Abstract
Reactome is an expert-authored, peer-reviewed knowledge base of human reactions and pathways that functions as a data-mining resource and electronic textbook. Its current release covers approximately 23 per cent of the complete human proteome from UniProt. The pathway browser, search and data-mining tools facilitate searching and visualising pathway data and the analysis of user-supplied high-throughput datasets. A catalogue of all the solute-carrier (SLC) class of transporters which have known ligands has been annotated in Reactome. Reactome provides a detailed and interactive view of this set of transport reactions. Using the example of the SLC class of transporters, we show how they can be overlaid with protein-protein interaction, protein-drug interaction and gene expression data and compared with equivalent pathways in other species, to facilitate over-representation, expression and other pathway analyses.
Keywords: solute transport, protein-ligand interaction, protein-protein interaction, pathway analysis, gene expression analysis
Reactome
Reactome (http://www.reactome.org) is a freely available and open-source database of biological pathways [1,2]. Expert scientists curate information into Reactome, which is then peer-reviewed to obtain a consensus representation of the process or pathway. The data are extensively cross-linked to major protein and nucleotide sequence databases, as well as to the Gene Ontology and PubMed databases. A new website for Reactome was recently released. This includes new functionality for interacting with curated pathways and analysing them with linked or user-supplied datasets. Tools in this new version of Reactome allow users to overlay interaction or expression data, further enriching the pathway information. The results of these analyses can then be downloaded in a range of formats.
Curated SLC-mediated transmembrane transport pathway in Reactome
We have recently completed a catalogue of the solute carrier (SLC) class of transmembrane transporters. These proteins are well conserved in all eukaryotes, as well as most prokaryotes, and play a gate-keeping role for cells and organelles, controlling the uptake and efflux of many types of substrates, such as sugars, inorganic cations and anions, organic anions and carboxylates, amino acids and oligopeptides, fatty acids and lipids, and neurotrans-mitters and vitamins. The SLC superfamily comprises 55 gene families, with 362 putatively functional protein-coding genes reported [3,4]. Of these, 231 with the criterion that the transporter has a substrate which it transports across the membrane have been catalogued in Reactome. The remainder are orphan proteins, with no characterised substrates at this time. This pathway will be used as the basis for describing the use of Reactome's analysis tools.
Method
Reactome's website can be viewed on PCs, Macs or Linux computers with later versions of the Internet Explorer (IE), Firefox or Safari browsers. Reactome's whole content can be downloaded as a mysql datadump (http://www.reactome.org/download/index.html). Other Reactome datasets and code that can be downloaded can be found here too.
Briefly, a query was constructed in the Universal Protein Resource (UniProt) (http://www.uniprot.org/). UniProt is a comprehensive resource for protein sequence and annotation data [5]. Reactome uses UniProt identifiers as the primary reference for proteins used in their database. The query searched for manually annotated and reviewed human SLC transporters.
gene: SLC* AND organism:human AND reviewed:yes
This query returned 367 results, the 362 proteins reviewed by He et al.[4] and five newly characterised ones. These combined results were used as the basis of the information entered into Reactome. Transporters for which substrates were experimentally identified were catalogued in Reactome using an in-house graphical user interface (GUI) called the Curator Tool, an interface which allows curators to structure data around Reactome's data model and commit to a central repository [1]. The base unit of Reactome is the reaction. The basic set of attributes of a reaction that are captured are details of the input and output molecule(s), the modulating protein(s), compartments for these entities, supporting literature reference(s), a textual summary describing this reaction and the species (eg human).
Visualisation
A new feature of Reactome is the pathway diagram (Figure 1). Here, reactions in a pathway are represented as connected, interactive objects. Each reaction displays the entities (proteins or other molecules) that make up the inputs and outputs of the reaction, together with the modulator protein, where appropriate, joined via a central 'reaction node'. Reactions are also compartmentalised on the diagram, to indicate cellular location. This is an important feature, especially for this pathway, where entities are transported from one side of a membrane to another. The entity and reaction nodes are interactive; clicking on them provides context-sensitive information, such as details of the overall reaction or specific details of the entities involved in the reaction.
Figure 1.
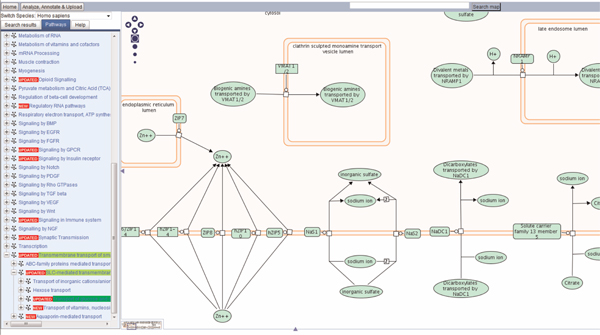
Pathway diagram of a section of the solute carrier (SLC) family of the transmembrane transporter pathway. A hierarchy is visible on the left and the diagram in the main window to the right. 'Mousing' over names shows the name and cellular location. The navigation section to the top left of the diagram allows the user to scroll around the whole pathway, as well as zooming options.
Tools in Reactome
Interaction data
Interaction data can be overlaid on pathway diagrams to enrich the data already in Reactome. For example, overlaying protein-protein or protein-drug/chemical interaction data on the solute carrier pathway provides a powerful extension to the existing data for identifying heterodi-meric partners. This molecular interaction overlay (MIO) feature may be useful -- for example, to the pharmaceutical industry, for identifying novel targets that otherwise may have remained elusive -- however, finding potential candidates to modulate a pathway experimentally is more likely. An example of protein-protein interactions sourced from the IntAct database (http://www.ebi.ac.uk/intact/)[6] overlaid across the SLC transporter pathway via the PSIQUIC web service (http://code.google.com/p/psicquic/) (Aranda et al., in preparation) is shown in Figure 2. The user clicks on the 'Analyze, Annotate & Upload' button, located at the top left of the page. By default, IntAct is the database showing in the 'Interaction Database' pull-down list. To see all interactors for the pathway, the user clicks on 'Display table of all interactors for pathway' button, and a table appears, displaying the proteins in the pathway and the interactors found. Clicking on the blue box next to a protein in the list takes the user to that protein in the diagram and displays all the interactors for it. For clarity, only ten interactors are displayed. A small white box with a number appears next to the protein, to indicate how many interactors there are. A more detailed description can be found in Reactome's user guide (http://www.reactome.org/userguide/Usersguide.html#Molecular_Interaction_Overlay). The MIO feature may also help in predicting the function or location of an unknown protein through 'guilt by association', a term used to describe annotation transfer based on analysis of interacting partners.
Figure 2.
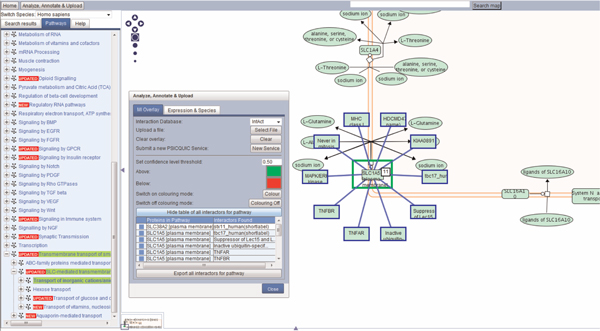
The protein SLC1A5 in the pathway is showing 11 interactions, overlaid with blue outlines. This interaction information can be exported as a tsv file by clicking the 'export all interactors for pathway' button.
The MIO feature using the PSIQUIC server function can be used to overlay protein-drug/chemical interactors on a pathway diagram (Figure 3). ChEMBL (http://www.ebi.ac.uk/chembldb/) is a database of bioactive drug-like small molecules. Again, the user clicks on the 'Analyze, Annotate & Upload' button, located at the top left of the page. By default, IntAct is the database showing in the 'Interaction Database' pull-down list. Clicking on the pull-down menu displays all interactor databases available. The user can choose ChEMBL and click on the 'Display table of all interactors for pathway' button. A list appears, showing the proteins in the pathway and the inter-actors found for them. Clicking on the blue box next to a protein takes the user to that protein in the diagram and displays all interactors for it. For clarity, only ten interactors are displayed. A small white box with a number appears next to the protein to indicate how many interactors there are. A more detailed description can be found in Reactome's Userguide (http://www.reactome.org/userguide/Usersguide.html#Molecular_Interaction_Overlay). The example shows drug/chemical interactors of the sodium-dependent dopamine transporter. Neuronal reuptake of dopamine is the primary means of regulating synaptic availability of dopamine. Altering the function of this transporter may be implicated in the pathophysiology or treatment of several neuropsychiatric disorders. The various small molecules displayed here may be useful starting points for novel drug design.
Figure 3.
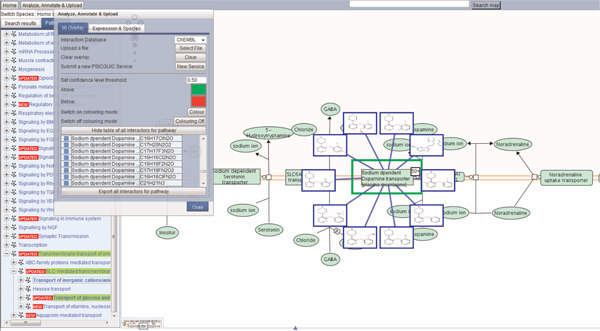
The sodium-dependent dopamine transporter in the pathway interacts with more than 50 chemical compounds, overlaid with blue outlines. The table can be exported as a tsv file by clicking on the 'Export all interactors for pathway' button.
Expression data
Gene expression datasets derived from microarray studies can be overlaid on pathway diagrams. Proteins which have expression values are coloured on the pathway diagram, allowing the user to visualise expression over the whole pathway at a glance. Proteins which are not 'hit' by the dataset are highlighted in grey. Figure 4 shows part of a diagram in which a dataset from the Gene Expression Atlas database (http://www.ebi.ac.uk/gxa/) [7]. A query was performed in the Gene Expression Atlas which searched for SLC genes which are upregulated in the brain and act as symporters. The results can be downloaded as a tab separated file, which is a format that Reactome can accept. The columns of identifiers and their expression values were extracted and used in the 'Analyze, Annotate & Upload' toolbar. There, the 'Expression & Species' tab is opened and the file containing the expression values uploaded. Check the box 'Expression painting on' and click the submit button. The pathway is painted according to the expression values from the submitted file. The most upregulated gene from this dataset is SLC1A1 (also known as excitatory amino acid carrier 1 [EAAC1], or excitatory amino acid transporter 3 [EAAT3]), which is particularly abundant in the brain.
Figure 4.
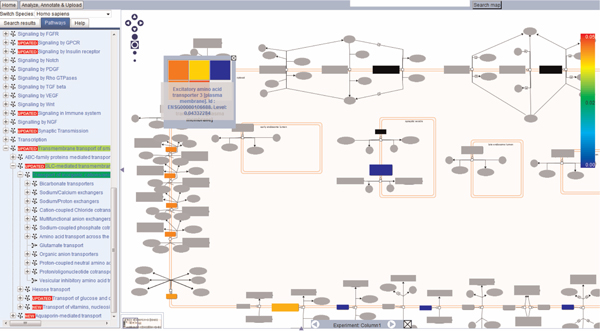
Gene expression dataset overlaid upon an SLC transporter pathway. The data indicate those SLC proteins which are upregulated in the human brain and act as symporters. The colour chart on the right indicates the range of regulation, from blue (downregulation) to red (upregulation). Blacked-out hits indicate complexes. Right-clicking on the complex will show all proteins in that complex, individually coloured. Here, the group of excitatory amino acid transporters show varying expression in the brain, indicated by the colour scheme. Proteins not hit by the dataset are greyed out.
Species comparison
Reactome uses manually curated human pathways electronically to 'infer' their equivalents in 20 other species. The 'Expression & Species' tab on the control panel allows a user to view these predicted pathways to see what is common to the human pathway or perhaps missing in the model organism. This may be a useful way to determine the extent of conservation of biological processes across species. Figures 5A and 5B show a human pathway compared with that of Mus musculus (mouse) and Escherichia coli. As one might expect, there is far more conservation between human and M. musculus than between human and E. coli.
Figure 5.
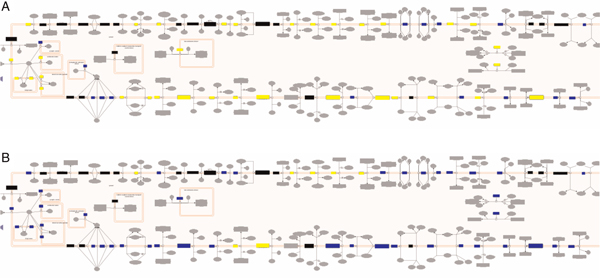
(A) Human pathway compared with that of Mus musculus. (B) Human pathway compared with that of Escherichia coli. Key: yellow: orthologous protein in other species; blue: protein only known in human; black: complex, right-click to open revealing grid representing components of that complex; grey: either inference not possible or small molecules.
Conclusion
Reactome is a freely available database of pathways. The SLC family of transporters plays a vital role in mediating the movement of essential metals, ions, drugs and many endogenous compounds into and out of the cell and cellular organelles. Information about the SLC family of transporters has been systematically annotated in Reactome and this provides a basis for a number of analyses which can be performed on these data. These analyses include interactions, expression data, over-representation analysis and species comparison. The results of such analyses can be starting points for further investigations using systems biology. The value of the database to users should continue to grow as additional pathways are annotated and new software for data analysis and integration are developed. Work is now under way to improve the visual overview of expression data and provide closer integration with Cytoscape (http://www.cytoscape.org/). Cytoscape is an open-source platform for the visualisation and data integration of biological pathways and networks. Tools are being developed to support additional analysis of interactors, including functional interactors; to pull data from other omics sources, such as expression data or transcription factors; and to support integration with medical data.
Acknowledgements
Development of the Reactome data model and database is a collaborative project and this work benefited greatly from interactions with fellow Reactome team members (http://www.reactome.org/about.html), as well as colleagues at the EBI from GO, ChEBI, ChEMBL, UniProt, ArrayExpress and IntAct. This work was supported by grants from the US National Human Genome Research Institute, NIH (P41 HG003751) and the EU 6th Framework Programme 'ENFIN' (LSHG-CT-2005-518254).
References
- Vastrik I, D'Eustachio P, Schmidt E, Gopinath G. et al. Reactome: A knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L, Gopinath G, Gillespie M, Caudy M. et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger MA, Romero MF, Peng JB, Rolfs A. et al. The ABCs of solute carriers: Physiological, pathological and therapeutic implications of human membrane transport proteins Introduction. Pflugers Arch. 2004;447:465–468. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- He L, Vasiliou K, Nebert DW. Analysis and update of the human solute carrier (SLC) gene superfamily. Hum Genomics. 2009;3:195–206. doi: 10.1186/1479-7364-3-2-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda B, Achuthan P, Alam-Faruque Y, Armean I. et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2009;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapushesky M, Emam I, Holloway E, Kurnosov P. et al. Gene expression atlas at the European bioinformatics institute. Nucleic Acids Res. 2010;38:D690–D698. doi: 10.1093/nar/gkp936. [DOI] [PMC free article] [PubMed] [Google Scholar]


