Abstract
This study investigated epigenetic mechanisms by which DNA methylation affects the function of bovine adaptive immune system cells, particularly during the peripartum period, when shifts in type 1 and type 2 immune response (IR) biases are thought to occur. Stimulation of CD4+ T-lymphocytes isolated from 5 Holstein dairy cows before and after parturition with concanavalin A (ConA) and stimulation of CD4+ T-lymphocytes isolated from 3 Holstein dairy cows in mid-lactation with ConA alone or ConA plus dexamethasone (Dex) had significant effects on production of the cytokines interferon gamma (IFN-γ, type 1) and interleukin 4 (IL-4, type 2) that were consistent with DNA methylation profiles of the IFN-γ gene promoter region but not consistent for the IL-4 promoter region. ConA stimulation increased the production of both cytokines before and after parturition. It decreased DNA methylation in the IFN-γ promoter region but increased for IL-4 promoter region. Parturition was associated with an increase in IFN-γ production in ConA-stimulated cells that approached significance. Overall, DNA methylation in both promoter regions increased between the prepartum and postpartum periods, although this did not correlate with secreted cytokine concentrations. Dexamethasone treated cells acted in a manner consistent with the glucocorticoid’s immunosuppressive activity, which mimicked the change at the IFN-γ promoter region observed during parturition. These results support pregnancy as type 2 IR biased, with increases of IFN-γ occurring after parturition and an increase in IL-4 production before calving. It is likely that these changes may be epigenetically controlled.
Résumé
Cette étude avait pour objectif d’investiguer les mécanismes par lesquels la méthylation de l’ADN affecte la fonction des cellules du système immunitaire adaptatif bovin, en particulier durant la période péri-partum, lorsque les changements des biais des réponses immunes (IR) de type 1 et type 2 sont sensés se produire. La stimulation de lymphocytes T CD4+ isolés de cinq vaches laitières de race Holstein avant et après la parturition avec de la concanavaline A (ConA) et la stimulation de lymphocytes T CD4+ isolés de trois vaches laitières de race Holstein en milieu de période de lactation avec de la ConA seule ou de la ConA plus de la dexaméthasone (Dex) a eu des effets significatifs sur la production des cytokines interféron gamma (IFN-γ, type 1) et interleukine 4 (IL-4, type 2) qui étaient conformes avec les profils de méthylation de l’ADN de la région du gène promoteur IFN-γ mais pas pour la région du promoteur IL-4. La stimulation par la ConA augmenta la production des deux cytokines avant et après la parturition. Elle diminua la méthylation de l’ADN dans la région du promoteur IFN-γ mais augmenta celle dans la région du promoteur IL-4. La parturition était associée avec une augmentation presque significative de production d’IFN-γ par les cellules stimulées par la ConA. De manière générale, on nota une augmentation de la méthylation de l’ADN dans les régions des deux promoteurs entre la période pré-partum et la période post-partum, sans que cela ne soit corrélée avec les concentrations des cytokines sécrétées. Les cellules traitées avec de la dexaméthasone se sont comportées d’une manière conforme à l’activité immunosuppressive des glucocorticoïdes, qui simulait les changements observés dans la région du promoteur IFN-γ durant la parturition. Ces résultats soutiennent le fait que la gestation amène un biais de type 2 de la réponse immunitaire, avec une augmentation d’IFN-γ se produisant après la parturition et une augmentation de production d’IL-4 avant le vêlage. Il est probable que ces changements peuvent être contrôlés de manière épigénétique.
(Traduit par Docteur Serge Messier)
Introduction
The peripartum period, generally defined as 3 wk before to 3 wk after calving, is a time of stress, transition, high energy demand, and suboptimal immune response (IR) (1). At this stage dairy cows experience hormonal, management, behavior, and feed changes, as well as the stress of the transition into high-volume lactation (2,3). Some specific hormonal changes include a decrease in progesterone production and an increase in estrogen and glucocorticoid (GC) production just before calving (4,5). These changes and stressors have marked this period as one of a high incidence of disease, both infectious and metabolic (6,7), which can be costly in terms of disease treatment, decreased milk production, and animal welfare.
Changes in IR during pregnancy are correlated with alterations of leukocyte populations and their function (5,8–10). A study by Van Kampen (11) showed that cells of the immune system change before and after calving. Meglia (12) found large changes in cell populations and function, including decreased lymphocyte and eosinophil counts just before calving, the counts returning to normal by 2 wk after calving. Meglia also observed an increase in the monocyte count just before calving and in the B-cell count early in the postpartum period. Others have noted a decline in the proliferation of T-lymphocytes during the peripartum period, along with a shift between a type 1 and a type 2 IR (5,8). This effect on T-lymphocyte proliferation and T-helper (Th)-cell phenotypes can be simulated in vitro in a dose-dependent manner by the synthetic GC dexamethasone (Dex), which tends to promote a type 2 IR (13,14). As such, Dex treatment of bovine CD4+ cells in vitro can offer insight into the role of GCs in the differentiation of Th-cells and hence the IR during the peripartum period (15).
Epigenetic modifications in the form of histone modifications and DNA methylation are highly involved in the induction, maintenance, and heritability of gene expression and subsequent protein production in various cell types (16). Epigenetic modifications play a role in cell differentiation throughout development and play a crucial role in immune cell phenotypes. In general, DNA methylation typically represses gene expression, whereas DNA demethylation typically increases gene expression by opening the chromatin structure. The extent to which DNA methylation has an effect on gene expression depends not only on the gene of interest but also on which regulatory factors may control subsequent transcription of that gene. For this study, the Th lineage decisions of key interest were those in which DNA methylation appears to have modulating effects on the promoters of the genes for interferon gamma (IFN-γ) and interleukin 4 (IL-4) in humans and several other species, including mice and cattle (17–19). Expression of these cytokines is critical for the induction and maintenance of Th-cell subtypes. Epigenetics is now commonly thought to represent a critical connection between gene expression and the environment contributing to sustained changes in cellular phenotypes that are acquired during development (20). As such, DNA methylation serves as an important subject for investigation of molecular, cellular, and physiological responses to internally and externally induced biochemical states.
There is still some debate concerning the exact changes that occur in bovine Th-cell populations during the peripartum period, particularly in relation to the shifts in type 1 and type 2 IR before, during, and after calving. For instance, in a study by Shafer-Weaver, et al (8), mid-gestation was characterized by a type 1 IR bias, whereas cows sampled 3 d after parturition showed a type 2 IR bias. These results were in contrast to others in the literature indicating that pregnancy tends to be dominated by type 2 IRs, as shown in several species, including humans and mice (21,22). Additionally, although there are numerous studies that focus on changes in peripartum IR, the causal mechanism of the observed immuno-depression, particularly any epigenetic contribution, remains largely unknown.
The objectives of this study, therefore, were: 1) to assess the effects of the T-cell mitogen concanavalin A (ConA) and the synthetic GC Dex on 2 key type 1 and 2 cytokines, IFN-γ and IL-4, respectively, as secreted from bovine CD4+ T-cells before and after parturition; and 2) to evaluate any changes in DNA methylation patterns within the IFN-γ and IL-4 promoters of bovine CD4+ T-cells before and after ConA stimulation, with and without Dex treatment. Our hypotheses were as follows: i) Dex treatment of CD4+ T-cells in vitro will abrogate any stimulatory effects of ConA and promote DNA methylation, consequently reducing cytokine expression in a manner consistent with a type 2 IR bias; and ii) changes in IFN-γ and IL-4 production by CD4+ T-cells during the peripartum period will be associated with changes in DNA methylation of the gene promoters of these cytokines.
Materials and methods
Animals and blood collection
Holstein dairy cows were housed at the University of Guelph dairy research farm. All animal handling was approved by the Animal Care Committee of the University of Guelph (AUP #04R063). Blood was collected from 5 of the cows 4 wk before calving and 4 d after calving for peripartum analysis. The dates for prepartum collection were chosen according to the predicted calving dates. The Dex-treatment blood samples were collected from 3 cows in mid-lactation (at ~100 d in milk); these cows were different from those used for the peripartum analysis. The blood (80 to 100 mL) was collected by caudal vein venepuncture into 10-mL Vacutainer tubes (BD, Franklin Lakes, New Jersey, USA) containing ethylene diamine tetraacetic acid.
Isolation of blood mononuclear cells (BMCs)
Cells were isolated by carefully overlaying 15 mL of Histopaque 1107 (Sigma, Oakville, Ontario), according to the manufacturer’s instructions, with an equal volume of blood. After centrifugation in a 50-mL conical Falcon tube [400 × g, 30 min, room temperature (RT)], cells were collected at the gradient interface. Phosphate-buffered saline (PBS; 0.01 M, pH 7.4) was added to the cells, for a total volume of 45 mL. After washing by centrifugation (250 × g, 10 min, RT) the remaining erythrocytes were lysed with 2 mL of sterile water. The BMCs were pelleted and washed again with 40 mL of PBS (0.01 M, pH 7.4) and centrifuged (250 × g, 15 min, RT). Viable BMCs were counted by means of a hemocytometer with the use of trypan blue exclusion dye (Sigma).
Selection of CD4+ T-lymphocytes
CD4+ T-cells were isolated with the MiniMACS system (Miltnyi Biotech, Auburn, California, USA) according to the manufacturer’s instructions. The cells (1 × 107) were incubated with 100 μL of mouse antibody against bovine CD4+ T-lymphocytes (ILA-11, VMRD, diluted 500-fold) for 30 min at 4°C. The cells were washed with PBS (300 × g, 10 min, RT), resuspended, and incubated in 80 μL of MiniMACS buffer (PBS, 2 mM EDTA, 0.5% bovine serum albumin) and 20 μL of magnetic microbeads coated with goat against mouse IgG (Miltnyi Biotech) per 1 × 107 cells (15 min, 4°C). Cells were counted as previously described, washed in 40 mL of PBS (250 × g, 10 min, RT), and then resuspended in 1 mL of MiniMACS buffer or, if necessary, an increased volume proportional to 2 × 108 cells/μL of MiniMACS buffer. The cell suspension (500 μL) was added to the magnet-bound column. The column was washed 3 times with 500 μL of MiniMACS buffer, and the bovine CD4+ cells were eluted in 1 mL of the buffer. The column separation was repeated to improve purity. Purity was confirmed by flow cytometry as greater than 99% (data not shown).
CD4+ T-cell cultures
For peripartum analysis
The isolated CD4+ T-cells were cultured in 200 μL of RPMI medium with 300 mg/L of glutamine, 10% fetal calf serum (FCS), and a 1/250 dilution of penicillin and streptomycin (Invitrogen Canada, Burlington, Ontario), at a concentration of 2.5 × 106 cells/mL, in a Costar 96-well round-bottom plate (Sigma, Oakville, Ontario) at 37°C for 24 h in an atmosphere of 5% CO2. Half of the plated cells were stimulated with ConA (2.5 μg/mL); the other half of the cells served as unstimulated controls.
For Dex analysis
CD4+ T-lymphocytes were cultured in phenol-red- free + glutamine RPMI medium and 10% charcoal-stripped FCS (Invitrogen Canada) on a similar plate, at the same concentration, under the same conditions, for 72 h. All the cells were stimulated with ConA (2.5 μg/mL); half were also stimulated with 10 μM of Dex, a dose shown in preliminary experiments to cause the maximum decrease in CD4+ T-cell proliferation in vitro (data not shown). The T-cell medium was made with phenol-red-free RPMI because phenol red can act as a weak estrogenic agonist (23), which was pertinent for projected downstream applications. For the same reason, the FCS was charcoal-stripped.
Enzyme-linked immunosorbent assay (ELISA)
An aliquot (150 μL) of supernatant was collected from each culture well after the designated incubation period to evaluate production of the cytokines IFN-γ and IL-4 when the cells were unstimulated, stimulated with ConA, or stimulated with ConA and Dex. The aliquots were pooled and then stored at −20°C for ELISA.
The IFN-γ concentration was determined with a bovine IFN-γ ELISA kit (Mabtech, Cincinnati, Ohio, USA) according to the manufacturer’s instructions. For the IL-4 ELISA, Immulon 2HB 96-well flat-bottom plates (Fisher Canada, Nepean, Ontario) were coated with a 1 μg/μL dilution of mouse antibody against bovine IL-4 (AbD Serotec, MorphoSys US, Raleigh, North Carolina, USA) in carbonate–bicarbonate buffer (pH 9.6), 100 μL/well, and incubated for 48 h at 4°C. The coating solution was aspirated and 200 μL of blocking buffer [PBS (pH 7.4) + 3% Tween 20] added to each plate. The plates were incubated for 90 min at RT. Samples and standards were added after removal of the blocking buffer. A recombinant bovine IL-4 (AbD Serotec) was used as the positive control starting with a dilution of 40 000 pg/mL to prepare a working dilution of 2000 pg/mL that was serially diluted from 1/2 to 1/256. Blocking buffer was used as the negative control. All controls and sample dilutions were added to the plates in duplicate and incubated for 150 min at RT on a shaker. The plates were washed 4 times with washing buffer [PBS (pH 7.4) + 0.05% Tween 20], 300 μL/well, in an ELx405 Autoplate Washer (BioTek Instruments, Winooski, Vermont, USA). For antibody detection a 1/8000 dilution of biotinylated mouse monoclonal antibody against bovine IL-4 (AbD Serotec) in washing buffer was added, 100 μL/well, and the plates were incubated for 60 min at RT on a shaker. Next, the plates were washed 4 times as previously described, 100 μL of a 1/10 000 dilution of a conjugate of streptavidin and horseradish peroxidase (Invitrogen Canada) in washing buffer was added to each well, and the plates were incubated for 45 min at RT on a shaker. After incubation the plates were washed 5 times as previously described, 100 μL of 3,3′,5,5′-tetramethylbenzidine substrate (IDEXX Laboratories, Westbrook, Maine, USA) was added to each well, and the plates were incubated in the dark for 45 min on a shaker. After incubation 100 μL of 1 M H2SO4 was added to each well to stop the reaction. The optical density of the individual wells was obtained at 450 nm with an EL808 plate reader (BioTek Instruments) and the KCjunior software package (Bio-Tek Instruments).
Genomic DNA (gDNA) extraction
After collection of the culture supernatant, 200 μL of PBS was added to the CD4+ T-cells remaining in the culture plate. The cell suspension was mixed and washed (300 × g, 5 min, RT) and either stored at −80°C for future DNA extraction or used immediately for DNA extraction with the DNeasy Tissue Kit (Qiagen, Mississauga, Ontario) according to the manufacturer’s instructions.
Bisulfite treatment and DNA amplification
To evaluate DNA methylation, gDNA was bisulfite-treated with the EZ DNA methylation kit (Zymo Research, Orange, California, USA) according to the manufacturer’s instructions. Specific primers (Table I) for both converted and unconverted promoter regions of the bovine IFN-γ (GI:23821137) and IL-4 (GI:555892) genes were designed with the use of BiSearch Software (24). The promoter region selected for IFN-γ contained 5 CpG sites, and the promoter region selected for IL-4 contained 5 CpG sites. The selected regions were amplified by polymerase chain reaction (PCR) with the use of Platinum Taq polymerase (Invitrogen Canada) in 20-μL reactions with 2 μL of template converted or unconverted DNA, 2 μL of 10× PCR buffer, 0.6 μL of 50 mM MgCl2, 0.5 μL of 10 mM deoxynucleotide triphosphates (Invitrogen Canada), and 1 μL of the respective forward and reverse primers at a concentration of 15 mM. For both analyses a touchdown PCR program was used with annealing temperatures going from 60°C to 54°C in the first part of the program after a denaturation step at 95°C for 2 min, 6 cycles of 95°C for 30 s, 60°C for 30 s, and then 72°C for 45 s, 23 cycles of 95°C for 30 s, 54°C for 30 s, and then 72°C for 45 s, and a final extension step of 72°C for 20 min. The PCR products were run on a 2% agarose gel to verify band size. For IFN-γ and IL-4, gel extraction was done with the PureLink Quick Gel Extraction Kit (Invitrogen Canada) on the band corresponding to the IFN-γ or the IL-4 promoter region, whose lengths were 609 and 684 base pairs (bp), respectively.
Table I.
Primers for promoters of the bovine genes for interferon gamma (IFN-γ) and interleukin 4 (IL-4)
| Primer | ID | 5′-3′ sequence | Band size (number of base pairs) | Annealing temperature (°C) |
|---|---|---|---|---|
| IFNG-B | Forward | CCACAAAAGAATGATCAATG | 548 | 55 |
| Reverse | CATAAGAACCAAGAAAAACC | |||
| IFNG-Y, converted | Forward | TTTGGATGAGGAGTTAATAT | 609 | 55 |
| Reverse | TTCAATCACAAAAAATACTA | |||
| IL-4-B | Forward | GAAGCCAAGGTGAAATACTA | 698 | 58 |
| Reverse | GAGCTGTTGTGTTCTTTCA | |||
| IL-4-E, converted | Forward | GGAAGAAGTTAAGGTGAAAT | 684 | 50 |
| Reverse | CACAATCTAAAAAATAAACAC |
Cloning and sequence analysis
Cloning was conducted with the TOPO TA Cloning Kit (Invitrogen Canada) according to the manufacturer’s instructions. Two Luria– Bertani (LB) plates per sample were prepared at different concentrations (suspensions of 20 and 40 μL of cells) and incubated at 37°C overnight. Ten individual colonies were selected from the 2 plates for each treatment and cultured in 5 mL of LB liquid broth overnight. Plasmids were extracted with the GenElute Plasmid Miniprep Kit (Sigma), and insertion of IFN-γ and IL-4 was verified by PCR and 1.5% agar gel electrophoresis. The PCR conditions were as follows: denaturation at 95°C for 10 min, 34 cycles of 95°C for 45 s, 59°C for 45 s, and then 72°C for 45 s, and extension at 72°C for 20 min. The verified insert-containing plasmid preparations were sequenced at the Robarts Research Institute, London, Ontario. The sequences were annotated and aligned in BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and analyzed with BiQ Analyzer software (25). Seven clones per condition were collected for the peripartum period and 10 clones per condition for the Dex treatment analysis.
Statistical analysis
Statistical differences were reported as significant at P ≤ 0.05, highly significant at P ≤ 0.01, and a trend at P ≤ 0.1. The treatment effect of Dex on the cytokine production of ConA-stimulated CD4+ T-cells, as measured by ELISA, was calculated with a 2-tailed, paired t-test with the program R 2.11.1 (http://www.r-project.org). The significance of the ELISA data was determined with analysis of variance between the 4 treatment groups (prepartum unstimulated, prepartum ConA-stimulated, postpartum unstimulated, postpartum ConA-stimulated) with R 2.11.1 for both IFN-γ and IL-4. The significance of differences in DNA methylation between unstimulated and stimulated cells from prepartum and postpartum samples was also calculated with a 2-tailed, paired t-test with R 2.11.1.
The percentage methylation in IFN-γ was analyzed by comparing the 6 CpG sites within the promoter region for each of the treatments. The overall change in methylation from prepartum to postpartum samples was calculated for both unstimulated and stimulated cells. The same procedure was completed for the 5 CpG sites of the IL-4 promoter region. Bioinformatic analysis for DNA element identification and conservation estimates was conducted in MultiTF (26). Subsequent CpG island prediction was done with the use of CpG Island Searcher (27).
Results
Effect of parturition on cytokine production
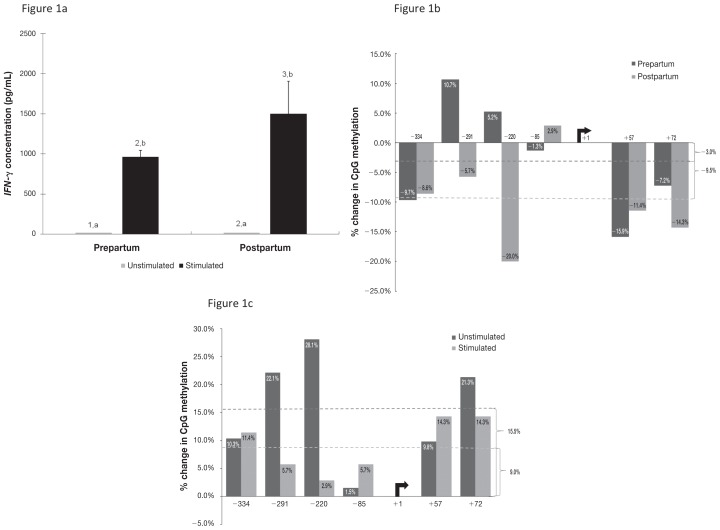
The isolated bovine CD4+ T-cells showed a significant increase in IFN-γ production upon ConA stimulation in both prepartum and postpartum samples (Figure 1a). The comparisons for peripartum effects were determined for ConA-stimulated cells only, as there was no detectable level of IFN-γ in the supernatant of the unstimulated CD4+ T-cells at 24 h. There was an overall increase in the concentration of IFN-γ in the supernatant of the ConA-stimulated CD4+ T-cells between the prepartum (mean 961 pg/mL) and postpartum (1498.7 pg/mL) blood samples, which were obtained from the same cows, and this difference approached significance (P = 0.08).
Figure 1.
Data for cultured CD4+ T-lymphocytes from enzyme-linked immunosorbent assay (ELISA) of the cytokine interferon gamma (IFN-γ) and from polymerase chain reaction and gel electrophoresis to determine DNA methylation of the cytokine’s gene promoter region before and after stimulation with the T-cell mitogen concanavalin A (ConA). Blood samples were collected from 5 dairy cows 4 wk before parturition (Prepartum) and 4 d after parturition (Postpartum). a) Concentration of IFN-γ in culture supernatant; significant differences between unstimulated and stimulated cells (P < 0.05) are indicated by different letters above the bars, and significant differences between the prepartum and postpartum periods (P < 0.001) are indicated by different numbers above the bars. b) Percentage change in methylation between the prepartum and postpartum periods. c) Percentage change in methylation after stimulation with ConA. For panels b and c, positive values represent a gain in methylation, whereas negative values represent a loss of methylation.
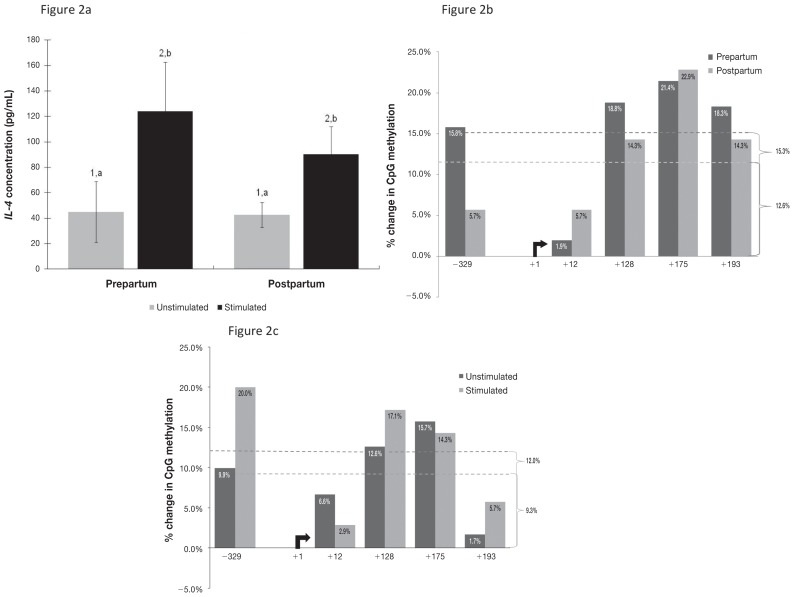
For IL-4 there was also an overall increase in production upon ConA stimulation in both prepartum and postpartum samples (Figure 2a). There was a higher concentration of IL-4 in the prepartum stimulated samples (mean 124 pg/mL) than in the postpartum stimulated samples (90.2 pg/mL), but this difference was not significant (P = 0.36).
Figure 2.
Corresponding data for the cytokine interleukin 4 (IL-4) and its gene promoter region.
Effect of parturition on DNA methylation of cytokine gene promoters
Upon stimulation with ConA, DNA methylation in the IFN-γ promoter region of isolated CD4+ T-cells decreased by 3.0% for prepartum samples and 9.5% for postpartum samples, as determined from the overall difference in the number of DNA-methylated sites divided by the total number of clones for each sample (Figure 1b). Overall, there was a 9.0% increase (P = 0.010) in DNA methylation from the prepartum to the postpartum period for stimulated cells and a 15.5% increase for unstimulated cells (P = 0.017). Although the change in methylation was uniform for each promoter and each treatment for many of the CpG sites, 3 of the 6 sites were noteworthy: site 2 [−291 bp from the transcription start site (TSS)], site 3 (−220 bp from the TSS), and site 4 (−85 bp from the TSS) (Figure 1c). As opposed to the general decrease in methylation upon stimulation, these sites increased in methylation, site 2 increasing 10.7% prepartum, site 3 increasing 5.3% prepartum, and site 4 increasing 2.8% postpartum (Figure 1c).
DNA methylation in the IL-4 promoter region increased upon ConA stimulation by 15.3% (P = 0.021) for prepartum samples and 12.6% (P = 0.017) for postpartum samples, as averaged for the 5 CpG sites in this location (Figure 2b). From the prepartum to the postpartum period there was an increase in methylation for both unstimulated (12.0%; P = 0.022) and stimulated (9.3%; P = 0.003) cells (Figure 2c). Unlike IFN-γ, IL-4 did not possess any sites in which methylation changed drastically. In addition, methylation increased more in the IFN-γ promoter region than in the IL-4 promoter region after parturition in both unstimulated and stimulated cells. Methylation differences were more marked in unstimulated cells for both promoters, with 15.5% and 12.0% increases for IFN-γ and IL-4, respectively, compared with 9.0% and 9.3%, respectively, in stimulated cells.
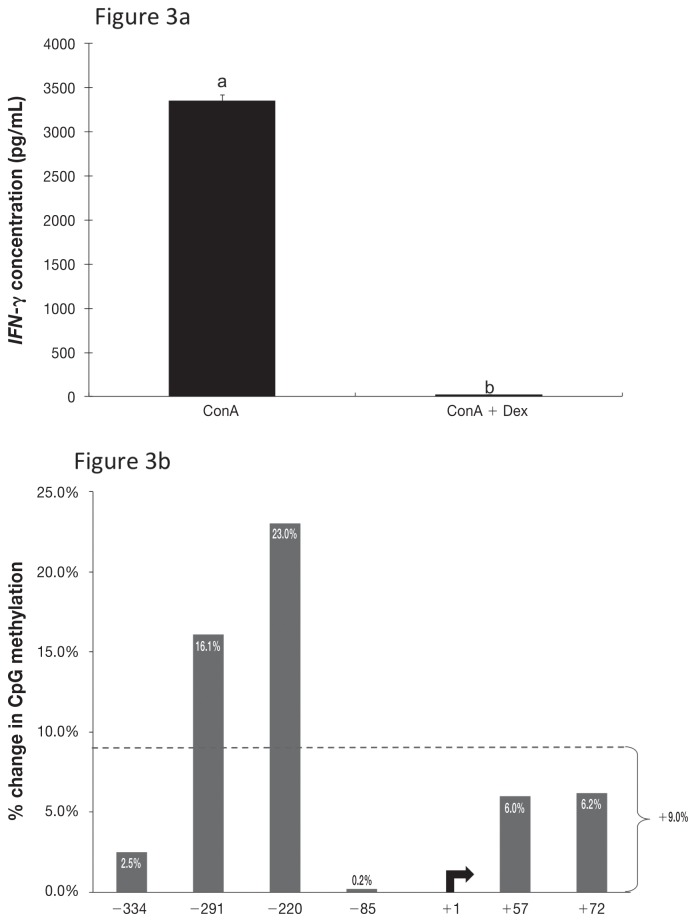
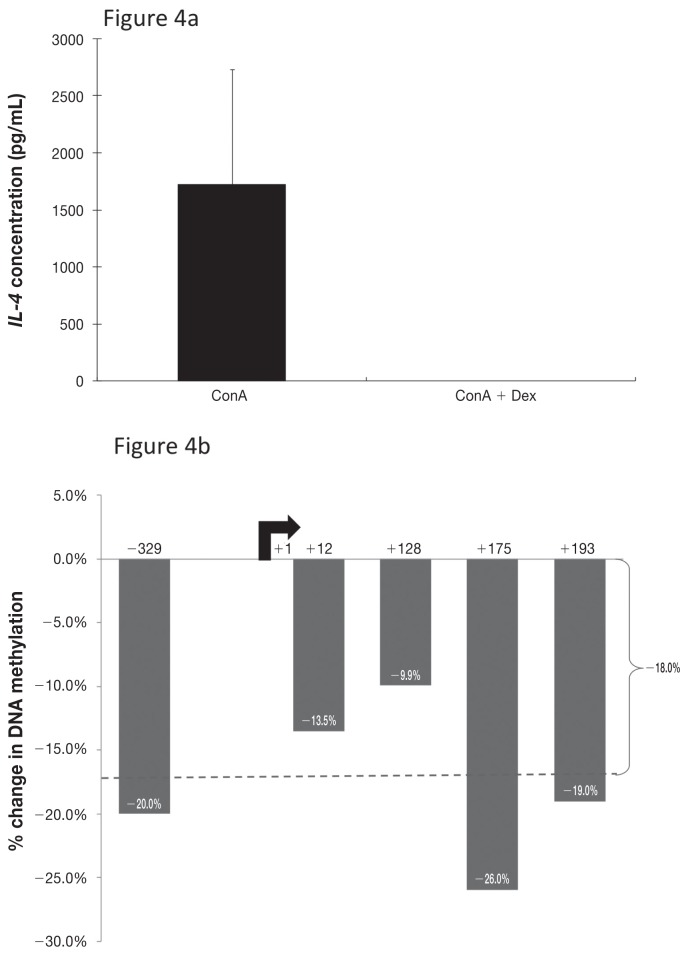
Effect of Dex on cytokine production
The unstimulated cells produced very little IFN-γ or IL-4 according to the results of ELISA of the supernatant from cultured cells. As expected, ConA stimulation caused a significant increase in cytokine production by cultured CD4+ T-cells, to mean concentrations of 3351 pg/mL for IFN-γ and 1726 pg/mL for IL-4. Treatment with 10 μM of Dex completely abrogated this effect of ConA, reducing the cytokine concentrations of both IFN-γ (P < 0.001) and IL-4 (P = 0.23) to nearly zero (Figures 3a and 4a).
Figure 3.
Corresponding data for IFN-γ and its gene promoter region in CD4+ T-lymphocytes isolated from blood collected from 3 dairy cows in mid-lactation (~100 d in milk). a) Concentration of IFN-γ in culture supernatant after stimulation with ConA alone or ConA plus the synthetic glucocorticoid dexamethasone (Dex); significant differences between the 2 treatments (P < 0.05) are indicated by different letters above the bars. b) Percentage change in methylation when Dex stimulation was added to ConA stimulation.
Figure 4.
Corresponding data for IL-4 and its gene promoter region.
Effect of Dex on DNA methylation of cytokine gene promoters
The addition of Dex resulted in a 9.0% increase in IFN-γ promoter methylation and an 18.0% decrease in IL-4 promoter methylation (Figures 3b and 4b) compared with ConA stimulation alone. Methylation patterns within the IFN-γ promoter region were consistent with the treatment effects on IFN-γ production, but this was not true for IL-4. The methylation profiles for IFN-γ and IL-4 were consistently inverse.
Discussion
Despite substantial implications for dairy production, the peripartum period remains relatively uncharacterized, particularly in terms of endocrine-associated immune regulation. Previously, GCs were shown to have immunosuppressive capabilities, the levels of these major regulatory hormones fluctuating around the time of parturition (15). Pregnancy is generally referred to as a “type 2 phenomenon,” as it is dominated by a type 2 IR bias through middle to late pregnancy in mice and humans (28–30). At parturition there is a shift to a type 1 IR bias, and in dairy cattle there may be a switch back to a type 2 dominance for a short period after parturition (8).
This was the first study, as far as the authors are aware, to initiate investigation of the possible epigenetic mechanism of DNA methylation on the function of cells of the bovine adaptive immune system, particularly during the peripartum period. IFN-γ and IL-4 production and DNA methylation of the gene promoters of these cytokines were used to investigate changes in the blood CD4+ T-cell populations of Holstein cows before and after parturition. IFN-γ and IL-4 were used as indicators of type 1 and type 2 IRs, respectively (8). Additionally, Dex treatment was administered to CD4+ T-cells isolated from nonparturient cows to simulate the effects of GC regulatory hormones that can occur in the peripartum period. The production of both IFN-γ and IL-4 increased upon ConA stimulation of CD4+ T-cells isolated from PBMCs in both prepartum and postpartum blood samples. The postpartum increase in IFN-γ production was not consistent with the previous observation in dairy cows of an apparent depression in IFN-γ levels of PBMCs and CD4+ T-cells isolated from samples taken around parturition (8,10). However, it was consistent with the previous findings in mice and humans of an overall type 1 bias around the time of parturition (28–30). The IL-4 levels in our study were much lower than the IFN-γ levels, and no difference in cytokine levels between the prepartum and postpartum periods was observed in the CD4+ T-cells. This contradicts the report of Shafer-Weaver et al (8) of an increase in IL-4 production on day 3 after calving, as determined by ELISA and reverse-transcription PCR. The discrepancy may be due to a difference in sampling periods: the previous investigators compared cows in mid- to late lactation with cows at day 3 after parturition, whereas the current study sampled cows 4 wk before calving (outside the peripartum period) and the same cows 4 d after parturition.
The results of the current study tend to strengthen the argument that pregnancy is dominated by a type 2 IR bias, with an increase in the production of a major type 1 cytokine (IFN-γ) at parturition and an increase in the production of a Th2 cytokine (IL-4) before calving, as in most mammalian species (28–30).
DNA methylation plays a crucial role in the regulation of cytokine gene expression: changes in DNA methylation patterns can either enable or repress gene expression (19). In this study changes in DNA methylation from the prepartum to the postpartum period were observed for the IFN-γ and IL-4 promoter regions. The regulator regions for bovine cytokine genes, specifically IFN-γ and IL-4, have not been established. ConA stimulation caused a general decrease in IFN-γ promoter methylation (3.0% prepartum, 9.5% postpartum), as was hypothesized and as was consistent with the increased cytokine production, and an overall increase in IL-4 promoter methylation (15.3% prepartum, 12.6% postpartum), which was not consistent with the increase in cytokine production. Methylation increased in both the IFN-γ and the IL-4 promoter regions from the prepartum to the postpartum period, by 9.0% for ConA-stimulated cells and 15.5% for unstimulated cells for IFN-γ and by 9.3% for stimulated cells and 12% for unstimulated cells for IL-4; the percentages were averaged over all CpGs for the stimulated and unstimulated cells. This finding is consistent with previously reported epigenetic regulation of T-cells by immunoregulatory cytokines, including IFN-γ (31).
Interestingly, CpG site 3 (−220 bp from the TSS) and site 4 (−85 bp from the TSS) within the IFN-γ promoter region are conserved putative binding sites for T-bet and CREB, respectively, and may be important for transcriptional regulation relative to the other CpGs analyzed. T-bet is the master regulator for Th1-cell differentiation and is essential for Th1-cell commitment (19). The conserved CREB binding site containing CpG 4 in dairy cows corresponds to the mouse IFN-γ promoter at −53 bp, which has been shown to determine IFN-γ transcriptional activity and the methylation of which inhibits CREB binding (32). Further investigation into the importance of these specific CpG sites and other enhancer regions to IFN-γ expression is needed for bovine CD4+ T-cells.
The results of this study were not necessarily consistent with production of both IFN-γ and IL-4 as hypothesized. This may be due to overriding treatment effects or epigenetic changes in other DNA regulatory regions for these cytokines. Subsequent bioinformatic analysis of the IL-4 promoter region during the current study suggested that the CpG sites selected for study did not possess any putative transcription-factor-binding motifs known to regulate IL-4. No CpG islands were predicted in the IL-4 region used in this study, which may explain the discordance between the IL-4 concentration data and the DNA methylation data for the IL-4 promoter. For example, Gata3, the master regulator for IL-4 locus transcription, does not bind specifically to the IL-4 promoter. According to results in other species, it binds to enhancers surrounding the IL-4 locus and is involved in regulating transcription of the IL-4, IL-5, and IL-13 loci (19,33–35). For future studies this region should be explored for DNA methylation patterns of putative transcription-factor-binding sites and their influence on the expression of IL-4, as the promoter of the IL-4 gene does not appear to play a role in direct transcriptional regulation.
The increase in production of IFN-γ and IL-4 caused by ConA stimulation was completely abrogated by treatment with Dex in vitro. This finding was similar to that in studies in vivo of the immunosuppressive effects of Dex on leukocyte function in Holstein cows (27,36). The expected Th2-promoting effect of Dex was not clearly demonstrated in the current study, as the secretion of IFN-γ was significantly reduced, as would be expected in Th2 response, there was also a reduction in IL-4 secretion, which was not expected in a Th2 promoting environment. In addition, treatment with Dex antagonized the effect of ConA stimulation and, as expected, increased IFN-γ promoter methylation (by 9%). IL-4 promoter methylation decreased by 18% upon Dex treatment of ConA-stimulated CD4+ cells, which was inconsistent with the observed decrease in IL-4 concentration in the culture supernatant.
In the current study the IFN-γ and IL-4 methylation patterns were inversely related. An inverse epigenetic relationship between the IFN-γ and IL-4 loci has been reported for other species (19). In general, the lack of correlation between DNA methylation and cytokine levels may be the result of numerous features, such as discordances between transcription of cytokine mRNA and secreted cytokine levels, the passive and the time dependent nature of DNA demethylation at the cytokine loci in T-cells, and the impact of enhancer and other extragenic regions on transcription, in which the DNA methylation status may be more influential. When comparing real-time PCR with ELISA data, especially for periods longer than 24 h, it is evident with mammalian cytokine loci such as IL-4 that mRNA levels may decrease rapidly, but secreted cytokine levels decrease much less or even increase, creating a discordance between transcriptional activity and secreted cytokine levels (37). Furthermore, demethylation at 5′ IL-4 loci is not required for early IL-4 transcript production, as shown in other species, meaning that the changes in the IL-4 promoter region may not be important for transcriptional activity (38). DNA demethylation, which is essential for T-cell development (17–19,32), is a passive process, relying on the semiconservative nature of DNA replication for the dispersal of hemimethylated DNA in dividing daughter cells (32,38). As such, cell division is important for lineage specificity, and at least 4 cell cycles may be required for CD4+ cell-line specificity in lineage-promoting conditions in vitro(39). Discordances between promoter methylation and cytokine secretion under Dex stimulation may be due to chromosomal activity that precedes DNA methylation. Specifically, GCs inhibit T-lymphocyte differentiation by inhibiting the production of lineage-specific transcription factors, cytokines, and receptors, albeit more potently for Th1 than for Th2 lineages (40). These considerations may explain not only the individual variation but also the discrepancy between total and site-specific promoter methylation and cytokine concentration in Dex treatment of CD4+ T-cells in vitro. Interestingly, though, IL-4 promoter methylation decreased in this study, whereas IFN-γ promoter methylation increased, a result consistent with a Th2 bias, as hypothesized. More specifically, Dex treatment mirrored the effect of parturition on the IFN-γ promoter, with an increase in methylation of 9.0% in both cases, although this similarity was not observed for IL-4.
This study has indicated that ConA and Dex treatment effects, and possibly parturition effects, on bovine CD4+ T-cells may be partially controlled through epigenetic modifications, specifically DNA methylation. Further work needs to be done to determine the effects of an individual inherent bias in type 1 or type 2 IR and how this can influence Th-cell populations around the time of parturition. It is also important to establish the exact DNA methylation profiles of key cytokine genes of naive T-cells and of Th1- and Th2-cells of the bovine species through site-specific bisulfite sequencing (32,41) or genome-wide analysis (42) in order to establish reference populations. Additionally, this study has provided evidence, similar to that from other species, that prepartum and postpartum dairy cows show differential cytokine secretion responses consistent with pregnancy having a type 2 IR bias that changes toward type 1 at parturition. Finally, treatment with Dex was able to reproduce the observed immunosuppressive effects on stimulated cells, although further characterization of its epigenetic mechanism is needed. This opens the door for the study of other major regulatory hormones, such as progesterone, oxytocin, and estrogen, and how they may contribute to our growing understanding of genetically and epigenetically regulated peripartum immune responsiveness.
Acknowledgments
The authors acknowledge the contributions of Sophia Lim, Heather Sanderse, and Salome Ndungu in technical support, William Sears in statistical analysis, and the Elora Research Station staff in assistance with animal handling. This research was financially supported by a grant from the Natural Sciences and Engineering Research Council of Canada to Dr. Mallard.
References
- 1.Sordillo LM, Contreras GA, Aitken SL. Metabolic factors affecting the inflammatory response of periparturient dairy cows. Anim Health Res Rev. 2009;10:53–63. doi: 10.1017/S1466252309990016. [DOI] [PubMed] [Google Scholar]
- 2.Huzzey JM, von Keyserlingk MAG, Weary DM. Changes in feeding, drinking, and standing behavior of dairy cows during the transition period. J Dairy Sci. 2005;88:2454–2461. doi: 10.3168/jds.S0022-0302(05)72923-4. [DOI] [PubMed] [Google Scholar]
- 3.Doepel L, Lapierre H, Kennelly JJ. Peripartum performance and metabolism of dairy cows in response to prepartum energy and protein intake. J Dairy Sci. 2002;85:2315–2334. doi: 10.3168/jds.S0022-0302(02)74312-9. [DOI] [PubMed] [Google Scholar]
- 4.Weber PS, Madsen SA, Smith GW, Ireland JJ, Burton JL. Pretranslational regulation of neutrophil L-selectin in glucocorticoid-challenged cattle. Vet Immunol Immunopathol. 2001;83:213–240. doi: 10.1016/s0165-2427(01)00381-6. [DOI] [PubMed] [Google Scholar]
- 5.Kimura K, Goff JP, Kehrli ME, Harp JA. Phenotype analysis of peripheral blood mononuclear cells in periparturient dairy cows. J Dairy Sci. 1999;82:315–319. doi: 10.3168/jds.S0022-0302(99)75238-0. [DOI] [PubMed] [Google Scholar]
- 6.Drackley JK. Biology of dairy cows during the transition period: The final frontier? J Dairy Sci. 1999;82:2259–2273. doi: 10.3168/jds.s0022-0302(99)75474-3. [DOI] [PubMed] [Google Scholar]
- 7.Kim IH, Na KJ, Yang MP. Immune responses during the peripartum period in dairy cows with postpartum endometritis. J Reprod Dev. 2005;51:757–764. doi: 10.1262/jrd.17036. [DOI] [PubMed] [Google Scholar]
- 8.Shafer-Weaver KA, Corl CM, Sordillo LM. Shifts in bovine CD4+ subpopulations increase T-helper2 compared with T-helper1 effector cells during the postpartum period. J Dairy Sci. 1999;82:1696–1706. doi: 10.3168/jds.S0022-0302(99)75399-3. [DOI] [PubMed] [Google Scholar]
- 9.Karcher EL, Beitz DC, Stabel JR. Modulation of cytokine gene expression and secretion during the periparturient period in dairy cows naturally infected with Mycobacterium avium subsp. paratuberculosis. Vet Immunol Immunopathol. 2008;123:277–288. doi: 10.1016/j.vetimm.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Ishikawa Y, Nakada K, Hagiwara K, Kirisawa R, Iwai H, Moriyoshi M, Sawamukai Y. Changes in interleukin-6 concentration in peripheral blood of pre- and post-partum dairy cattle and its relationship to postpartum reproductive diseases. J Vet Med Sci. 2004;66:1403–1408. doi: 10.1292/jvms.66.1403. [DOI] [PubMed] [Google Scholar]
- 11.Van Kampen CV, Mallard BA. Effects of peripartum stress and health on circulating bovine lymphocyte subsets. Vet Immunol Immunopathol. 1997;59:79–91. doi: 10.1016/s0165-2427(97)00069-x. [DOI] [PubMed] [Google Scholar]
- 12.Meglia GE, Johannisson A, Holtenius K, Waller KP. Effects of feeding intensity during the dry period on leukocyte. Vet J. 2005;169:376–384. doi: 10.1016/j.tvjl.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Ramirez F, Fowell D, Puklavec M, Simmonds S, Mason D. Glucocorticoids promote a Th2 cytokine response by CD4+ T cells in vitro. J Immunol. 1996;156:2406–2412. [PubMed] [Google Scholar]
- 14.De A, Blotta HM, Mamoni RL, et al. Effects of dexamethasone on lymphocyte proliferation and cytokine production in rheumatoid arthritis. J Rheumatol. 2002;29:46–51. [PubMed] [Google Scholar]
- 15.Preisler MT, Weber PSD, Tempelman RJ, Erskine RJ, Hunt H, Burton JL. Glucocorticoid receptor downregulation in neutrophils of periparturient cows. Am J Vet Res. 2000;61:14–19. doi: 10.2460/ajvr.2000.61.14. [DOI] [PubMed] [Google Scholar]
- 16.Karrow NA, Sharma BS, Fisher RE, Mallard BA. Epigenetics and animal health. In: Moo-Young M, Butler M, Webb C, et al., editors. Comprehensive Biotechnology. 2nd ed. Amsterdam: Elsevier; 2011. pp. 381–394. [Google Scholar]
- 17.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper cell differentiation. Nature Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 18.Reiner SL. Epigenetic control in the immune response. Hum Mol Genet. 2005;14:41–46. doi: 10.1093/hmg/ddi115. [DOI] [PubMed] [Google Scholar]
- 19.Sanderse H, Nino-Soto M, Mallard BA. Epigenetic influences on promoter regions of bovine interleukin-4 (IL4) and interferon-gamma (IFN-g) genes during peripartum period of dairy cows. Proceedings of the 2nd International Symposium on Animal Functional Genomics; Michigan State University, East Lansing, Michigan. May 16–19, 2006. [Google Scholar]
- 20.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–727. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 21.Shimaoka Y, Hidaka Y, Tada H, Nakamura T, Mitsuda N, Morimoto Y. Changes in cytokine production during and after normal pregnancy. Am J Reprod Immunol. 2000;44:143–147. doi: 10.1111/j.8755-8920.2000.440303.x. [DOI] [PubMed] [Google Scholar]
- 22.Marzi M, Vigano A, Trabattoni D, Villa ML, Salvaggio A, Clerici E. Characterization of type 1 and type 2 cytokine production profile in physiologic and pathologic human pregnancy. Clin Exp Immunol. 1996;106:127–133. doi: 10.1046/j.1365-2249.1996.d01-809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berthois Y, Katzenellenbogen J, Katzenellenbogen BS. Phenol red in tissue culture media is a weak estrogen: Implications concerning the study of estrogen-responsive cells in culture. Proc Natl Acad Sci U S A. 1986;83:2496–2500. doi: 10.1073/pnas.83.8.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tusnady GE, Simon I, Varadi A, Aranyi T. BiSearch: Primer-design and search tool for PCR on bisulfite-treated genomes. Nucleic Acids Res. 2005;33:e9. doi: 10.1093/nar/gni012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: Visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 26.Ovcharenko I, Loots GG, Giardine BM, et al. Mulan: Multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 2005;15:184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takai D, Jones PA. The CpG island searcher: A new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- 28.Aris A, Lambert F, Bessette P, Moutquin JM. Maternal circulating interferon-gamma and interleukin-6 as biomarkers of Th1/Th2 immune status throughout pregnancy. J Obstet Gynaecol Res. 2008;34:7–11. doi: 10.1111/j.1447-0756.2007.00676.x. [DOI] [PubMed] [Google Scholar]
- 29.Raghupathy R. Pregnancy: Success and failure within the Th1/Th2/Th3 paradigm. Semin Immunol. 2001;13:219–227. doi: 10.1006/smim.2001.0316. [DOI] [PubMed] [Google Scholar]
- 30.Lim KJ, Odukoya OA, Ajjan RA, Li TC, Weetman AP, Cooke ID. The role of T-helper cytokines in human reproduction. Fertil Steril. 2000;73:136–142. doi: 10.1016/s0015-0282(99)00457-4. [DOI] [PubMed] [Google Scholar]
- 31.Janson PCJ, Winerdal ME, Winqvist O. At the crossroads of T helper lineage commitment: Epigenetics points the way. Biochim Biophys Acta. 2009;1790:906–919. doi: 10.1016/j.bbagen.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Jones B, Chen J. Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J. 2006;25:2443–2452. doi: 10.1038/sj.emboj.7601148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee DU, Rao A. Molecular analysis of a locus control region in the T helper 2 cytokine gene cluster: A target for STAT6 but not GATA3. Proc Natl Acad Sci U S A. 2004;101:16010–16015. doi: 10.1073/pnas.0407031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and IL4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 35.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nonnecke BJ, Burton JL, Kehrli ME. Associations between function and composition of blood mononuclear leukocyte populations from Holstein bulls treated with dexamethasone. J Dairy Sci. 1997;80:2403–2410. doi: 10.3168/jds.S0022-0302(97)76192-7. [DOI] [PubMed] [Google Scholar]
- 37.Mena A, Ioannou XP, Van Kessel A, et al. Th1/Th2 biasing effects of vaccination in cattle as determined by real-time PCR. J Immunol Methods. 2002;263:11–21. doi: 10.1016/s0022-1759(02)00029-7. [DOI] [PubMed] [Google Scholar]
- 38.Lee DU, Agarwal S, Rao A. Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity. 2002;16:649–660. doi: 10.1016/s1074-7613(02)00314-x. [DOI] [PubMed] [Google Scholar]
- 39.Grogan JL, Mohrs M, Harmon B, Lacy DA, Sedat JW, Locksley RM. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity. 2001;14:205–215. doi: 10.1016/s1074-7613(01)00103-0. [DOI] [PubMed] [Google Scholar]
- 40.Liberman AC, Druker J, Perone MJ, Arzt E. Glucocorticoids in the regulation of transcription factors that control cytokine synthesis. Cytokine Growth Factor Rev. 2007;18:45–56. doi: 10.1016/j.cytogfr.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 41.Winders BR, Schwartz RH, Bruniquel D. Hypomethylated from early T cell development through mature naïve and Th1 cell differentiation, but is hypermethylated in Th2 cells. J Immunol. 2011;173:7377–7384. doi: 10.4049/jimmunol.173.12.7377. [DOI] [PubMed] [Google Scholar]
- 42.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nature Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]