Abstract
Aldehyde oxidases (EC 1.2.3.1) are a small group of structurally conserved cytosolic proteins represented in both the animal and plant kingdoms. In vertebrates, aldehyde oxidases constitute the small sub-family of molybdo-flavoenzymes, along with the evolutionarily and structurally related protein, xanthine oxidoreductase. These enzymes require a molybdo-pterin cofactor (molybdenum cofactor, MoCo) and flavin adenine dinucleotide for their catalytic activity. Aldehyde oxidases have broad substrate specificity and catalyse the hydroxylation of N-heterocycles and the oxidation of aldehydes to the corresponding acid. In humans, a single aldehyde oxidase gene (AOX1) and two pseudogenes clustering on a short stretch of chromosome 2q are known. In other mammals, a variable number of structurally conserved aldehyde oxidase genes has been described. Four genes (Aox1, Aox3, Aox4 and Aox3l1), coding for an equivalent number of catalytically active enzymes, are present in the mouse and rat genomes. Although human AOX1 and its homologous proteins are best known as drug metabolising enzymes, the physiological substrate(s) and function(s) are as yet unknown. The present paper provides an update of the available information on the evolutionary history, tissue- and cell-specific distribution and function of mammalian aldehyde oxidases.
Keywords: aldehyde oxidases, molybdo-enzymes, molybdenum cofactor, xanthine oxidase
Introduction
Aldehyde oxidases (EC 1.2.3.1) are proteins belonging to the family of molybdo- and tungsten-enzymes,[1] which is represented in both eukaryotes and prokaryotes. In mammals, no tungsten-containing proteins are known, while three other types of molybdo-enzymes, besides aldehyde oxidases -- that is, xanthine oxidoreductase (XOR), sulphite oxidase (SO) and the recently discovered mitochondrial amidoxime reductase component (mARC) -- have been described [2]. Mammalian catalytically active molybdo-enzymes require a specific form of organic molybdenum, known as the molybdenum cofactor (MoCo) [3]. Unlike that observed in SO and mARC, the holo-enzymatic forms of aldehyde oxidases and XORs contain a sulphurated species of MoCo [3-5].
Aldehyde oxidases and XORs are further sub-classified as molybdo-flavoenzymes (MFEs), if they require flavin adenine dinucleotide (FAD) as a cofactor. Both enzymes are homodimers, with each subunit (approximately 150 kDa) consisting of three distinct domains. The 25 kDa amino- terminal domain contains two non-identical iron/sulphur clusters, the 40 kDa intermediate domain consists of the FAD-binding region, while the 85 kDa carboxy-terminal domain is characterised by the presence of the substrate-binding pocket, which lies in close proximity to the MoCo site. One of the main differences between aldehyde oxidases and XORs is represented by a NAD+ binding site,[6] which is absent in aldehyde oxidases. XORs, in their dehydrogenase form, transfer the reducing equivalents generated by the oxidation of the substrate to NAD [7]. By contrast, aldehyde oxidases [8,9] and XORs, in their oxidase form, use molecular oxygen as the final electron acceptor, producing hydrogen peroxide.
The similarity between aldehyde oxidases and XORs is not limited to their general characteristics, and extends to the primary structure. The overall amino acid identity between aldehyde oxidases and XORs from the same animal species is approximately 40 per cent. In addition, the primary structures of plant, insect and vertebrate aldehyde oxidases show a remarkable degree of similarity to XORs of bacterial origin. This is in line with the idea that the two enzymes are evolutionarily conserved and originated through at least two asynchronous duplication events of the corresponding genes [1,8,10].
Some years ago, it was believed that the family of mammalian MFEs consisted of only two members -- XOR and a single aldehyde oxidase (annotated in the National Center for Biotechnology Information [NCBI] database as AOX1). The first complete amino acid sequence of a mammalian aldehyde oxidase was deduced from the molecular cloning of the corresponding bovine cDNA in our laboratory [11]. Based on the high level of amino acid similarity, it soon became apparent to us that this sequence corresponded to a human cDNA that had been originally identified as XOR [12]. Our original inference was subsequently confirmed by the cloning of the human AOX1 gene [13].
The first hint of the presence of multiple aldehyde oxidases in certain animal species came from an early analysis of the limited number of mouse expressed sequence tags (ESTs) available in the NCBI database. This allowed us to identify and isolate two other mouse cDNAs [14,15] encoding catalytically active proteins highly related to bovine, mouse and rat AOX1, the primary structure of which had been elucidated in the meantime [16-18]. In a subsequent study, we identified a fourth murine functional enzyme and demonstrated that rats are characterised by the same complement of four aldehyde oxidases as mice [19]. Each novel rodent aldehyde oxidase was shown to be the product of distinct loci, mapping with the original AOX1 orthologous genes to a small region (approximately 350 kilobases) of mouse chromosome 1 and rat chromosome 9. The three novel proteins present in rodents were named aldehyde oxidase homologues 1, 2 and 3 (AOH1, AOH2 and AOH3), and the corresponding genes were referred to as Aoh1, Aoh2 and Aoh3, respectively. The current nomenclature adopted by the NCBI is different, as Aoh1 is referred to as Aox3, while Aoh2 and Aoh3 have been renamed Aox4 and Aox3l1, respectively (Table 1). We will conform to this nomenclature throughout the paper, and refer to the various aldehyde oxidases present in different mammalian species as AOX1, AOX3, AOX4 and AOX3L1. The term 'aldehyde oxidase(s)' will be used in a general sense, whenever no distinction between the various isoforms of the family is meant.
Table 1.
Vertebrate aldehyde oxidase nomenclature.
| Organism | Gene and corresponding protein | ||||
|---|---|---|---|---|---|
| Common name | Specific name | Abbrev. | Gene symbol | Our original symbol | Accession no. |
| Human |
Homo sapiens |
Hs | AOX1 | AOX1 | NP_001150 |
| Chimpanzee |
Pan troglodytes |
Pt | AOX1 | AOX1 | XR_024408 |
| Orangutan |
Pongo pygmaeus |
Pp | AOX1 | AOX1 | ENSPTRP0000021853 |
| Rhesus monkey |
Macaca mulatta |
Mam | AOX1 | AOX1 | XP_001089327 |
| Rhesus monkey |
Macaca mulatta |
Mam | AOX4* | AOH2 | XP_001089798 |
| Rhesus monkey |
Macaca mulatta |
Mam | AOX3L1 | AOH3 | XP_001089912 |
| Macaque |
Macaca fascicularis |
Mf | AOX1 | AOX1 | AB201545 |
| Rabbit |
Oryctolagus cuniculus |
Oc | AOX1 | AOX1 | BAA81726 |
| Mouse |
Mus musculus |
Mm | Aox1 | Aox1 | NP_033806 |
| Mouse |
Mus musculus |
Mm | Aox3 | Aoh1 | NP_076106 |
| Mouse |
Mus musculus |
Mm | Aox4 | Aoh2 | NP_076120 |
| Mouse |
Mus musculus |
Mm | Aoxl | Aoh3 | NP_001008419 |
| Rat |
Rattus norvegicus |
Rn | Aox1 | Aox1 | NP_062236 |
| Rat |
Rattus norvegicus |
Rn | Aox3 | Aoh1 | NP_001008527 |
| Rat |
Rattus norvegicus |
Rn | Aox4 | Aoh2 | NP_001008523 |
| Rat |
Rattus norvegicus |
Rn | Aoxl1 | Aoh3 | NP_001008522 |
| Dog | Canis lupus familiaris | Cf | AOX4 | AOH2 | DQ150104 |
| Dog | Canis lupus familiaris | Cf | AOXRL1 | AOH3 | DQ150105 |
| Cow | Bos taurus | Bt | AOX1 | AOX1 | NP_788841 |
| Cow | Bos taurus | Bt | AOX4 | AOH2 | XP_596585 |
| Cow | Bos taurus | Bt | AOX3L1 | AOH3 | XP_610199 |
| Opossum | Monodelphis domestica | Md | AOX1 | AOX1 | XP_0011379598 |
| Opossum | Monodelphis domestica | Md | AOX3 | AOH1 | XP_0011379605 |
| Opossum | Monodelphis domestica | Md | AOX4 | AOH2 | XP_0011379630 |
| Opossum | Monodelphis domestica | Md | AOX3L1 | AOH3 | XP_0011370277 |
| Chicken | Gallus gallus | Gg | AOX1 | AOX1 | DQ150102 |
| Chicken | Gallus gallus | Gg | AOH | AOH | DQ150103 |
| Zebra finch | Taeniopygia guttata | Tg | AOX1 | AOX1 | XP_002191075 |
| Fugu | Takifugu rubripes | Tr | AOX1 | AOX1 | SINFRUP000000131133 |
| Pufferfish | Tetraodon nigroviridis | Tn | AOX1 | AOX1 | GSTENP00031889001 |
| Zebrafish | Danio rerio | Dr | AOX1 | AOX1 | XP_699030 |
The table lists the proteins mentioned in this paper. The names of the different organisms are shown on the left. The accession numbers of the proteins or predicted translation products of the corresponding genes present in the GenBank or Ensembl databases are indicated in the rightmost column. The official or proposed gene symbol ('Gene symbol') along with the acronym originally suggested in our previous references ('Our original symbol') are also indicated.
*At present, it is unclear whether Macaca mulatta AOX4 is a functional gene or a pseudogene.
Evolution of aldehyde oxidases
The dendrogram shown in Figure 1 summarises our current knowledge of the phylogenesis of aldehyde oxidases. The primary structure of these proteins is relatively well conserved throughout evolution, and the most primitive organism showing evidence for one such enzyme is the flat worm, Caenorhabditis elegans.
Figure 1.
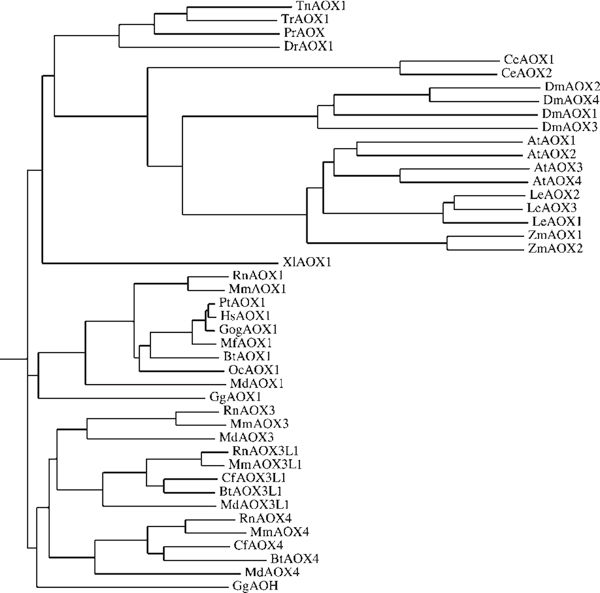
Phylogeny of eukaryotic aldehyde oxidases. A rooted dendrogram was obtained by the Phylip method after CLUSTAL-W computer-aided alignment of the indicated proteins. Poere AOX1 = Poecilia reticulata (guppy) AOX1; Coeel AOX1/AOX2 = Caenorhabditis elegans AOX1 and AOX2; Drome AOX1/AOX2/AOX3/AOX4 = Drosophila melanogaster AOX1, AOX2, AOX3 and AOX4; Arath AOX1/AOX2/AOX3/AOX4 = Arabidopsis thaliana AOX1, AOX2, AOX3 and AOX4; Lyces AOX1/AOX2/AOX3 = Lycopersicon esculentum (tomato) AOX1, AOX2 and AOX3; Zeama AOX1/AOX2 = Zea mays AOX1 and AOX2; Xenla AOX1 = Xenopus laevis AOX1; Gorgo AOX1 = Gorilla gorilla AOX1. All other abbreviations are as in Table 1. At present, it is unclear whether Macaca mulatta AOX4 is a functional gene product.
Bona fide aldehyde oxidase homologous proteins do not seem to be represented in prokaryotes, although many molybdenum and tungsten enzymes with low levels of amino acid similarity to vertebrate aldehyde oxidases are known (eg see Blase' et al.[20]). This is at variance with what has been observed for XOR, the ancestors of which can be easily traced back to prokaryotes and very primitive eukaryotes, like Aspergillus nidulans [21].
As already discussed, aldehyde oxidases and XORs are highly related proteins and have a common origin. Aldehyde oxidases are indeed the products of duplication events from an ancestral eukaryotic XOR gene; however, two structurally different families of aldehyde oxidases can be recognised on the basis of the amino acid sequences (Figure 1). A first cluster consists of nematode, insect and plant enzymes, while a second group contains all vertebrate aldehyde oxidases. Vertebrate aldehyde oxidases are closer to XORs than to aldehyde oxidases from more primitive animals and plants [8]. This finding is suggestive of two separate and evolutionarily divergent duplication events from an ancestral XOR gene. The first event produced the aldehyde oxidase precursor to worm, insect and plant enzymes. The second led to the appearance of the vertebrate counterpart. This is supported by the intron/exon structure of all the vertebrate aldehyde oxidase and XOR genes, which are strikingly conserved and much more complex than those of homologues from the lower eukaryotes and plants [1,8]. The two original duplications were followed by a number of other such events that led to the extant complement of aldehyde oxidase genes in the plant and animal kingdoms.
The evolution of vertebrate aldehyde oxidases (Figure 2) is characterised by a first phase of asynchronous gene multiplication events, which started in certain birds. Fishes (Danio rerio) and amphibians (Xenopus laevis) are endowed with a single functional aldehyde oxidase gene.
Figure 2.
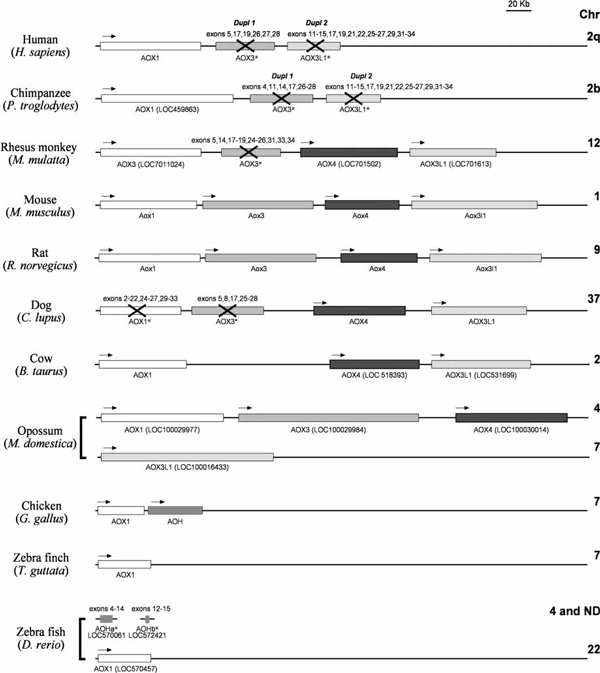
Aldehyde oxidase genes in vertebrates. The figure contains a schematic representation of the aldehyde oxidase genes in selected vertebrates for which complete or almost complete genomic sequence data are available. Orthologous genes are indicated with the same shadowing. The direction of transcription is indicated by arrows. Pseudogenes are crossed through and asterisked. The exons identified in the pseudogenes are also shown. The chromosomal location is indicated on the right (ND, not determined). Whenever the structure of the gene is predicted solely on the basis of the genomic sequence, and the corresponding cDNAs have not been characterised, the GenBank locus number (LOC) is indicated. At present, it is unclear whether Macaca mulatta AOX4 is a functional gene or a pseudogene.
The corresponding gene products show the highest level of amino acid identity to the rodent AOX1 isoenzyme, supporting an orthologous relationship. D. rerio or X. laevis AOX1 has the conserved 35/36 exon structure typical of all vertebrate aldehyde oxidase genes. In addition, fish AOX1 and XOR map to different chromosomes, which is also characteristic of all vertebrates except primates. Some avians (chicken) show evidence of a gene duplication event involving AOX1 and resulting in the production of a new synthenic gene (on chromosome 7), which we named aldehyde oxidase homologue (AOH) [22]. AOH is characterised by an identical exon/intron structure, with perfect conservation of exon/intron junctions along the coding sequence. The amino acid sequences of chicken AOX1 and AOH protein products are approximately 60 per cent identical and only more distantly related to the corresponding XOR enzyme (40 per cent identity). The presence of two aldehyde oxidase genes in birds does not seem to be a general phenomenon. In fact, a BLAST search indicates that the zebra finch, Taeniopygia guttata, has a single AOX1 locus on chromosome 7 (Table 1 and Figure 2).
Moving up along the evolutionary ladder to marsupials (Monodelphis domestica, opossum), whole-genome sequencing data provide evidence of four functionally active aldehyde oxidase genes. The exon-intron structure of all the genes is strictly conserved, indicative of two further gene duplication events. Three of the loci (Aox1, Aox3 and Aox4) map to chromosome 1, whereas the fourth gene (Aox3l1) is located on chromosome 7. The genes identified in M. domestica are the orthologues of the four loci present in rodents (mouse and rats).
The evolutionary process of the aldehyde oxidase gene cluster in mammals is characterised by a sudden and species-specific shift from multiplication to suppression/deletion. Bos taurus (cow) seems to have maintained three active aldehyde oxidase genes (AOX1, AOX4 and AOX3L1) on chromosome 2. The absence of nucleotide sequences with similarity to AOX3 strongly suggests that this gene has been deleted. Deletion of the AOX3 gene seems to be a conserved feature in another herbivore, the horse, although our present view of the aldehyde oxidase cluster in this animal species is still incomplete. Functional inactivation of AOX3 seems to be a common theme. The genome of dogs is characterised by two seemingly active AOX4 and AOX3L1 loci and two inactive AOX1 and AOX3 pseudogenes clustering on chromosome 37. The vestiges of numerous exons with nucleotide similarity to the rodent Aox1 and Aox3 genes are easily identified on two separate regions slightly upstream of the dog AOX4 and AOX3L1 loci. It is interesting to notice that the dog is currently the only mammalian species that seems to be lacking AOX1, in addition to AOX4. This observation has important implications, as this mammal is devoid of aldehyde oxidase activity in the liver [22].
Humans are endowed with a single functional aldehyde oxidase gene, namely AOX1, consisting of the canonical 35 conserved coding exons. This is the result of the persistence of the AOX3 deletion and the simultaneous transformation of AOX4 and AOX3L1 into inactive, albeit transcribed, pseudogenes. AOX1 and the two pseudogenes map to a short segment on chromosome 2q. Functional inactivation of the AOX4 and AOX3L1 genes occurred before the appearance of the human species, as chimpanzees (Pan troglodytes) are endowed with the same complement of aldehyde oxidase genes and pseudogenes as humans. Functional suppression of AOX4 and AOX3L1 seems to be the result of two recent, distinct and asynchronous events, based on the results obtained in the old-world monkeys (Macaca fascicularis and Macaca mulatta). In fact, the genomes of these monkeys seem to contain two functional (AOX1 and AOX3L1) as well as one or two inactive (AOX3 and AOX4) pseudogenes clustering on chromosome 12 (M. Terao, unpublished result).
In summary, two phylogenetically distinct branches of the evolutionary process led to the extant complement of worm/insect/plant and vertebrate aldehyde oxidases through an equivalent number of primitive and distinct gene duplication events involving the XOR ancestor. These initial events were followed by a divergent series of species-specific and more recent gene multiplication events that occurred not only in vertebrates but also in plants, worms and insects. Maize, tomato and Arabidopsis thaliana are respectively characterised by two, three and four structurally very similar aldehyde oxidase genes [8]. Similarly, the flatworm, C. elegans, and the fruitfly, Drosophila melanogaster, are endowed, respectively, with two and four aldehyde oxidases [8].
The most ancient member of the vertebrate aldehyde oxidase family is AOX1. AOX1 was duplicated into AOH. At present, the evolutionary order of appearance of AOX3, AOX4 and AOX3L1 is unknown; however, a number of considerations led us to propose that AOX4 is more ancient than AOX3, which, in turn, appeared earlier than AOX3L1 [8].
Tissue-specific expression of aldehyde oxidases in humans, primates and rodents
The data available on the tissue distribution of human AOX1 are limited [23-25]. The richest source of the enzyme is the liver, however, where AOX1 serves an important role in the metabolism of xenobiotics. AOX1 is also present in the respiratory system, where it can be identified in the epithelial cells lining the trachea and bronchi, as well as in the alveolar cells of the lung. In the digestive system, AOX1 was identified in the epithelia of the small and large intestine. The kidney is another source of AOX1, with synthesis of the protein occurring in the proximal distal and collecting tubules. Finally, the prostate and adrenal glands seem to contain detectable amounts of the protein. The localisation of AOX1 in the central nervous system is largely unknown, although it was reported that the corresponding transcript is expressed in the glial component of the anterior horns of the spinal cord [25]. In general, the tissue distributions of AOX1 in humans and baboons, the only other primate for which studies are available, are concordant, particularly in relation to the presence of the enzyme in the livers and the respiratory systems [26].
Two mouse aldehyde oxidases (AOX1 and AOX3) have an overlapping tissue distribution, which is largely superimposable with that of human AOX1. While this is expected in the case of AOX1, given its orthologous relationship with the human enzyme, the observation is important in the case of AOX3. By far the richest source of the two enzymes is the liver, where AOX1 and AOX3 are present exclusively in the cytosolic fraction of the hepatocyte compartment [15,27]. Different mouse strains are characterised by different relative amounts of the two hepatic enzymes, however [28]. High levels of AOX3 and much lower levels of AOX1 are present in the outbred CD1 and the inbred C57BL/6J strains. By contrast, the two inbred strains DBA/2 and CBA have an almost complete deficit of expression of AOX3. This deficit is the result of epigenetic silencing of the corresponding gene by methylation of the regulatory sequences [28]. Significant expression of both Aox1 and Aox3 is observed also in the lung, which is the second richest source. Limited amounts of AOX1 and AOX3 are also believed to be present in the central nervous system, on the basis of in situ hybridisation experiments [29]http://www.brainatlas.org/aba/. Expression is limited to the choroid plexus, which is the organ devoted to the production and re-absorption of the cephalorachidian fluid, and the motor neuronal cell population in the brain and spinal cord. While cellular co-localisation of AOX3 in the choroid plexus is clear, similar evidence is not available for the motor neurones. Low levels of AOX1 and/or AOX3 are also inferred from the expression data available in the Online Mendelian Inheritance in Man (OMIM) section of the NCBI website http://http./www.ncbi.nlm.nih.gov/. Of particular interest is the presence of AOX1 in the skin (which was recently confirmed),[27] the mammary gland and the genitourinary tract.
The expression of mouse AOX4 and AOX3L1 is much more restricted. The richest source of AOX4 is the Harderian gland,[27] a prominent intra-orbital exocrine gland involved in diverse homeostatic functions, such as thermoregulation, lubrication of the eye surface, control of pheromonal cues and regulation of the circadian rhythm [30-32]. The gland is present in various vertebrate species, but is absent in humans and primates. Far lower amounts of AOX4 have also been identified in the epidermal layer of the skin and the keratinised epithelia lining the oral cavity, oesophagus and proximal portion of the stomach. In the skin, the epidermis is not the only structure containing AOX4, as sebaceous glands are also enriched in the corresponding mRNA [27]. Interestingly, sebaceous and Harderian glands share common characteristics, producing a lipid-rich secretion, which is involved in thermoregulation and pheromone release.
Finally, AOX3L1 is unique, being limited to the secretory cells of the Bowman's gland, the most important exocrine gland of the olfactory mucosa [19]. These structures are present in all vertebrates, including humans, with the exception of fish. The serous/mucous product of Bowman's glands is important in shaping the microenvironment necessary for a proper stimulation of the nasal neuroepithelium by odorants. Bowman's glands are so rich in AOX3L1 that they were used as the source for the purification of the enzyme to homogeneity [19].
Exogenous and endogenous substrates of mammalian aldehyde oxidases
Mammalian aldehyde oxidases are best known as drug metabolising enzymes. Most of the literature on aldehyde oxidases has focused on this aspect of their biology [33-38]. All the members of the family are characterised by broad substrate specificity, oxidising numerous types of molecule. In spite of their name, aldehyde oxidases catalyse not only the oxidation of aldehydes into the corresponding carboxylic acid, but they also hydroxylate N-heterocyclic molecules with high efficiency. Human AOX1 and rodent AOX1/AOX3 may actually represent the major cytosolic enzymes metabolising xenobiotics in the liver. The enzymes are involved in the phase I metabolism of numerous compounds of both medical and toxicological relevance, potentially acting in concert with the microsomal cytochrome P450 system [33-39]. Extensive discussion of the compounds metabolised by human or rodent aldehyde oxidases is beyond the scope of this paper, and the reader is referred to specific reviews for more details [34,40,41]. It is worth mentioning that aldehyde oxidases have an important role in the metabolism of the anti-tumour and immunosuppressive agents, methotrexate,[42] 6-mercaptopurine and azathioprine,[43,44] and aldophosphamide, the active metabolite of cyclophosphamide [45,46]. Aldehyde oxidases have also been reported to metabolise the antimalarial agent, quinine [47] and the anti-viral drug famcyclovir [48]. Finally, human AOX1 is known to cause the inactivation of the hypnotic, zaleplon [49,50]. As evident from this short and incomplete list, the drug metabolising activity of aldehyde oxidases is predominantly related to the ability of these enzymes to hydroxylate N-heterocyclic rings. From a toxicological perspective, the aldehyde oxidase activity present in mammalian liver has been reported to play a major role in the oxidation of environmental pollutants such as phthalazines [8]. By contrast, it is unlikely that mouse AOX1 and AOX3 are of any relevance for the oxidation of the ethanol metabolite, acetaldehyde, into acetic acid [28].
Although a wealth of data are available on exogenous substrates, endogenous substrates of aldehyde oxidases are still being sought. The Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.kegg.jp) contains a number of metabolic pathways listing potential physiological substrates of aldehyde oxidases, such as the serotonin metabolite, 5-hydroxyindoleacetaldehyde (pathway: ko00380), and the amino acid catabolites (S)-methylmalonate semi-aldehyde (pathway: ko00280) and gentisate aldehyde (pathway: ko00350). At present, however, there is no direct evidence of the significance of human AOX1 or any of the other mammalian homologues in the oxidation of these substrates. Aldehyde oxidases are also purported to play a role in the metabolism of vitamins B3 (nicotinamide), B6 (pyridoxal phosphate) and A (retinol). The vitamin B3 metabolite, N1-methylnicotinamide is a good substrate for semi-purified human and monkey AOX1 preparations [51]. Pyridoxal, the vitamin B6 precursor, is oxidised to 4-pyridoxic acid by the human enzyme [52] and by the two mouse liver aldehyde oxidases, AOX1 and AOX3 (Terao M., unpublished results) [27]. Pyridoxal is currently the sole example of a substrate that shows a certain degree of selectivity for a specific aldehyde oxidase protein as it is not recognised by purified mouse AOX4 [27].
Vitamin A metabolism is probably the endogenous pathway for which more stringent information on the involvement of aldehyde oxidases is available. The ability of mammalian aldehyde oxidases to catalyse the oxidation of retinaldehyde (a physiological precursor) into retinoic acid (the active metabolite of vitamin A) was discovered in rabbit liver cytosol [53] and confirmed using purified preparations of mouse liver AOX1 [28,54]. In our hands, retinaldehyde has been one of the best substrates not only of mouse AOX1, but also of AOX3, AOX4 and AOX3L1 [14,19,27]. The Km and Vmax measured for the purified forms of AOX1, AOX3 and AOX4 compare well [8] with the same parameters reported for the three aldehyde dehydrogenases, ALDH1A1, ALDH1A2 and ALDH1A3,[55,56] which have long been known to catalyse the oxidation of retinaldehyde and to play a critical role in the morphogenetic activity of retinoic acid during the development of the vertebrate embryo [57]. Support for the relevance of AOX4 for the local synthesis and accumulation of retinoic acid in the Harderian gland has recently been provided by the phenotypic, genetic and biochemical characterisation of the first aldehyde oxidase knockout mouse to be generated [27].
Conclusion and future perspectives
Although a substantial amount of information on the structure and evolution of mammalian aldehyde oxidases is available, little is known about the physiological function of these enzymes in mammals, with particular reference to humans and relevant animal models. Progress in establishing the significance of this class of enzymes for the homeostasis of the mammalian organism is clearly a priority.
To this end, it will be necessary to identify the endogenous substrate(s) of human AOX1 and all the other mammalian isoenzymes. This is likely to require an integrated approach based on the definition of the substrate(s) using purified enzyme preparations, and validation of the results in vivo. It is envisaged that in vivo studies in humans will require the measurement of relevant molecules in biological fluids,[58] using cohorts of individuals genotyped for the presence of AOX1 allelic variants with alterations in the catalytic activity of the corresponding protein products. It will therefore be important to increase the number of population studies [59] designed to discover new AOX1 single nucleotide polymorphisms (SNPs) (see, for instance, our reference rs41309768, in the Single Nucleotide Polymorphism Database [dbSNP] section of the NCBI database). Similar studies will also have to be performed in experimental animals, with the caveat that mice and rats are characterised by the presence of multiple aldehyde oxidase forms, as discussed above. In this context, strains of mice characterised by a different profile of aldehyde oxidases,[28] as well as specific knockout animals,[27] will represent useful tools to this end. These knockout mouse lines would be great candidates for urinary metabolite profile analysis. UPLC/Q-tof-MS/MS, followed by chemometrics and analysis of principle components, would very likely identify one or more metabolic pathways that are perturbed in the absence of each one of the mouse AOX enzymes.
Association studies aimed at defining the relevance of AOX1 for the aetio-pathogenesis and progression of specific human diseases are also needed, as they may provide insights into the functional significance of the enzyme. These studies will integrate the knowledge that is likely to be generated from the phenotypic analysis of the genetically engineered mice that are already available or will be available in the near future. Along these lines, our studies on the Aox4 knockout mouse have already provided evidence that the corresponding protein is involved in the control of skin homeostasis and in the postnatal development of the Harderian gland. Whether the two phenomena are linked to a decrease in the local metabolism of retinaldehyde to retinoic acid is still a matter of speculation. Further knowledge is likely to be gained soon by phenotypic analysis of the newly generated Aox3l1 knockout animal (M. Terao, unpublished results). Nevertheless, the data already gathered strongly suggest that the various aldehyde oxidases are unlikely to play a vital function in the homeostasis of the mouse, as both types of knockout animal develop normally and are fertile.
Three final considerations are worth making. The first two concern, again, the basic problem of the physiological significance of aldehyde oxidases. In this regard, a key point relates to the reason(s) why the evolution of vertebrate organisms is associated with a first wave of multiplication and a subsequent phase of deletion/suppression of the aldehyde oxidase genes. These phenomena are likely to be related to the necessity for developing specific and possibly tissue-related functions in certain animal species, which must have become dispensable in humans and primates. The idea is readily acceptable for an enzyme like AOX4, which is highly enriched in the Harderian gland, as the structure is absent in humans and primates. It may be also viable in the case of AOX3l1, if the protein serves a function in the recognition of odorants. Indeed, it is well known that olfaction is much more developed and sophisticated in rodents than in humans. The presence of an extra enzyme (AOX3) in the liver of the mouse, relative to the human, is more complicated to explain. Nevertheless, in generating hypotheses as to the real function of the various aldehyde oxidases, an open mind should be always kept, since the function of these proteins may not necessarily be related to their enzymatic activity [60,61]. The third consideration is of a more practical nature and is relevant for the role exerted by aldehyde oxidases in drug metabolism. With respect to this, it is clear from what has been reported here that caution should be exercised in using rodents, dogs and possibly Rhesus monkeys as good proxies for the human situation.
Acknowledgements
This work was made possible by unrestricted grants from the Fondazione Monzino, the Associazione Italian per la Ricerca contro il Cancro (AIRC) and the Fondo di Investimento per la Ricerca di Base (FIRB). We would also like to thank Mr F. Deceglie and Mr A. Soave for the artwork.
References
- Garattini E, Mendel R, Romao MJ, Wright R. et al. 'Mammalian molybdo-flavoenzymes, an expanding family of proteins: Structure, genetics, regulation, function and pathophysiology'. Biochem J. 2003;372:15–32. doi: 10.1042/BJ20030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havemeyer A, Bittner F, Wollers S, Mendel R. et al. 'Identification of the missing component in the mitochondrial benzami- doxime prodrug-converting system as a novel molybdenum enzyme'. J Biol Chem. 2006;281:34796–34802. doi: 10.1074/jbc.M607697200. [DOI] [PubMed] [Google Scholar]
- Mendel RR, Bittner F. 'Cell biology of molybdenum'. Biochim Biophys Acta. 2006;1763:621–635. doi: 10.1016/j.bbamcr.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Wollers S, Heidenreich T, Zarepour M, Zachmann D. et al. 'Binding of sulfurated molybdenum cofactor to the C-terminal domain of ABA3 from Arabidopsis thaliana provides insight into the mechanism of molybdenum cofactor sulfuration'. J Biol Chem. 2008;283:9642–9650. doi: 10.1074/jbc.M708549200. [DOI] [PubMed] [Google Scholar]
- Bittner F, Oreb M, Mendel RR. 'ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana'. J Biol Chem. 2001;276:40381–40384. doi: 10.1074/jbc.C100472200. [DOI] [PubMed] [Google Scholar]
- Nishino T, Nishino T, Schopfer LM, Massey V. 'Reactivity of chicken liver xanthine dehydrogenase containing modified flavins'. J Biol Chem. 1989;264:6075–6085. [PubMed] [Google Scholar]
- Nishino T, Okamoto K, Eger BT, Pai EF. 'Mammalian xanthine oxidoreductase -- Mechanism of transition from xanthine dehydrogenase to xanthine oxidase'. FEBS J. 2008;275:3278–3289. doi: 10.1111/j.1742-4658.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- Garattini E, Fratelli M, Terao M. 'Mammalian aldehyde oxidases: Genetics, evolution and biochemistry'. Cell Mol Life Sci. 2008;65:1019–1048. doi: 10.1007/s00018-007-7398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann S, Terao M, Garattini E, Saggu M. et al. 'Site directed mutagenesis of amino acid residues at the active site of mouse aldehyde oxidase AOX1'. PLoS One. 2009;4:e5348. doi: 10.1371/journal.pone.0005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Trelles F, Tarrio R, Ayala FJ. 'Convergent neo-functionalization by positive Darwinian selection after ancient recurrent duplications of the xanthine dehydrogenase gene'. Proc Natl Acad Sci USA. 2003;100:13413–13417. doi: 10.1073/pnas.1835646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzi ML, Raviolo C, Ghibaudi E, de Gioia L. et al. 'Purification, cDNA cloning, and tissue distribution of bovine liver aldehyde oxidase'. J Biol Chem. 1995;270:31037–31045. doi: 10.1074/jbc.270.52.31037. [DOI] [PubMed] [Google Scholar]
- Wright RM, Vaitaitis GM, Wilson CM, Repine TB. et al. 'cDNA cloning, characterization, and tissue-specific expression of human xanthine dehydrogenase/xanthine oxidase'. Proc Natl Acad Sci USA. 1993;90:10690–10694. doi: 10.1073/pnas.90.22.10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao M, Kurosaki M, Demontis S, Zanotta S. et al. 'Isolation and characterization of the human aldehyde oxidase gene: Conservation of intron/exon boundaries with the xanthine oxidoreductase gene indicates a common origin'. Biochem J. 1998;332:383–393. doi: 10.1042/bj3320383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao M, Kurosaki M, Marini M, Vanoni MA. et al. 'Purification of the aldehyde oxidase homolog 1 (AOH1) protein and cloning of the AOH1 and aldehyde oxidase homolog 2 (AOH2) genes. Identification of a novel molybdo-flavoprotein gene cluster on mouse chromosome 1'. J Biol Chem. 2001;276:46347–46363. doi: 10.1074/jbc.M105744200. [DOI] [PubMed] [Google Scholar]
- Terao M, Kurosaki M, Saltini G, Demontis S. et al. 'Cloning of the cDNAs coding for two novel molybdo-flavoproteins showing high similarity with aldehyde oxidase and xanthine oxidoreductase'. J Biol Chem. 2000;275:30690–30700. doi: 10.1074/jbc.M005355200. [DOI] [PubMed] [Google Scholar]
- Kurosaki M, Demontis S, Barzago MM, Garattini E. et al. 'Molecular cloning of the cDNA coding for mouse aldehyde oxidase: Tissue distribution and regulation in vivo by testosterone'. Biochem J. 1999;341:71–80. doi: 10.1042/0264-6021:3410071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis S, Kurosaki M, Saccone S, Motta S. et al. 'The mouse aldehyde oxidase gene: Molecular cloning, chromosomal mapping and functional characterization of the 5'-flanking region'. Biochim Biophys Acta. 1999;1489:207–222. doi: 10.1016/S0167-4781(99)00174-8. [DOI] [PubMed] [Google Scholar]
- Wright RM, Clayton DA, Riley MG, McManaman JL. et al. 'cDNA cloning, sequencing, and characterization of male and female rat liver aldehyde oxidase (rAOX1). Differences in redox status may distinguish male and female forms of hepatic APX'. J Biol Chem. 1999;274:3878–3886. doi: 10.1074/jbc.274.6.3878. [DOI] [PubMed] [Google Scholar]
- Kurosaki M, Terao M, Barzago MM, Bastone A. et al. 'The aldehyde oxidase gene cluster in mice and rats. Aldehyde oxidase homologue 3, a novel member of the molybdo-flavoenzyme family with selective expression in the olfactory mucosa'. J Biol Chem. 2004;279:50482–50498. doi: 10.1074/jbc.M408734200. [DOI] [PubMed] [Google Scholar]
- Bruntner C, Tshisuaka B, Fetzner S. et al. 'Cloning, expression, and sequence analysis of the three genes encoding quinoline 2-oxidoreductase, a molybdenum-containing hydroxylase from Pseudomonas putida 86'. J Biol Chem. 1996;271:23068–23079. doi: 10.1074/jbc.271.38.23068. [DOI] [PubMed] [Google Scholar]
- Glatigny A, Scazzocchio C. 'Cloning and molecular characterization of hxA, the gene coding for the xanthine dehydrogenase (purine hydroxylase I) of Aspergillus nidulans'. J Biol Chem. 1995;270:3534–3550. doi: 10.1074/jbc.270.8.3534. [DOI] [PubMed] [Google Scholar]
- Terao M, Kurosaki M, Barzago MM, Varasano E. et al. 'Avian and canine aldehyde oxidases. Novel insights into the biology evolution of molybdo-flavoenzymes'. J Biol Chem. 2006;281:19748–19761. doi: 10.1074/jbc.M600850200. [DOI] [PubMed] [Google Scholar]
- Moriwaki Y, Yamamoto T, Takahashi S, Tsutsumi Z. et al. 'Widespread cellular distribution of aldehyde oxidase in human tissues found by immunohistochemistry staining'. Histol Histopathol. 2001;16:745–753. doi: 10.14670/HH-16.745. [DOI] [PubMed] [Google Scholar]
- Duley JA, Harris O, Holmes RS. 'Analysis of human alcohol- and aldehyde-metabolizing isozymes by electrophoresis and isoelectric focusing'. Alcohol Clin Exp Res. 1985;9:263–271. doi: 10.1111/j.1530-0277.1985.tb05747.x. [DOI] [PubMed] [Google Scholar]
- Berger R, Mezey E, Clancy KP, Harta G. et al. 'Analysis of aldehyde oxidase and xanthine dehydrogenase/oxidase as possible candidate genes for autosomal recessive familial amyotrophic lateral sclerosis'. Somat Cell Mol Genet. 1995;21:121–131. doi: 10.1007/BF02255787. [DOI] [PubMed] [Google Scholar]
- Holmes RS, Vandeberg JL. 'Aldehyde dehydrogenases, aldehyde oxidase and xanthine oxidase from baboon tissues: Phenotypic variability and subcellular distribution in liver and brain'. Alcohol. 1986;3:205–214. doi: 10.1016/0741-8329(86)90046-7. [DOI] [PubMed] [Google Scholar]
- Terao M, Kurosaki M, Barzago MM, Fratelli M. et al. 'Role of the molybdoflavoenzyme aldehyde oxidase homolog 2 in the biosynthesis of retinoic acid: Generation and characterization of a knockout mouse'. Mol Cell Biol. 2009;29:357–377. doi: 10.1128/MCB.01385-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila R, Kurosaki M, Barzago MM, Kolek M. et al. 'Regulation and biochemistry of mouse molybdo-flavoenzymes. The DBA/2 mouse is selectively deficient in the expression of aldehyde oxidase homologues 1 and 2 and represents a unique source for the purification and characterization of aldehyde oxidase'. J Biol Chem. 2004;279:8668–8683. doi: 10.1074/jbc.M308137200. [DOI] [PubMed] [Google Scholar]
- Bendotti C, Prosperini E, Kurosaki M, Garattini E. et al. 'Selective localization of mouse aldehyde oxidase mRNA in the choroid plexus and motor neurons'. Neuroreport. 1997;8:2343–2349. doi: 10.1097/00001756-199707070-00048. [DOI] [PubMed] [Google Scholar]
- Buzzell GR. 'Sexual dimorphism in the Harderian gland of the Syrian hamster is controlled and maintained by hormones, despite seasonal fluctuations in hormone levels: Functional implications'. Microsc Res Tech. 1996;34:133–138. doi: 10.1002/(SICI)1097-0029(19960601)34:2<133::AID-JEMT6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Buzzell GR. 'The Harderian gland: Perspectives'. Microsc Res Tech. 1996;34:2–5. doi: 10.1002/(SICI)1097-0029(19960501)34:1<2::AID-JEMT2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Buzzell GR, Hida A, Fu S, Seyama Y. 'Effect of the photoperiod in modulating the androgenic control of 1-alkyl-2,3-diacylglycerol composition in the Harderian gland of the golden hamster, Mesocricetus auratus'. J Exp Zool. 1997;277:99–105. doi: 10.1002/(SICI)1097-010X(19970201)277:2<99::AID-JEZ2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Austin NE, Baldwin SJ, Cutler L, Deeks N. et al. 'Pharmacokinetics of the novel, high-affinity and selective dopamine D3 receptor antagonist SB-277011 in rat, dog and monkey: In vitro/in vivo correlation and the role of aldehyde oxidase'. Xenobiotica. 2001;31:677–686. doi: 10.1080/00498250110056531. [DOI] [PubMed] [Google Scholar]
- Beedham C. 'The role of non-P450 enzymes in drug oxidation'. Pharm World Sci. 1997;19:255–263. doi: 10.1023/A:1008668913093. [DOI] [PubMed] [Google Scholar]
- Beedham C, Miceli JJ, Obach RS. 'Ziprasidone metabolism, aldehyde oxidase, and clinical implications'. J Clin Psychopharmacol. 2003;23:229–232. doi: 10.1097/01.jcp.0000084028.22282.f2. [DOI] [PubMed] [Google Scholar]
- Clarke SE, Harrell AW, Chenery RJ. 'Role of aldehyde oxidase in the in vitro conversion of famciclovir to penciclovir in human liver'. Drug Metab Dispos. 1995;23:251–254. [PubMed] [Google Scholar]
- Ishigai M, Ishitani Y, Orikasa Y, Kamiyama H. et al. 'Metabolism of 2(R, S)-1,2-bis(nicotinamido)propane, a new agent with anti-vasospasm activity, in rats and rabbits'. Arzneimittelforschung. 1998;48:429–435. [PubMed] [Google Scholar]
- Rodrigues AD, Kukulka MJ, Ferrero JL, Cashman JR. 'In vitro hepatic metabolism of ABT-418 in chimpanzee (Pan troglodytes). A unique pattern of microsomal flavin-containing monooxygenase-dependent stereoselective N'-oxidation'. Drug Metab Dispos. 1995;23:1143–1152. [PubMed] [Google Scholar]
- Strolin Benedetti M, Whomsley R, Baltes E. 'Involvement of enzymes other than CYPs in the oxidative metabolism of xenobiotics'. Expert Opin Drug Metab Toxicol. 2006;2:895–921. doi: 10.1517/17425255.2.6.895. [DOI] [PubMed] [Google Scholar]
- Beedham C. 'Molybdenum hydroxylases: biological distribution and substrate-inhibitor specificity'. Prog Med Chem. 1987;24:85–127. doi: 10.1016/s0079-6468(08)70420-x. [DOI] [PubMed] [Google Scholar]
- Obach RS, Huynh P, Allen MC, Beedham C. 'Human liver aldehyde oxidase: Inhibition by 239 drugs'. J Clin Pharmacol. 2004;44:7–19. doi: 10.1177/0091270003260336. [DOI] [PubMed] [Google Scholar]
- Moriyasu A, Sugihara K, Nakatani K, Ohta S. et al. 'In vivo-in vitro relationship of methotrexate 7-hydroxylation by aldehyde oxidase in four different strain rats'. Drug Metab Pharmacokinet. 2006;21:485–491. doi: 10.2133/dmpk.21.485. [DOI] [PubMed] [Google Scholar]
- Ding TL, Benet LZ. 'Comparative bioavailability and pharmacokinetic studies of azathioprine and 6-mercaptopurine in the rhesus monkey'. Drug Metab Dispos. 1979;7:373–377. [PubMed] [Google Scholar]
- Rooseboom M, Commandeur JN, Vermeulen NP. 'Enzyme-catalyzed activation of anticancer prodrugs'. Pharmacol Rev. 2004;56:53–102. doi: 10.1124/pr.56.1.3. [DOI] [PubMed] [Google Scholar]
- Cates LA, Jones GS Jr, Good DJ, Tsai HY. et al. 'Cyclophosphamide potentiation and aldehyde oxidase inhibition by phosphorylated aldehydes and acetals'. J Med Chem. 1980;23:300–304. doi: 10.1021/jm00177a018. [DOI] [PubMed] [Google Scholar]
- Tochino Y, Sugeno Y. 'Proceedings: Relationship between aldehyde oxidase activity and biological activity of cyclophosphamide'. Jpn J Pharmacol. 1974;24:s:81. [PubMed] [Google Scholar]
- Beedham C, Al-Tayib Y, Smith JA. 'Role of guinea pig and rabbit hepatic aldehyde oxidase in oxidative in vitro metabolism of cinchona antimalarials'. Drug Metab Dispos. 1992;20:889–895. [PubMed] [Google Scholar]
- Rashidi MR, Smith JA, Clarke SE, Beedham C. 'In vitro oxidation of famciclovir and 6-deoxypenciclovir by aldehyde oxidase from human, guinea pig, rabbit, and rat liver'. Drug Metab Dispos. 1997;25:805–813. [PubMed] [Google Scholar]
- Lake BG, Ball SE, Kao J, Renwick AB. et al. 'Metabolism of zaleplon by human liver: Evidence for involvement of aldehyde oxidase'. Xenobiotica. 2002;32:835–847. doi: 10.1080/00498250210158915. [DOI] [PubMed] [Google Scholar]
- Renwick AB, Ball SE, Tredger JM, Price RJ. et al. 'Inhibition of zaleplon metabolism by cimetidine in the human liver: In vitro studies with subcellular fractions and precision-cut liver slices'. Xenobiotica. 2002;32:849–862. doi: 10.1080/00498250210158221. [DOI] [PubMed] [Google Scholar]
- Sugihara K, Kitamura S, Tatsumi K, Asahara T. et al. 'Differences in aldehyde oxidase activity in cytosolic preparations of human and monkey liver'. Biochem Mol Biol Int. 1997;41:1153–1160. doi: 10.1080/15216549700202241. [DOI] [PubMed] [Google Scholar]
- Schwartz R, Kjeldgaard NO. 'The enzymic oxidation of pyridoxal by liver aldehyde oxidase'. Biochem J. 1951;48:333–337. doi: 10.1042/bj0480333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Tsujita M, Ichikawa Y. 'Retinal oxidase is identical to aldehyde oxidase'. FEBS Lett. 1993;336:272–274. doi: 10.1016/0014-5793(93)80818-F. [DOI] [PubMed] [Google Scholar]
- Huang DY, Furukawa A, Ichikawa Y. 'Molecular cloning of retinal oxidase/aldehyde oxidase cDNAs from rabbit and mouse livers and functional expression of recombinant mouse retinal oxidase cDNA in Escherichia coli'. Arch Biochem Biophys. 1999;364:264–272. doi: 10.1006/abbi.1999.1129. [DOI] [PubMed] [Google Scholar]
- Duester G. 'Families of retinoid dehydrogenases regulating vitamin A function: Production of visual pigment and retinoic acid'. Eur J Biochem. 2000;267:4315–4324. doi: 10.1046/j.1432-1327.2000.01497.x. [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Nebert DW. 'Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family'. Hum Genomics. 2005;2:138–143. doi: 10.1186/1479-7364-2-2-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G. 'Retinoic acid synthesis and signaling during early organogenesis'. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara K, Tayama Y, Shimomiya K, Yoshimoto D. et al. 'Estimation of aldehyde oxidase activity in vivo from conversion ratio of N1-methylnicotinamide to pyridones, and intraspecies variation of the enzyme activity in rats'. Drug Metab Dispos. 2006;34:208–212. doi: 10.1124/dmd.105.006544. [DOI] [PubMed] [Google Scholar]
- Steinberg KK, Relling MV, Gallagher ML, Greene CN. et al. 'Genetic studies of a cluster of acute lymphoblastic leukemia cases in Churchill County, Nevada'. Environ Health Perspect. 2007;115:158–164. doi: 10.1289/ehp.9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graessler J, Fischer S. 'The dual substrate specificity of aldehyde oxidase 1 for retinal and acetaldehyde and its role in ABCA1 mediated efflux'. Horm Metab Res. 2007;39:775–776. doi: 10.1055/s-2007-992126. [DOI] [PubMed] [Google Scholar]
- Weigert J, Neumeier M, Bauer S, Mages W. et al. 'Small-interference RNA-mediated knock-down of aldehyde oxidase 1 in 3T3-L1 cells impairs adipogenesis and adiponectin release'. FEBS Lett. 2008;582:2965–2972. doi: 10.1016/j.febslet.2008.07.034. [DOI] [PubMed] [Google Scholar]


