Abstract
To investigate the possible role of cpb2-positive type A Clostridium perfringens in neonatal diarrheal illness in pigs, the jejunum and colon of matched normal and diarrheic piglets from 10 farms with a history of neonatal diarrhea were examined grossly and by histopathology, and tested for C. perfringens, for C. perfringens beta2 (CPB2) toxin, as well as for Clostridium difficile toxins, Salmonella, enterotoxigenic Escherichia coli, rotavirus, transmissible gastroenteritis (TGE) virus, and coccidia. Clostridium perfringens isolates were tested using a multiplex real-time polymerase chain reaction (PCR) to determine the presence of cpa, consensus and atypical cpb2, and other virulence-associated genes. The numbers of C. perfringens in the intestinal contents were lower in diarrheic piglets (log10 5.4 CFU/g) compared with normal piglets (log10 6.5 CFU/g) (P < 0.05). The consensus cpb2 was present in 93% of isolates in each group, but atypical cpb2 was less common (56% healthy, 32% diarrheic piglets isolates, respectively, P < 0.05). The presence of CPB2 toxin in the intestinal contents of normal and diarrheic piglets did not differ significantly. Clostridium difficile toxins and rotavirus were each detected in 7 of the 21 (33%) diarrheic piglets. Rotavirus, C. difficile toxins, Salmonella, or enterotoxigenic E. coli were concurrently recovered in different combinations in 4 diarrheic piglets. The cause of diarrhea in 8 of the 21 (38%) piglets on 6 farms remained unknown. The etiological diagnosis of diarrhea could not be determined in any of the piglets on 2 of the farms. This study demonstrated that the number of cpb2-positive type A C. perfringens in the intestinal contents was not a useful approach for making a diagnosis of type A C. perfringens enteritis in piglets. Further work is required to confirm whether cpb2-carrying type A C. perfringens have a pathogenic role in enteric infection in neonatal swine.
Résumé
Dans le but d’étudier le rôle possible de Clostridium perfringens type A possédant le gène cpb2 dans les cas de diarrhée néonatale chez les porcs, le jéjunum et le côlon de porcelets provenant de 10 fermes avec une histoire de diarrhée néonatale et pairés en fonction qu’ils aient ou non de la diarrhée ont été examinés macroscopiquement et en histopathologie, et testés pour C. perfringens, la toxine bêta2 de C. perfringens (CBP2), ainsi que pour les toxines de Clostridium difficile, Salmonella, Escherichia coli entérotoxinogène, rotavirus, le virus de la gastro-entérite transmissible (TGE) et les coccidies. Les isolats de C. perfringens ont été testés par réaction d’amplification en chaîne par la polymérase (PCR) multiplex pour déterminer la présence de cpa, de cpb2 consensus et atypiques, ainsi que d’autres gènes associés à la virulence. Le nombre de C. perfringens dans le contenu intestinal des porcelets diarrhéiques étaient plus faible (log10 5,4 UFC/g) que dans celui des porcelets en santé (log10 6,5 UFC/g) (P < 0,05). Le cpb2 consensus était présente chez 93 % des isolats dans chaque groupe, mais le cpb2 atypique était moins fréquent (56 % des isolats de porcelets en santé, et 32 % des isolats provenant de porcelets diarrhéiques, respectivement, P < 0,05). La présence de la toxine CPB2 dans le contenu intestinal de porcelets avec ou sans diarrhée ne différait pas de manière significative. Les toxines de C. difficile et les rotavirus ont chacun été détectés chez 7 des 21 (33 %) des porcelets diarrhéiques. Des rotavirus, les toxines de C. difficile, Salmonella ou des E. coli enterotoxinogènes ont été retrouvés de manière concomitante en différentes combinaisons chez 4 porcelets diarrhéiques. Chez 8 de 21 (38 %) porcelets provenant de 6 fermes, la cause de la diarrhée est demeurée inconnue. Le diagnostic étiologique de la diarrhée n’a pu être déterminé chez aucun des porcelets de 2 fermes. Cette étude démontre que le nombre d’isolats de C. perfringens de type A positifs pour cpb2 dans le contenu intestinal n’était pas une approche utile pour établir un diagnostic d’entérite à C. perfringens type A chez les porcelets. Des études supplémentaires sont nécessaires pour confirmer si les isolats de C. perfringens de type A porteurs de cpb2 ont un rôle pathogène dans les infections entériques chez les porcelets nouveau-nés.
(Traduit par Docteur Serge Messier)
Introduction
Mortality and morbidity among newborn piglets through enteric disease is a substantial problem in the swine industry. Type A Clostridium perfringens with other microbial pathogens, including enterotoxigenic E. coli (ETEC), Salmonella spp., rotavirus, transmissible gastroenteritis virus (TGE), and Isospora suis, have been associated with diarrhea in suckling piglets (1).
Neonatal enteritis attributed to C. perfringens type A infection associated with pre-weaning mortality has been reported previously (1,2). Disease is typically seen in piglets within 48 h of birth and is associated with creamy or pasty feces lasting several days. Necropsy shows flaccid, thin-walled, gaseous, or watery small intestine and pasty colonic contents (3,4). Microscopically, intestinal sections may have superficial villous tip necrosis, with fibrin and neutrophils, or appear normal or perhaps slightly congested (4).
The cpb2 gene and its expressed protein, a 27.6 kD toxin (CPB2 toxin), was first described in an isolate from a piglet with necrotic enteritis (5). Subsequently, cpb2 was demonstrated by polymerase chain reaction (PCR) in isolates from a variety of animals with diarrheic illness or fatal enteritis (6). The CPB2 toxin is encoded by either a “consensus” gene or by an “atypical” gene with 80.4% similarity between the 2 proteins (7,8). Type A, consensus cpb2-positive and CPB2 toxin-expressing isolates from swine form a homogenous genetic cluster (9).
The association between cpb2-carrying type A C. perfringens and neonatal diarrhea in piglets remains unclear. Some studies have reported an association between the presence of the consensus cpb2 gene and type A-associated diarrheal illness of neonatal piglets compared with healthy controls (10–13). In one report, the presence of cpb gene in isolates from piglets with type C C. perfringens infection was a confounding variable that makes it difficult to make an association between cpb2 and diarrhea in piglets (10). Reports of this association have not, however, been designed to systematically investigate differences, if any, between healthy and diarrheic neonatal piglets. A proportion of C. perfringens recovered from healthy piglets also carry cpb2(6). Attributing a pathogenic role to type A C. perfringens in neonatal enteritis of piglets is potentially problematic because of the marked colonization of the stomach and small intestine of neonatal piglets by C. perfringens that occurs within the first hours of life (14). In addition, the gross and histopathological changes described in piglets diagnosed with type A C. perfringens are strikingly variable (2–4). Therefore, the diagnosis of type A C. perfringens-associated porcine neonatal diarrhea requires the isolation of large numbers of cpb2-positive bacteria and the exclusion of other causes (4,15).
The objective of this study was to detect and quantify cpb2-positive type A C. perfringens in piglets with diarrhea compared with unaffected cohort piglets and to determine its association with neonatal diarrheal illness in pigs.
Materials and methods
Study farms and sample collection
Ten farms with a consistent history of diarrheal illness attributed to type A C. perfringens neonatal diarrhea were identified. From each farm, 2 to 4 neonatal diarrheic piglets with liquid or watery feces and 1 to 2 healthy, non-diarrheic, piglets from a different litter were selected. In total, 46 piglets were selected (36 with diarrhea and 12 without diarrhea), of which 44 piglets were aged 1 to 4 d and 4 pigs were aged between 6 to 13 d. The piglets were submitted to Animal Health Laboratory, University of Guelph. Live animals were euthanized, necropsied, and intestinal samples were collected for histopathology and microbiologic testing (outline follows). Samples from small intestine and colon were stored at −70°C to be tested for the presence of C. perfringens CPB2 toxin by enzyme-linked immunosorbent assay (ELISA). Approval of this study was obtained from the Animal Care Committee, University of Guelph, in accordance with guidelines of the Canadian Committee on Animal Care.
Histopathology and microbiological analysis
Intestinal samples were collected immediately following euthanasia for histopathologic examination and for microbiological testing. For histopathology, samples were collected from 32 piglets (21 with diarrhea and 11 healthy) from duodenum, jejunum, ileum, and colon, and fixed in 10% neutral buffered formalin for 24 h. Following histologic processing, hematoxylin and eosin-stained slides were examined by light microscopy. For cases in which histologic lesions of atrophic enteritis were identified, immunohistochemistry (IHC) was carried out for TGE viral antigen using anti-porcine corona-virus monoclonal antibody (Custom Monoclonals International, Sacramento, California, USA). For each of the 32 piglets, mucosal scrapings from fresh samples of ileum and spiral colon were cultured on trypticase soy agar containing 5% sheep blood and MacConkey’s agar (Difco, Detroit, Michigan, USA), and E. coli isolates were serotyped using polyvalent ‘O,K’ antiserum pool to detect the most common serotypes of enterotoxigenic E. coli, as well as antisera against F4 (K88) and F5 (K99) fimbriae. The ELISA for C. difficile toxins A and B, and rotavirus A latex agglutination (RLA) testing were carried out on colonic content. For those RLA-negative cases with histologic evidence of atrophic enteritis and negative TGE IHC results, further testing for porcine rotavirus was carried out by Rotavirus A/C RT-PCR and a conventional gel based PCR for Rotavirus B RT-PCR (Animal Health Laboratory, University of Guelph). The presence of intestinal coccidia was evaluated histologically and, in some animals, by sucrose wet mount examination of colonic content. Samples from the small intestine and colon were taken from all 48 piglets to quantify C. perfringens, to genotype the C. perfringens isolates, and to test for CPB2 toxin. Histopathological examination and microbial identification was done on 21 piglets with diarrhea and 11 normal cohorts from the 10 farms (on 9 farms 2 piglets with diarrhea and 1 normal piglet each, and on 1 farm 3 piglets with diarrhea and 2 normal piglets).
Clostridium perfringens quantification
Clostridium perfringens was recovered quantitatively from fresh colonic and jejunal contents obtained at necropsy from piglets. Samples were delivered to the laboratory at 4°C and then immediately weighed and serially diluted in phosphate-buffered saline (PBS), pH 7.2. A 100 μL aliquot of each serial dilution was plated onto Shahidi-Ferguson Perfringens (SFP) medium (Difco) containing cycloserine and 5% egg yolk, and incubated anaerobically at 37°C overnight. Colonies with the characteristic C. perfringens morphology and lecithinase activity were counted and the log10 number of colony-forming units (CFU) per gram of intestinal content determined.
Genotyping by multiplex real-time PCR
This method and primers were described by Albini et al (16) except that 3 new primers were designed for atypical cpb2, netB, and tpeL (Table I). The lysed bacteria were subjected to Taqman real-time PCR (Roche LC480II instrument; Roche Canada, Mississauga, Ontario). A volume of 1 × Quanta PCR Master mix and primers were loaded onto a 384-well PCR plate (Roche Canada) and 2 μL of the lysed bacterial samples and reference strains were subjected to PCR in a final volume of 15 μL. The control isolates with known toxin genotype profiles were treated exactly the same way as the untested bacterial isolates both in the lysis and PCR stages. The resulting PCR curves for the reference isolates were checked for consistency with the known genotype control strains and the template controls were checked for absence of amplification. A software package (Roche LightCycler 480 SW 1.5) was used to analyze the data to present the results with P-values. When available, 4 isolates were typed per intestinal sample.
Table I.
Primers and probes used for fluorogenic (TaqMan) polymerase chain reaction (PCR)
| Target gene | Nucleic acid sequences of primers and probes (5′–3′) with probe dyes | Product length (bp) |
|---|---|---|
| cpa | AAGAACTAGTAGCTTACATATCAACTAGTGGTG (54°C) | 124 |
| TTTCCTGGGTTGTCCATTTCC (51°C) | ||
| TEX-TTGGAATCAAAACAAAGGATGGAAAAACTCAAG-BHQ2 (55°C) | ||
| cpb | TGGAGCGTGAAAGAAACTGTTATTA (51°C) | 85 |
| GGTATCAAAAGCTAGCCTGGAATAGA (52°C) | ||
| FAM-CTTAATTGGAATGGTGCTAACTGGGTAGGACAA-BHQ1 (57°C) | ||
| cpb2 | TATTTCAAAGTTTACTGTAATTTTTATGTTTTCA (49°C) | 127 |
| CCATTACCTTTCTATAAGCGTCGATT (65°C) | ||
| CalFluor Orange 560-TGCACTTGCTTTCATTGGACTTATTGCTCC-BHQ-1 (58°C) | ||
| cpe | AGCTGCTGCTACAGAAAGATTAAATTT (52°C) | 88 |
| TGAGTTAGAAGAACGCCAATCATATAA (50°C) | ||
| FAM-ACTGATGCATTAAACTCAAATCCAGCT-BHQ1 (53°C) | ||
| etx | TTTGATAAGGTTACTATAAATCCACAAGGA (50°C) | 121 |
| AGAGAGCTTTTCCAACATAAACATCTTC (52°C) | ||
| CalFluor Orange 560-TAATCCTAAAGTTGAATTAGATGGAGAACCA-BHQ-1 (52°C) | ||
| iap | GCATTAAAGCTCACACCTATTCCA (51°C) | 85 |
| GAGATGTGAGAGTTAATCCAAATTCTTG (51°C) | ||
| FAM-CTAACTTAATTGTATATAGAAGGTCTGGTCC-BHQ1 (51°C) | ||
| netB | GGCGGTAATATATCTGTTGAAGG (53°C) | 168 |
| ACCGTCCTTAGTCTCAAC (51°C) | ||
| CalFluor Orange 560-ACTGCTGGTGCTGGAATAAATGCTTCA-BHQ1 (56°C) | ||
| Atypic cpb2 | GTAGTAGACCATTGGATGTGGG (54°C) 114 | |
| GAAAGTTTCTCCTGAACCTAGA (51°C) | ||
| CalFluor Orange 560-GAGCTTCTCAATGGGTATATGGTG-BHQ1 (55°C) | ||
| tpeL | GGAATTAGCAGCTAGAATTGG (51°C) | 120 |
| CCTGCAATTGGCACTA (49°C) | ||
| FAM-TGACAGCAGCTATTAGTTCT-BHQ1 (51°C) |
ELISA for C. perfringens CPB2 toxin
The ELISA plates (96-well, MaxiSorp C: Nunc, Roskilde, Denmark) were coated with 100 μL per well of capture antibody (rabbit polyclonal anti-CPB2 antibodies in carbonate/bicarbonate buffer, pH 9.6). After 2 h at 37°C, incubation was continued overnight at 4°C. Plates were washed twice using washing buffer (PBS, 0.05% Tween 20, 0.5% fish skin gelatin) and once using PBS, pH 7.4, which was followed by blocking with the blocking buffer (PBS, 0.05% Tween 20, 2% bovine serum albumen (BSA) for 2 h at 37°C. Plates were washed 3 times with the washing buffer, and 100 μL per well of purified recombinant CPB2 (used as a positive control) and processed intestinal content samples from neonatal piglets were added into the plate. Washing and the addition of the antigen were done on ice. The washing buffer was used as a negative control. Incubation at 4°C overnight was followed by 5 washings on ice using cold washing buffer, after which the detecting antibody (mouse monoclonal anti-CPB2 antibody, labeled with horseradish peroxidise, mAb-HRP) was applied (100 μL per well). The plate was then incubated 1 h at room temperature and washed 3 times with the washing buffer. An hour after addition of the chromogenic substrate (100 μL per well, incubation at room temperature), the reaction was stopped [0.5% sodium dodecyl sulfate (SDS), 50 μL per well] and the optical density was measured in an ELISA spectrophotometer at 405 and 490 nm.
Data analysis
Data were entered into a spreadsheet (Microsoft Excel 2003; Microsoft Corporation, Redmond, Washington, USA) and imported into another program for analysis (Stata 11 Intercooled for Windows XP; StataCorp LP, College Station, Texas, USA). A mixed multivariable analyzing method was used to investigate the association between log10 of C. perfringens numbers as continuous variable and neonatal diarrhea. In addition, a logistic regression method was applied in order to find relationships between categorical parameters including C. perfringens genes, CPB2 toxin, C. difficile toxin, enterotoxigenic E. coli, rotavirus, and histopathologic findings with diarrhea in piglets. In both methods, farm was included as random effect in order to take clustering within piglets on each farm into consideration. In addition, for each model, other variables were tested for any possible confounding.
Results
Quantification of intestinal C. perfringens
Ninety-seven (48 jejunal, 49 colonic) samples from 48 piglets (36 diarrheal, 12 normal) were examined. Type A C. perfringens was not isolated from 2 out of 24 (8.3%) and 11 out of 72 (15.1%) of intestinal samples from normal and diseased pigs, respectively; it was present in either the jejunum or colon contents of all 48 piglets but not in both sites from 3 piglets with diarrhea (Table II). The mean number (log10 CFU/g) of C. perfringens is shown in Table III; larger numbers of C. perfringens were isolated from the colon than from the jejunum (P < 0.001). In regression analysis with farm as the random effect, the log10 colony-forming units (CFU/g) of C. perfringens in the intestinal contents of healthy piglets was 0.8 higher than that in the diarrheic piglets (P = 0.02) (Table IV). Moreover, the log10 CFU of C. perfringens increased by 1.7 in the colonic contents compared to the small intestine (P < 0.001). Age of piglets was not significantly associated with log10 CFU of C. perfringens.
Table II.
Colony-forming unit (CFU) of C. perfringens and etiological diagnosis in 36 diarrheic and 12 healthy piglets on from 10 farms
| Farm | Pig | Age (day) | Status | Log10 of colony- forming unit (CFU/g) | Etiological diagnosis | |
|---|---|---|---|---|---|---|
| Small intestine | Colon | |||||
| 1 | 1 | 4 | D | 5.2 | 6.3 | Unknown |
| 2 | 4 | D | 4.2 | 6.0 | Rota | |
| 3 | 4 | H | 7.7 | 8.9 | Unknown | |
| 2 | 1 | 1 | D | 7.0 | 7.5 | Rota |
| 2 | 1 | D | 6.6 | 7.1 | Unknown | |
| 3 | 1 | D | 7.6 | 8.2 | C. difficile | |
| 4 | 1 | H | 7.3 | 8.4 | Unknown | |
| 5 | 3 | H | 6.4 | 8.3 | Unknown | |
| 3 | 1 | 1 | D | 5.8 | 7.5 | Unknown |
| 2 | 1 | D | 5.7 | 7.5 | Unknown | |
| 3 | 2 | H | 5.1 | 7.8 | Unknown | |
| 4 | 1 | 3 | D | 6.9 | 7.0 | C. difficile |
| 2 | 3 | D | ND | ND | Unknown | |
| 3 | 3 | H | 7.8 | 7.2 | Unknown | |
| 5 | 1 | 3 | D | 6.8 | 7.5 | C. difficile |
| 2 | 4 | H | 5.9 | 7.0 | Unknown | |
| 3 | 1 | D | 1.9 | 7.0 | C. difficile | |
| 4 | 3 | H | 6.5 | 7.9 | NT | |
| 5 | 3 | D | 7.7 | 7.8 | NT | |
| 6 | 1 | D | 7.3 | 8.1 | NT | |
| 6 | 1 | 1 | D | 3.0 | 6.8 | Unknown |
| 2 | 6 | H | 2.2 | 5.4 | Unknown | |
| 3 | 3 | D | 2.0 | 7.0 | C. difficile | |
| 4 | 8 | D | 1.8 | 7.1 | NT | |
| 5 | 10 | D | 1.8 | 5.0 | NT | |
| 6 | 13 | D | 2.0 | 7.0 | NT | |
| 7 | 1 | 3 | H | ND | 5.2 | Unknown |
| 2 | 1 | D | 3.5 | 5.0 | Unknown | |
| 3 | 3 | D | ND | 8.2 | Unknown | |
| 4 | 3 | D | ND | 7.1 | NT | |
| 5 | 3 | D | ND | 5.9 | NT | |
| 6 | 2 | D | 6.5 | 6.3 | NT | |
| 8 | 1 | 2 | D | ND | 5.4 | Rota |
| 2 | 2 | D | ND | 3.8 | Rota/Salmonella | |
| 3 | 2 | D | ND | 5.2 | NT | |
| 4 | 2 | D | 3.7 | ND | NT | |
| 5 | 2 | D | 4.3 | 5.1 | NT | |
| 6 | 2 | H | ND | 4.6 | Unknown | |
| 9 | 1 | 3 | D | 1.0 | 2.0 | Rota |
| 2 | 3 | D | 2.3 | 7.2 | Rota | |
| 3 | 4 | H | 7.7 | 8.0 | ETEC | |
| 4 | 4 | D | ND | 6.0 | NT | |
| 5 | 3 | D | 7.3 | 7.5 | NT | |
| 10 | 1 | 1 | H | 4.0 | 4.0 | NT |
| 2 | 1 | D | 3.6 | 6.0 | C. difficile | |
| 3 | 4 | D | 2.0 | 3.0 | C. difficile/Rota | |
| 4 | 3 | D | 2.0 | 4.0 | NT | |
| 5 | 3 | D | 2.3 | ND | NT |
D — Diarrhea; H — Healthy; ND — Not detected; NT — Not tested for C. difficile, rotavirus, Salmonella, enterotoxigenic E. coli (ETEC), transmissible gastroenteritis (TGE), and coccidia.
Table III.
Mean of log10 colony-forming unit (CFU) and standard error of the C. perfringens in intestinal contents of 12 normal piglets and 36 piglets with diarrhea
| Log10 of CFU/g (SE) | |||||
|---|---|---|---|---|---|
|
|
|||||
| Number of positive samples | Normal pigs | Pigs with diarrhea | Total | P-value | |
| Jejunum | 38 | 6.1 (0.58) | 4.4 (0.42) | 4.8 (0.37) | 0.038 |
| Colon | 46 | 6.9 (0.48) | 6.3 (0.26) | 6.5 (0.23) | 0.28 |
| Total | 84 | 6.5 (0.37) | 5.4 (0.27) | 5.7 (0.22) | 0.035 |
SE — standard error.
Table IV.
Regression analysis for log10 of colony-forming units (CFU) of C. perfringens in the intestinal contents of normal piglets and piglets with diarrhea with farm as random effect
| Tissue | Parameter | Coefficient | Standard error | 95% confidence interval | P-value |
|---|---|---|---|---|---|
| Group | Normal | 0.8 | 0.40 | 0.02, 1.6 | 0.04 |
| Diarrhea | Reference | ||||
| Sample | Colon | 1.7 | 0.36 | 3.9, 5.3 | < 0.001 |
| Small intestine | Reference |
Genotyping by multiplex RT PCR
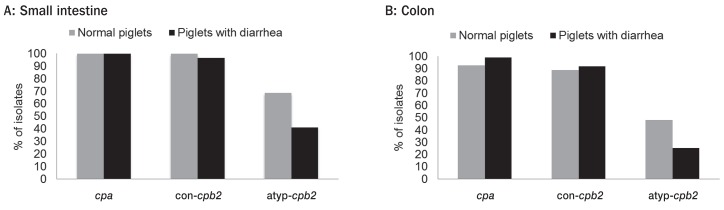
Genotyping was done on 1 to 4 isolates per jejunal and colonic samples so that 201 isolates (43 isolates healthy, 158 diarrheic piglets) were genotyped. Results are shown in Figures 1A and 1B, respectively. Of the 9 genes used in the PCR, only cpa, consensus cpb2, and atypical cpb2 were identified. No significant difference in distribution of the cpa and cpb2 was observed between isolates in healthy and diarrheic piglets. The consensus cpb2 was present in 93% of isolates in both groups. A greater proportion of isolates from healthy pigs carried the atypical cpb2 gene (P < 0.05).
Figure 1.
Distribution of cpa, consensus cpb2, and atypical cpb2 genes among C. perfringens isolates recovered from jejunum (A) and colon (B) of normal piglets and piglets with diarrhea.
Clostridium perfringens CPB2 toxin
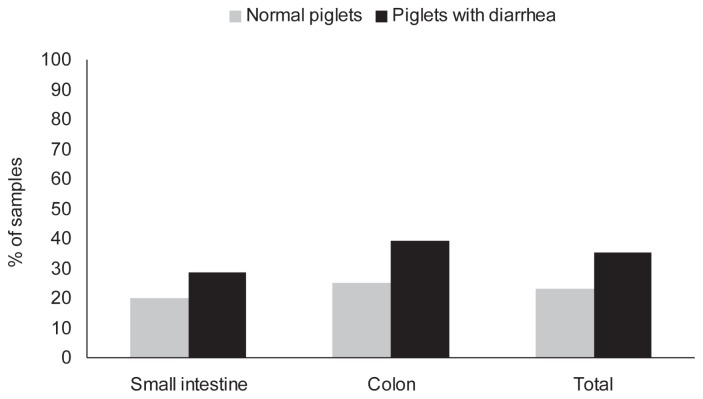
Some samples were unusable because of “noise” attributed to intestinal protease activity. Overall, 32% of 50 samples (from 8 normal, 24 diarrheic piglets) tested positive for CPB2; a piglet was defined as positive if CPB2 could be detected by ELISA in either the small intestinal or colonic contents. Three of 8 normal (37.5%) and 9 of 24 diarrheal (37.5%) and piglets were positive for CPB2. The positive proportion did not differ significantly in samples collected from normal and diarrheic piglets (Figure 2). The numbers of C. perfringens were higher in CPB2 toxin-positive piglets than in piglets that tested negative (log10 6.8 versus 5.1) (P = 0.02).
Figure 2.
Proportion of intestinal contents of normal piglets and piglets with diarrhea testing positive for cpb2 toxin using ELISA.
Other infectious agents
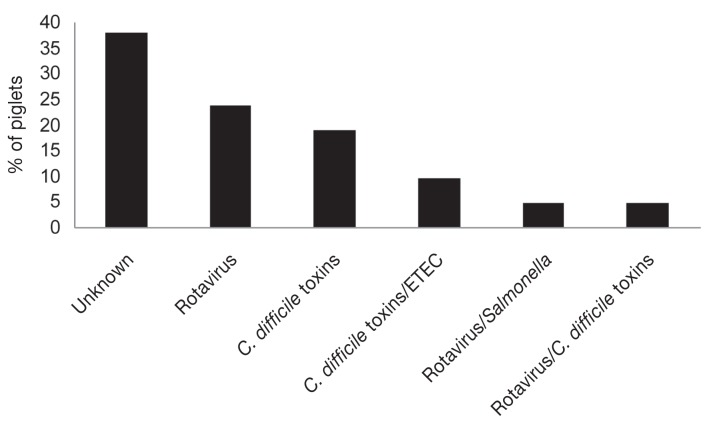
The final etiologic diagnosis of 21 piglets with diarrhea on 10 farms is shown in Figure 3. The etiology of the diarrhea in 8 of the 21 (38%) of piglets remained unknown. On 2 farms, the etiology of diarrhea could not be determined (Table II).
Figure 3.
Etiological diagnosis of cause of diarrhea in piglets on 10 farms with a history of neonatal diarrheal illness.
The intestine of 11 normal and 21 diarrheal piglets from the 10 farms were examined for histopathologic changes, and by isolation procedures for Salmonella spp. and enterotoxigenic E. coli. All 21 piglets with diarrhea and 2 normal piglets were tested for C. difficile toxins; these were detected in the intestinal contents of 7 of 21 (33%) diarrheic piglets from 4 farms but were not identified in the 2 healthy piglets tested. Twelve diarrheic piglets with histologic evidence of atrophic enteritis but negative RLA and TGE IHC results were tested by PCR for rotavirus. Rotavirus (groups A and C) was detected in 7 (33.3%) piglets on 4 farms. Rotavirus group C, Salmonella (Orion Var.15+34+), C. difficile toxins, or K88-positive enterotoxigenic E. coli were identified in different combinations in 4 piglets with diarrhea from 2 farms. Tissues from 8 diarrheic piglets examined immunohistochemically for TGE virus antigen were negative. Coccidia were not identified in any piglets using sucrose wet mount examination of feces or histologic examination of intestine. The ETEC or Salmonella spp. were not isolated from the small intestine or colon of any of the clinically normal (non-diarrheic) piglets. The log10 mean CFU/g of C. perfringens in the jejunum of diarrheic piglets that were positive for C. difficile toxins was slightly higher than in C. difficile-negative piglets (7.3 versus 6.7) (P = 0.07) but it did not differ based on rotavirus status.
Histopathological changes
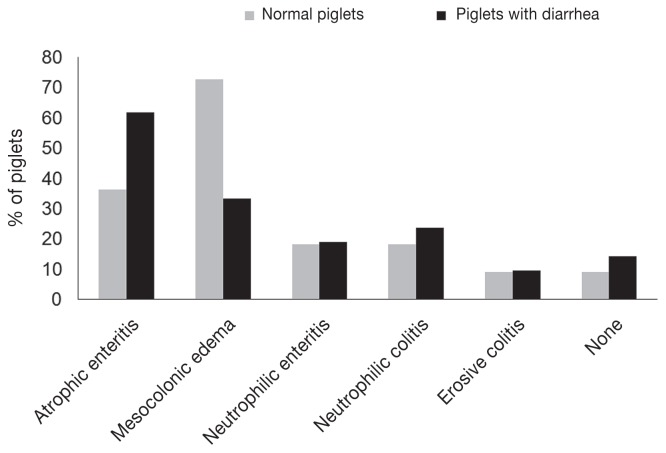
Histopathological findings in diarrheic (21) and normal (11) piglets are shown in Figure 4. Atrophic enteritis was the most frequent diagnosis in diarrheal (62%) and normal (36%) piglets. Other findings in diarrheal and normal piglets included neutrophilic enteritis (19% versus 18%), neutrophilic colitis (24% versus 18%), and erosive colitis (10% versus 9%), respectively. There was no significant difference in the presence of these histologic findings between diarrheic and normal piglets except that mesocolonic edema was more frequent in normal piglets (74% versus 33%) (P = 0.03). Of the diarrheic piglets with histologic evidence of neutrophilic colitis, one tested positive for C. difficile toxins A and B by ELISA. No histopathological changes were present in 14% and 9% of diarrheic and normal piglets, respectively. There was no correlation between histological changes observed and the presence of different toxin genes in C. perfringens isolates recovered from the intestinal contents (P > 0.05). No association was seen between histopathology findings and presence of other infectious agents at the level of P < 0.05. Other than atrophic enteritis, which seemed to be associated with a lower CFU of C. perfringens in jejunal contents (P = 0.06), no correlation between the CFU in the intestinal contents and histological changes was observed.
Figure 4.
Histopathological changes in 11 normal piglets and 21 piglets with diarrhea.
Discussion
If C. perfringens type A cause diarrhea in neonatal piglets, as is widely believed, most aspects of the disease are unclear and the diagnosis is equivocal, made only by exclusion of other enteric pathogens together with the detection of large numbers of cpb2-positive C. perfringens in the small intestine (4). Type A C. perfringens was recovered in large numbers from the small and large intestines of both diarrheic and normal piglets in this study, with lower numbers in diarrheic animals. In the absence of examination of control piglets, some of the piglets in this study, designated as having diarrhea of “unknown” etiology, might well have been assigned a diagnosis of type A C. perfringens enteritis based on the presence of “large numbers” of cpb2-positive C. perfringens and the exclusion of other causes (4). Most piglets had evidence of inflammation of some type.
Clostridium perfringens rapidly colonize the gastrointestinal lumen after birth and are present throughout the life of animals (14). The finding of large numbers of C. perfringens in piglets reported here are consistent with the report by Smith and Jones (14) on the development of the intestinal microflora of healthy piglets. In those studies intestinal colonization of the healthy newborn piglet by C. perfringens occurred initially in the stomach and small intestine, and followed that of E. coli. Clostridium perfringens were first found in these sites 12 h after birth, with numbers reaching log10 8.2 in stomach and 8.3 in the mid-small intestine. By 7 d after birth, numbers had fallen markedly, to log10 2.0 in the stomach and log10 2.7 in the mid-small intestine, whereas numbers in the large intestine were higher (log10 6.4). We report here that healthy neonatal piglets were commonly colonized with large numbers of C. perfringens, interestingly in numbers exceeding those observed in diarrheal piglets. It was more common to find these bacteria in the colon than the jejunum, also consistent with the earlier findings of Smith and Jones (14). These finding considerably complicate a diagnosis of type A C. perfringens enteritis, since the presence of large numbers of these bacteria in the stomach and small intestine is part of the normal intestinal colonization.
This is the first study measuring CPB2 toxin in the intestinal tract. The CPB2 was detected in only about 1/3 of samples with no significant difference between healthy and diarrheic piglets, although only just over half of the samples could be tested because of signal “noise” and sample size was low. The CPB2-positive pigs had a higher number of C. perfringens in jejunum, but the toxin was not detected in the 2/3 of piglets with a high number of C. perfringens. Although the consensus cpb2 gene is almost always expressed in porcine origin C. perfringens(7), its expression and the factors affecting its expression in vivo have not been investigated. Further studies are required to determine whether it is involved in neonatal enteritis in piglets; as noted by others, cpb2 may be a marker for virulence of porcine isolates rather than being directly involved in intestinal disease (4). One interesting feature of this study, that has not been described in detail previously, was the recognition that atypical cpb2 was also present in a proportion of isolates, and, interestingly, a significantly higher proportion of isolates from normal piglets carried this gene. The atypical gene is widespread and moderately common in isolates from different animals (6). We did not identify other toxin genes in the isolates.
The lack of a specific association between the presence of cpb2-positive C. perfringens(17) and neonatal diarrhea contrasts with previous studies (10–12) but is consistent with a report of the high prevalence of cpb2-positive isolates in piglets dying of mostly identified causes other than necrotic enteritis (and type A C. perfringens infection) (18). Garmory and others (11) found cpb2 in 82% of 33 isolates from piglets with diarrhea and it was absent in 7 control piglets. Bueschel and others (12) detected cpb2 in more than 90% of 381 isolates from porcine neonatal cases of enteritis but only in about 10% of isolates from 11 “normal pigs.” Waters et al (13) confirmed that 29 isolates from diarrheic pigs carried cpb2, but this gene was not found in 6 isolates from “healthy pigs.” The studies of these authors (11–13) were not, however, designed rigorously to address the hypothesis of an association between cpb2 and diarrheal illness in neonatal piglets and, by contrast to the current study, the relative numbers of healthy to diarrheic pigs were low, and the provenance of healthy pigs was unclear.
One possible explanation for the lack of observed association between cpb2-positive C. perfringens and diarrhea in piglets in the present study is that the normal and diarrheal piglets were selected during an outbreak of neonatal diarrhea. It is possible that cpb2-positive C. perfringens spread between piglets and that normal appearing piglets were infected but did not develop the diarrhea because of the lack of other disease contributing parameters (19). However, 62% of 21 diarrheic piglets had microbiological diagnoses other than type A C. perfringens and the etiology of diarrhea in other diarrheic piglets remained unknown. As noted, it is possible that, in the absence of recognition of rotavirus or C. difficile infection, diarrhea in these piglets might be erroneously attributed to type A C. perfringens on the basis of numbers of bacteria isolated and their possession of cpb2.
There have been reports of the reproduction of enteric disease by C. perfringens type A in colostrum-deprived, hysterectomy-derived, neonatal piglets (20) or in colostrum-deprived neonates (21–23). In one study, all piglets died or were euthanized in extremis within 72 h (20), whereas in the other study mortality was about 10% but morbidity was 55% in infected animals (21). A general loss of condition was common with diarrhea in about half of the affected animals (21). Small intestinal contents described as fluid with flecks of blood and necrotic debris (20) are not characteristic of descriptions of C. perfringens type A-associated enteritis, although the diffuse hyperemia and mild enteritis observed in some instances are more consistent with field descriptions (21). Because of the difficulty of rearing colostrum-deprived piglets, including their tendency to develop septicemia, reproduction of disease in hysterectomy-derived gnotobiotic piglets may be required to convincingly demonstrate a role for type A C. perfringens in neonatal piglet enteric disease.
Clostridium difficile toxins were detected in 7 scouring piglets and enterotoxigenic E. coli were detected in 2 of these. Clostridium difficile plays an important role in neonatal diarrhea in piglets (4). Rotavirus was also detected in 7 piglets with histopathological changes suggesting rotaviral infection. The co-incidence of rotavirus with C. difficile toxins in 2 piglets with diarrhea and with Salmonella in 1 diarrheal piglet indicates that the secondary infection with other infectious agents may cause more serious disease. Therefore, it is possible that other pathogens including C. difficile and rotavirus might form a multiple cause of the enteritis in neonatal piglets.
The etiologic diagnosis found in this study may not be extra-polatable to the larger population of Ontario swine farms since only a small number of farms were included in this study and the farms were not randomly selected. A future study with a larger sample size needs to be done to determine the causes of diarrhea in neonatal piglets and its associated risk factors.
In conclusion, this study highlights the complexity of diagnosis of porcine neonatal diarrhea associated with cpb2-positive type A C. perfringens(4). We were unable to distinguish between healthy and diarrheic piglets on the basis of bacterial numbers in the intestine, the presence of consensus cpb2 in C. perfringens isolates, the expression of CPB2 in the intestine of piglets, and between diarrheal piglets with known or unknown causes of diarrhea. No association was observed between histopathological findings and the presence of CPB2. This study adds urgency to the previously identified need for more definitive diagnostic criteria (4). Based on the current study, exclusion of other causative agents (4) is not an adequate diagnostic criterion for diagnosis of the disease. An alternative suggestion is that, in the absence of other criteria, the presence of large numbers of cpb2-positive type A C. perfringens in the intestine of neonatal piglets should be regarded as normal commensals.
Acknowledgments
The authors gratefully acknowledge the Ontario Ministry of Agriculture Food and Rural Affairs (OMAFRA) — University of Guelph Research Program, and the OMAFRA Animal Health Strategic Initiative for funding this project. We also acknowledge funding from the Natural Sciences and Engineering Council of Canada. The authors thank Drs. Charbonneau, Scorgie, and Templeton for recruiting the farms, as well as Vivian Nicholson and Yanlong Pei for laboratory assistance.
References
- 1.Straw BE, Dewey CE, Wilson MR. In: Diseases of Swine. 9th ed. Straw BE, Zimmermann JJ, D’Allaire WL, Taylor DJ, editors. Oxford, UK: Blackwell Science; 2006. pp. 241–286. [Google Scholar]
- 2.Lehe K. Successful control of clostridial enteritis: A report from the back forty. Am Assoc Swine Pract. 2009:413–414. [Google Scholar]
- 3.Schwartz KJ. Clostridium-associated diseases in swine. proceedings: The 40th American Association of Swine Veterinarians (AASV) annual meeting; 2009. pp. 415–422. [Google Scholar]
- 4.Songer JG, Uzal FA. Clostridial enteric infections in pigs. J Vet Diag Invest. 2005;17:528–536. doi: 10.1177/104063870501700602. [DOI] [PubMed] [Google Scholar]
- 5.Gibert M, Jolivet-Reynaud C, Popoff MR. Beta2 toxin, a novel toxin produced by Clostridium perfringens. Gene. 1997;203:65–73. doi: 10.1016/s0378-1119(97)00493-9. [DOI] [PubMed] [Google Scholar]
- 6.Van Asten AJ, Nikolaou GN, Gröne A. The occurrence cpb2-toxigenic Clostridium perfringens and the possible role of the Cpb2-toxin in enteric disease of domestic animals, wild animals and humans. Vet J. 2010;183:135–140. doi: 10.1016/j.tvjl.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Jost BH, Billington SJ, Trinh HT, et al. Atypical cpb2 genes encoding beta2-toxin in Clostridium perfringens isolates of non-porcine origin. Infect Immun. 2005;73:652–656. doi: 10.1128/IAI.73.1.652-656.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lebrun M, Filée P, Mousset B, et al. The expression of Clostridium perfringens consensus beta2 toxin is associated with bovine enterotoxaemia syndrome. Vet Microbiol. 2007;120:151–157. doi: 10.1016/j.vetmic.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 9.Jost BH, Trinh HT, Songer JG. Clonal relationships among Clostridium perfringens of porcine origin as determined by multilocus sequence typing. Vet Microbiol. 2006;116:158–165. doi: 10.1016/j.vetmic.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 10.Klaasen HL, Molkenboer MJ, Bakker J, et al. Detection of the cpb2-toxin gene of Clostridium perfringens in diarrhoeic pigs in The Netherlands and Switzerland. FEMS Immunol Med Microbiol. 1999;24:325–332. doi: 10.1111/j.1574-695X.1999.tb01301.x. [DOI] [PubMed] [Google Scholar]
- 11.Garmory HS, Chanter N, French NP, Bueschel D, Songer JG, Titball RW. Occurrence of Clostridium perfringens beta2-toxin amongst animals, determined using genotyping and subtyping PCR assays. Epidemiol Infect. 2000;124:61–67. doi: 10.1017/s0950268899003295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bueschel DM, Jost BH, Billington SJ, et al. Prevalence of cpb2 encoding beta2 toxin in Clostridium perfringens field isolates: A correlation of genotype with phenotype. Vet Microbiol. 2003;94:121–129. doi: 10.1016/s0378-1135(03)00081-6. [DOI] [PubMed] [Google Scholar]
- 13.Waters M, Savoie A, Garmory HS, et al. Genotyping and phenotyping of beta2-toxigenic Clostridium perfringens fecal isolates associated with gastrointestinal diseases in piglets. J Clin Microbiol. 2003;41:3584–3591. doi: 10.1128/JCM.41.8.3584-3591.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Smith HW, Jones JE. Observations on the alimentary tract and its bacterial flora in healthy and diseased pigs. J Pathol Bacteriol. 1963;86:387–412. [PubMed] [Google Scholar]
- 15.Collins JE, Bergeland ME, Bouley D, et al. Diarrhea associated with Clostridium perfringens enterotoxin in neonatal pigs. J Vet Diag Invest. 1989;1:351–353. doi: 10.1177/104063878900100414. [DOI] [PubMed] [Google Scholar]
- 16.Albini S, Brodard I, Jaussi A, et al. Real-time multiplex PCR assays for reliable detection of Clostridium perfringens toxin genes in animal isolates. Vet Microbiol. 2008;127:179–185. doi: 10.1016/j.vetmic.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Fisher DJ, Miyamoto K, Harrison B, et al. Association of beta2 toxin production with Clostridum perfringens type A human gastrointestinal disease carrying a plasmid enterotoxin gene. Mol Microbiol. 2005;56:747–762. doi: 10.1111/j.1365-2958.2005.04573.x. [DOI] [PubMed] [Google Scholar]
- 18.Jäggi U, Wollschläger N, Abril C, et al. Retrospective study on necrotizing enteritis in piglets in Switzerland. Schweiz Arch Tierheilk. 2009;151:369–375. doi: 10.1024/0036-7281.151.8.369. [DOI] [PubMed] [Google Scholar]
- 19.Schotte U, Truyen U, Neubauer H. Significance of cpb2-toxigenic Clostridium perfringens infections in animals and their predisposing factors — A review. J Vet Med B Infect Dis Vet Public Health. 2004;51:423–426. doi: 10.1111/j.1439-0450.2004.00802.x. [DOI] [PubMed] [Google Scholar]
- 20.Olubumni PA, Taylor DJ. Clostridium perfringens type A in enteric diseases of pig. Trop Vet. 1985;3:28–33. [Google Scholar]
- 21.Johannsen U, Arnold P, Kohler B, Selbitz HJ. Untersuchngen zur Experimentellen Clostridium perfringens-Typ A-Enterotoxameie der Saugferkel. Monat Vet Med. 1993a;48:129–136. [Google Scholar]
- 22.Johannsen U, Menger S, Arnold P, et al. Untersuchungen zur Experimentellen Clostridium perfringens-Typ A-Enterotoxameie der Saugferkel. Monat Vet Med. 1993b;48:267–273. [Google Scholar]
- 23.Johannsen U, Menger S, Arnold P, et al. Untersuchngen zur Experimentellen Clostridium perfringens-Typ A-Enterotoxameie der Saugferkel. Licht- und Elektronenmikrokopischen Untersuchungen zur Pathologie. Monat Vet Med. 1993c;48:299–306. [Google Scholar]