2004;75:1131–1135. doi: 10.1086/426403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufault MR, Betz B, Wappenschmidt B. et al. Limited relevance of the CHEK2 gene in hereditary breast cancer. Int J Cancer. 2004;110:320–325. doi: 10.1002/ijc.20073. [DOI] [PubMed] [Google Scholar]
- Gorski B, Cybulski C, Huzarski T. et al. Breast cancer predisposing alleles in Poland. Breast Cancer Res Treat. 2005;92:19–24. doi: 10.1007/s10549-005-1409-1. [DOI] [PubMed] [Google Scholar]
- Meijers-Heijboer H, van den Ouweland A, Klijn J. et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- Vahteristo P, Bartkova J, Eerola H. et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002;71:432–438. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ. et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D. et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/WNL.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Mayeux R, Stern Y, Ottman R. et al. The apolipoprotein epsilon 4 allele in patients with Alzheimer's disease. Ann Neurol. 1993;34:752–754. doi: 10.1002/ana.410340527. [DOI] [PubMed] [Google Scholar]
- Anon. Apolipoprotein E genotype and Alzheimer's disease. Alzheimer's Disease Collaborative Group. Lancet. 1993;342:737–738. doi: 10.1016/0140-6736(93)91728-5. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Roses AD. Apolipoprotein E and Alzheimer disease. Proc Natl Acad Sci USA. 1995;92:4725–4727. doi: 10.1073/pnas.92.11.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo RM, Scacchi R. Apolipoprotein E (APOE) allele distribution in the world Is APOE*4 a "thrifty' allele?". Ann Hum Genet. 1999;63:301–310. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
- Sayi JG, Patel NB, Premkumar DR. et al. Apolipoprotein E polymorphism in elderly east Africans. East Afr Med J. 1997;74:668–670. [PubMed] [Google Scholar]
- Lane KA, Gao S, Hui SL. et al. Apolipoprotein E and mortality in African-Americans and Yoruba. J Alzheimers Dis. 2003;5:383–390. doi: 10.3233/jad-2003-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JH, Lo SK, Wen MS, Kao JT. Characterization of apolipoprotein E genetic variations in Taiwanese: Association with coronary heart disease and plasma lipid levels. Hum Biol. 2002;74:25–31. doi: 10.1353/hub.2002.0012. [DOI] [PubMed] [Google Scholar]
- Gloyn AL, Weedon MN, Owen KR. et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52:568–572. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- Laukkanen O, Pihlajamaki J, Lindstrom J. et al. Polymorphisms of the SUR1 (ABCC8) and Kir6.2 (KCNJ11) genes predict the conversion from impaired glucose tolerance to type 2 diabetes. The Finnish Diabetes Prevention Study. J Clin Endocrinol Metab. 2004;89:6286–6290. doi: 10.1210/jc.2004-1204. [DOI] [PubMed] [Google Scholar]
- McCarthy MI. Progress in defining the molecular basis of type 2 diabetes mellitus through susceptibility-gene identification. Hum Mol Genet. 2004;13:R33–R41. doi: 10.1093/hmg/ddh057. [DOI] [PubMed] [Google Scholar]
- Dean M, Carrington M, Winkler C. et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- Huang Y, Paxton WA, Wolinsky SM. et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- Liu R, Paxton WA, Choe S. et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/S0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- Samson M, Libert F, Doranz BJ. et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- Zimmerman PA, Buckler-White A, Alkhatib G. et al. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: Studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]
- Martinson JJ, Chapman NH, Rees DC. et al. Global distribution of the CCR5 gene 32-basepair deletion. Nat Genet. 1997;16:100–103. doi: 10.1038/ng0597-100. [DOI] [PubMed] [Google Scholar]
- Shiffman D, Ellis SG, Rowland CM. et al. Identification of four gene variants associated with myocardial infarction. Am J Hum Genet. 2005;77:596–605. doi: 10.1086/491674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, O'Brien SJ. Mapping by admixture linkage disequilibrium: Advances, limitations and guidelines. Nat Rev Genet. 2005;6:623–632. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Ghosh D, Nichols TE. Linkage disequilibrium: Ancient history drives the new genetics. Hum Hered. 2005;59:118–124. doi: 10.1159/000085226. [DOI] [PubMed] [Google Scholar]
- Halder I, Shriver MD. Measuring and using admixture to study the genetics of complex diseases. Hum Genomics. 2003;1:52–62. doi: 10.1186/1479-7364-1-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisse C, Clement K, Durand E. et al. Melanocortin-4 receptor mutations are a frequent and heterogeneous cause of morbid obesity. J Clin Invest. 2000;106:253–262. doi: 10.1172/JCI9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A. et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman E. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faham M, Zheng J, Moorhead M. et al. Multiplexed variation scanning for 1,000 amplicons in hundreds of patients using mismatch repair detection (MRD) on tag arrays. Proc Natl Acad Sci USA. 2005;102:14717–14722. doi: 10.1073/pnas.0506677102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J. et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe'er I. et al. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Van Eerdewegh P, Little RD, Dupuis J. et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–430. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- Saleh M, Vaillancourt JP, Graham RK. et al. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature. 2004;429:75–79. doi: 10.1038/nature02451. [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Qu C. et al. Direct isolation and identification of promoters in the human genome. Genome Res. 2005;15:830–839. doi: 10.1101/gr.3430605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadi KR, Weale ME, Xue ZY. et al. A single-nucleotide polymorphism tagging set for human drug metabolism and transport. Nat Genet. 2005;37:84–89. doi: 10.1038/ng1488. [DOI] [PubMed] [Google Scholar]
- Evans DM, Cardon LR, Morris AP. Genotype prediction using a dense map of SNPs. Genet Epidemiol. 2004;27:375–384. doi: 10.1002/gepi.20045. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ. et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Schrodi SJ, Ross DA, Cargill M. Selecting tagging SNPs for association studies using power calculations from genotype data. Hum Hered. 2004;57:156–170. doi: 10.1159/000079246. [DOI] [PubMed] [Google Scholar]
- Ke X, Durrant C, Morris AP. et al. Efficiency and consistency of haplotype tagging of dense SNP maps in multiple samples. Hum Mol Genet. 2004;13:2557–2565. doi: 10.1093/hmg/ddh294. [DOI] [PubMed] [Google Scholar]
- Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/S0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Cox NJ. The allelic architecture of human disease genes: Common disease-common variant... or not? Hum Mol Genet. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- Gordon D, Finch SJ, Nothnagel M, Ott J. Power and sample size calculations for case-control genetic association tests when errors are present: Application to single nucleotide polymorphisms. Hum Hered. 2002;54:22–33. doi: 10.1159/000066696. [DOI] [PubMed] [Google Scholar]
- Fan JB, Oliphant A, Shen R. et al. Highly parallel SNP genotyping. Cold Spring Harb Symp Quant Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- Hardenbol P, Yu F, Belmont J. et al. Highly multiplexed molecular inversion probe genotyping: Over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005;15:269–275. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Goldstein DB. Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol. 2001;20:4–16. doi: 10.1002/1098-2272(200101)20:1<4::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HB, Faham M. Evidence and implications for multiplicative interactions among loci predisposing to human common disease. Hum Hered. 2005;59:176–184. doi: 10.1159/000086118. [DOI] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I. et al. Prediction of deleterious human allele. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: Server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland J, Carlton VE, Falkowski M. et al. Large-scale characterization of public database SNPs causing non-synonymous changes in three ethnic groups. Hum Genet. 2006;119:75–83. doi: 10.1007/s00439-005-0105-x. [DOI] [PubMed] [Google Scholar]
- Lin S, Chakravarti A, Cutler DJ. Exhaustive allelic transmission disequilibrium tests as a new approach to genome-wide association studies. Nat Genet. 2004;36:1181–1188. doi: 10.1038/ng1457. [DOI] [PubMed] [Google Scholar]
- Altshuler D, Hirschhorn JN, Klannemark M. et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- Haga H, Yamada R, Ohnishi Y. et al. Gene-based SNP discovery as part of the Japanese Millennium Genome Project 2002. Identification of 190,562 genetic variations in the human genome. J Hum Genet. 2002;47:605–610. doi: 10.1007/s100380200092. [DOI] [PubMed] [Google Scholar]
- Botstein D, Risch N. Discovering genotypes underlying human phenotypes: Past successes for Mendelian disease, future approaches for complex disease. Nat Genet. 2003;33:228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- Halushka MK, Fan J-B, Bentley K. et al. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J. et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DC, Akey DT, Nickerson DA. The patterns of natural variation in human genes. Annu Rev Genomics Hum Genet. 2005;6:287–312. doi: 10.1146/annurev.genom.6.080604.162309. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: Causes and consequences. Hum Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- Treisman R, Orkin SH, Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983;302:591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Mitchell GA, Labuda D, Fontaine G. et al. Splice-mediated insertion of an Alu sequence inactivates ornithine delta-aminotransferase: A role for Alu elements in human mutation. Proc Natl Acad Sci USA. 1991;88:815–819. doi: 10.1073/pnas.88.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani F, Buratti E, Stuani C. et al. A new type of mutation causes a splicing defect in ATM. Nat Genet. 2002;30:426–429. doi: 10.1038/ng858. [DOI] [PubMed] [Google Scholar]
- Min GL, Martiat P, Pu GA, Goldman J. Use of pulsed field gel electrophoresis to characterize BCR gene involvement in CML patients lacking M-BCR rearrangement. Leukemia. 1990;4:650–656. [PubMed] [Google Scholar]
- Zhang XH, Leslie CS, Chasin LA. Dichotomous splicing signals in exon flanks. Genome Res. 2005;15:768–779. doi: 10.1101/gr.3217705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Holste D, Burge CB, Sharp PA. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2004;2:E268. doi: 10.1371/journal.pbio.0020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P, Shapiro MB, Harris NL. Splice junctions, branch point sites, and exons: Sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal TD, Maniatis T. Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol Cell Biol. 1999;19:261–273. doi: 10.1128/mcb.19.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1113. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Callahan III, Balbinder E. Tryptophan operon: Structural gene mutation creating a 'promoter' and leading to 5-methyltryptophan dependence. Science. 1970;168:1586–1589. doi: 10.1126/science.168.3939.1586. [DOI] [PubMed] [Google Scholar]
- Roberts JW. Promoter mutation in vitro. Nature. 1969;223:480–482. doi: 10.1038/223480a0. [DOI] [PubMed] [Google Scholar]
- Kulozik AE, Bellan-Koch A, Bail S. et al. Thalassemia intermedia: Moderate reduction of beta globin gene transcriptional activity by a novel mutation of the proximal CACCC promoter element. Blood. 1991;77:2054–2058. [PubMed] [Google Scholar]
- Bosma PJ, Chowdhury JR, Bakkerm C. et al. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N Engl J Med. 1995;333:1171–1175. doi: 10.1056/NEJM199511023331802. [DOI] [PubMed] [Google Scholar]
- Trinklein ND, Aldred SJ, Saldanha AJ, Myers RM. Identification and functional analysis of human transcriptional promoters. Genome Res. 2003;13:308–312. doi: 10.1101/gr.794803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi T, Itoh T, Suzuki Y. et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Yamashita R, Sugano S, Nakai K. DBTSS, DataBase of Transcriptional Start Sites: Progress report 2004. Nucleic Acids Res. 2004;32:D78–D81. doi: 10.1093/nar/gkh076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Yamashita R, Shirota M. et al. Large-scale collection and characterization of promoters of human and mouse genes. In Silico Biol. 2004;4:429–444. [PubMed] [Google Scholar]
- Rodriguez-Jato S, Nicholls RD, Driscoll DJ, Yang TP. Characterization of cis- and trans-acting elements in the imprinted human SNURF-SNRPN locus. Nucleic Acids Res. 2005;33:4740–4753. doi: 10.1093/nar/gki786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA. et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyl. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004. pp. 636–640. [DOI] [PubMed]
- Kolbe D, Taylor J, Elnitski L. et al. Regulatory potential scores from genome-wide three-way alignments of human, mouse, and rat. Genome Res. 2004;14:700–707. doi: 10.1101/gr.1976004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L, Hardison RC, Li J. et al. Distinguishing regulatory DNA from neutral sites. Genome Res. 2003;13:64–72. doi: 10.1101/gr.817703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK. et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis ET, Reymond A, Lyle R. et al. Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature. 2002;420:578–582. doi: 10.1038/nature01251. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G. et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis ET, Reymond A, Antonarakis SE. Conserved non-genic sequences -- An unexpected feature of mammalian genomes. Nat Rev Genet. 2005;6:151–157. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- Margulies EH, Blanchette M, Haussler D, Green ED. Identification and characterization of multi-species conserved sequences. Genome Res. 2003;13:2507–2518. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli D, McAuliffe J, Ovcharenko D. et al. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Tao H, Osoegawa K. et al. Noncoding sequences conserved in a limited number of mammals in the SIM2 interval are frequently functional. Genome Res. 2004;14:367–372. doi: 10.1101/gr.1961204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Rubin EM. Genomic strategies to identify mammalian regulatory sequences. Nat Rev Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- Hardison RC. Comparative genomics. PLoS Biol. 2003;1:E58. doi: 10.1371/journal.pbio.0000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culi J, Modolell J. Proneural gene self-stimulation in neural precursors: An essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 1998;12:2036–2047. doi: 10.1101/gad.12.13.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renucci A, Zappavigna V, Zàkàny J. et al. Comparison of mouse and human HOX-4 complexes defines conserved sequences involved in the regulation of Hox-4.4. EMBO J. 1992;11:1459–1468. doi: 10.1002/j.1460-2075.1992.tb05190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM. et al. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Poulin F, Nobrega MA, Plajzer-Frick I. et al. In vivo characterization of a vertebrate ultraconserved enhancer. Genomics. 2005;85:774–781. doi: 10.1016/j.ygeno.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Kimura-Yoshida C, Kitajima K, Oda-Ishii I. et al. Characterization of the pufferfish Otx2 cis-regulators reveals evolutionarily conserved genetic mechanisms for vertebrate head specification. Development. 2004;131:57–71. doi: 10.1242/dev.00877. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Takemoto T, Kamachi Y, Kondoh H. Efficient identification of regulatory sequences in the chicken genome by a powerful combination of embryo electroporation and genome comparison. Mech Dev. 2004;121:1145–1158. doi: 10.1016/j.mod.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Ganley AR, Hayashi K, Horiuchi T, Kobayashi T. Identifying gene-independent noncoding functional elements in the yeast ribosomal DNA by phylogenetic footprinting. Proc Natl Acad Sci USA. 2005;102:11787–11792. doi: 10.1073/pnas.0504905102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ. et al. Systematic discovery of regulatory motifs in human promoters and 3'UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazko GV, Koonin EV, Rogozin IB, Shabalina SA. A significant fraction of conserved noncoding DNA in human and mouse consists of predicted matrix attachment regions. Trends Genet. 2003;19:119–124. doi: 10.1016/S0168-9525(03)00016-7. [DOI] [PubMed] [Google Scholar]
- Drake JA, Bird C, Nemesh J. et al. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat Genet. 2006;38:223–227. doi: 10.1038/ng1710. [DOI] [PubMed] [Google Scholar]
- Altshuler D, Brooks LD, Chakravarti A. et al. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli D, Nobrega MA, Rubin EM. Comparative genomics at the vertebrate extremes. Nat Rev Genet. 2004;5:456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R. et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Bustamante CD, Lancet D, Paabo S. Natural selection on the olfactory receptor gene family in humans and chimpanzees. Am J Hum Genet. 2003;73:489–501. doi: 10.1086/378132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M. et al. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML. et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- The International Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki H, Dong S, Loi H. et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- Fakhrai-Rad H, Zheng J, Willis TD. et al. SNP discovery in pooled samples with mismatch repair detection. Genome Res. 2004;14:1404–1412. doi: 10.1101/gr.2373904. [DOI] [PMC free article] [PubMed] [Google Scholar]


