Abstract
Changes in DNA copy number are one of the hallmarks of the genetic instability common to most human cancers. Previous micro-array-based methods have been used to identify chromosomal gains and losses; however, they are unable to genotype alleles at the level of single nucleotide polymorphisms (SNPs). Here we describe a novel algorithm that uses a recently developed high-density oligonucleotide array-based SNP genotyping method, whole genome sampling analysis (WGSA), to identify genome-wide chromosomal gains and losses at high resolution. WGSA simultaneously genotypes over 10,000 SNPs by allele-specific hybridisation to perfect match (PM) and mismatch (MM) probes synthesised on a single array. The copy number algorithm jointly uses PM intensity and discrimination ratios between paired PM and MM intensity values to identify and estimate genetic copy number changes. Values from an experimental sample are compared with SNP-specific distributions derived from a reference set containing over 100 normal individuals to gain statistical power. Genomic regions with statistically significant copy number changes can be identified using both single point analysis and contiguous point analysis of SNP intensities. We identified multiple regions of amplification and deletion using a panel of human breast cancer cell lines. We verified these results using an independent method based on quantitative polymerase chain reaction and found that our approach is both sensitive and specific and can tolerate samples which contain a mixture of both tumour and normal DNA. In addition, by using known allele frequencies from the reference set, statistically significant genomic intervals can be identified containing contiguous stretches of homozygous markers, potentially allowing the detection of regions undergoing loss of heterozygosity (LOH) without the need for a matched normal control sample. The coupling of LOH analysis, via SNP genotyping, with copy number estimations using a single array provides additional insight into the structure of genomic alterations. With mean and median inter-SNP euchromatin distances of 244 kilobases (kb) and 119 kb, respectively, this method affords a resolution that is not easily achievable with non-oligonucleotide-based experimental approaches.
Keywords: SNPs, genotypes, amplifications, deletions, copy number, LOH
Introduction
The underlying progression of genetic events which transform a normal cell into a cancer cell is characterised by a shift from the diploid to aneuploid state. [1,2] As a result of genomic instability, cancer cells accumulate both random and causal alterations at multiple levels, from point mutations to whole-chromosome aberrations. DNA copy number changes include, but are not limited to, loss of heterozygosity (LOH) and homozygous deletions, which can result in the loss of tumour suppressor genes, and gene amplification events, which can result in the activation of cellular proto-oncogenes. One of the continuing challenges to unravelling the complex karyotype of the tumour cell is the development of improved molecula methods that can globally catalogue LOH, gains and losses with both high resolution and accuracy.
Numerous molecular approaches have been described to identify genome-wide LOH and copy number changes within tumours. Classical LOH studies designed to identify allelic loss using paired tumour and blood samples have made use of restriction fragment length polymorphisms (RFLPs) and, more often, highly polymorphic microsatellite markers (short tandem repeats, variable number of tandem repeats). The demonstration of Knudson's two-hit tumourigenesis model using LOH analysis of the retinoblastoma gene, Rb1, showed that the copy number of the mutant allele can vary from one to three copies as the result of multiple second-hit mechanisms. [3] Thus, regions undergoing LOH do not necessarily contain DNA copy number changes. Approaches to measuring genome-wide increases or decreases in DNA copy number include comparative genomic hybridisation (CGH),[4] spectral karyotyping (SKY),[5] fluorescence in situ hybridisation (FISH),[6] molecular subtraction (such as representational difference analysis)[7,8] and digital karyotyping [9]. CGH, perhaps the most widely used and powerful approach, has limited resolution [10-20 megabases (Mb) in the case of metaphase spreads and 1-2 Mb for genomic clones] and is not well suited for identifying regions of the genome that have undergone LOH such that a single allele is present but there is no reduction in copy number. Recently, a method called RO MA, which uses digonucleotide probes (70 nucleotides in length) to assess copy number alterations, achieved a resolution of 30 kb throughout the genome. Like CGH, however, it does not provide genotype information and thus also cannot identify regions of LOH with no copy number change [10].
With the completion of the human genome, single nucleotide polymorphisms (SNPs), the most common sequence variations among individuals, are emerging as the marker of choice in large-scale genetic studies due to their abundance, stability and relative ease of scoring. These same characteristics make SNPs powerful markers for LOH studies. High-density DNA array technolog [11-13] has been applied for the identification of genomic alterations in tumour cells, most notably LOH [14-17]. We have recently developed a method termed 'whole genome sampling analysis' (WGSA) for large-scale SNP genotyping of complex DNA [18,19]. Here, we describe the development of an algorithm used in conjunction with WGSA which is capable of detecting genome-wide gains and losses from a single DNA sample. The median distance of 119 kilobases (kb) between markers provides high resolution for global surveying of DNA amplifications and deletions using a single array. Using a panel of ten human breast cancer cell lines, along with DNA samples with varying X chromosome copies, we show that the algorithm is both specific, sensitive and robust, even with mixed samples containing both normal and tumour DNA, suggesting its utility for bona fide tumour samples. Thus, the development of a molecular approach capable of identifying regions of allelic loss along with regions of amplification within a single experiment should have an impact on the basic understanding of the cancer genome, as well as potentially lead to improved clinical applications in both diagnostics and treatment regimens.
Material and methods
Cell lines and nucleic acid isolation
Nine human breast cancer cell lines (BT-20, MCF-7, MCF-12A, MDA-MB-157, MDA-MB-436, MDA-MB-468, SK-BR-3, ZR-75-1 and ZR-75-30) and two syngeneic human breast cancer cell lines (Hs-578T and Hs-578Bst) [20] were obtained from the American Type Culture Collection (ATCC). A normal human mammary epithelial cell line (HMEC) was obtained from Clonetics. All cells were grown under recommended culture conditions. Genomic DNA was isolated using a QIAGEN QIAamp DNA Blood Mini Kit. DNAs from cell lines containing 3X(NA04626), 4X(NA01416) and 5X(NA06061) chromosomes and DNAs for the normal reference set of 110 individuals (48 males and 62 females) were purchased from the National Institute of General Medical Sciences (NIGMS) Human Genetic Cell Repository, Coriell Institute for Medical Research (Camden, NJ).
WGSA
The assay was performed as described by Kenned et al. [18] except for modifications to the target amplification and DNA labelling steps. DNA amplification by polymerase chain reaction (PCR) was carried out under the following conditions: each 100 μl reaction contained 25 ng of adaptor-ligated genomic DNA, 0.7 μM primer, 250 μM deoxynucleotide triphosphates, 2.5 mM MgCl2 and 10U AmpliTaq Gold (Applied Biosystems (ABI)) in 1X PCR Buffer II (ABI). Cycling was performed as follows: 95°C/three minutes, followed by 35 cycles of 95°C/30 seconds, 59°C/30 seconds, 72°C/30 seconds and an extension at 72°C for seven minutes. The PCR products were purified and concentrated with QIAGEN MinElute PCR Purification kit, and DNA concentrations were determined by measuring absorbance at 260 nm. Fragmented DNA was labelled in 1X terminal transferase (TdT) buffer with 105 U TdT (Promega) and 0.15 mM DLR (a proprietory labelling agent from Affymetrix) at 37°C for two hours, followed by heat inactivation at 95°C for 15 minutes. All experimental samples were hybridised to the Affymetrix Gene Chip® 10K Mapping Xba_131 Array in duplicate, washing, staining and scanning were performed using the protocol specified in the manufacturer's instructions. Samples comprising the normal reference set were hybridised to a predecessor array which used the same probe sequences and tiling strategy to generate genotype calls as the commercially available array. The call rates of all samples were above 88 per cent and the average genotype concordance was 99.97 per cent. WGSA DNA mixing experiments were performed as follows: the concentrations of genomic DNA from Hs-578T and Hs-578Bst were determined by PicoGreen dsDNA Quantitation Assay (Molecular Probes) and Hs-578Bst DNA was added to Hs-578T DNA in 10 per cent increments.
Quantitative PCR
PCR was performed using the ABI Prism 7700 Sequence Detection System. PCR primers were designed by using Primer Express 1.5 software (ABI) and were synthesised by QIAGEN. Reactions (25 μl containing 25 ng DNA) were prepared using the SYBR-Green PCR Core Reagents kit (ABI). Conditions for amplification were as follows: one cycle of 50°C/two minutes, one cycle of 95°C/ten minutes, followed by 35 cycles of 95°C/20 seconds, 56°C/30 seconds and 72°C/30 seconds. Threshold cycle numbers (Ct) were obtained by using Sequence Detector v1.7a software. Human genomic DNA (Roche) was used as the normal control. All reactions were carried out in duplicate and Ct numbers were averaged. DNA amounts were measured by ultraviolet spectrophotometer and were normalised to LINE-1 elements. [9] Relative quantitation was carried out using the comparative Ct method (ABI User Bulletin #2, 1997). Primer pair sequence information for all 99 SNPs is available upon request. Quantitative PCR assays for c-MYC and p16 genes were carried out as described, except that the annealing temperature was 60°C.
Feature extraction
WGSA uses 20 probe pairs (25-mers) equally divided between the sense and anti-sense strands for each SNP, with ten probe pairs for allele A and ten probe pairs for allele B. A probe pair includes a perfect match cell and a single-base mismatch cell. The log [21] of the arithmetic average of the PM intensities across 20 probes () is used as the basic measurement for any given SNP. It has an approximate Gaussian distribution on each sample
where PMi is the intensity of the perfect match cell of probe pair. After S is calculated, it is scaled to have a mean of zero and a variance of one for all autosomal SNPs to increase the comparability across samples.
j = 1,...,J are the autosomal SNPs on the chip. In addition to log average intensity (S), discrimination ratio (DR) -- which measures the difference between perfect match and mismatch probes -- is used as a supplementary metric for regions of homozygous deletions [22].
Significance calculation
The significance of the copy number variation in the target cancer cell line is estimated by a comparison with a normal reference set. The SNP genotypes of the target cell line are considered prior to the comparison, such that, for each SNP, the cancer cell line is compared with only those normal samples that share the same genotype. This allows comparisons to be made within a homogeneous distribution instead of a mixture of several genotypes [23]. The basic assumption is that for any given SNP j with genotype g (g = AA, AB or BB), the standardised log intensity follows a Gaussian distribution [24]. The mean and variance are estimated using the normal reference samples.
where k = 1,...,Kg represents the normal samples that have the same genotype g as the target cell line. While the normal samples may contain isolated regions of gains and losses, outlier data points, defined as having values more than three standard deviations away from the mean, are excluded from the estimation of the reference distribution [25]. The significance of the difference of from the normal reference distribution is measured by the p-value:
where Φ is the quantile function for the standard Gaussian distribution.
Contiguous point analysi
For each SNP j, with genotype g, the individual test statistic for the significance calculation is:
As previously described, is assumed to have a standard Gaussian distribution and SNPs are assumed to be independent. Thus, for any given stretch in the genome starting at point m and ending at point n
This score, , can be converted to a probability by using the Φ function, which is called the contiguous point analysis (CPA) p-value and is substituted for single point analysis (SPA) p-values of each SNP when appropriate. CPA is most suitable when consecutive markers show the same direction of alterations. Accordingly, a candidate stretch is defined starting at point m and ending at point n as:
The starting point is from j = 1, ie the beginning of the chromosome, and a search is performed for such candidate stretches up to the end of the chromosome. For any given SNP, if the SPA p-value is less significant than the CPA p-value, the former is substituted by the latter.
LOH
For each individual SNP j, the probability of being homozygous is calculated:
If each SNP is treated independently, then the probability of a stretch of SNPs (from position m to position n) all being homozygous will be:
Results
Copy number estimation and significance calculation
Three main approaches were used to validate the copy number and significance estimations. They were: 1) X-chromosome dosage response experiments; 2) independent copy number estimates using quantitative PCR; and 3) confirmation of known true-positive regions using the cancer cell line panel. The dosage response between copy number and chip intensity was tested using samples with varying X chromosome copy numbers (1X to 5X). Using (I) to indicate chip intensity, the dosage response assumption Ia ≅ Cab × Ib, where Ia is the intensity for a region with copy number a, Ib is the intensity on the same region with copy number b and Cab is the intensity ratio determined by a and b. , defined in the above section 'Feature extraction', is an approximation of log intensity. Thus, a log transformation leads to . Also is the log of the intensity ratio determined by a and b. Results from DNA samples with 1, 3, 4 and 5X chromosomes were com pared with a 2X sample and are summarised in Figure 1a. There is a high linear correlation among the sample pairs; for any given pair, the linear trend is parallel to Y = X, confirming the equation . Using 2X as the baseline, the estimated log of the intensity ratio for each sample shows a strong linear relationship with the log of the copy number (Figure 1b). These X chromosome results are used to generalise to autosomes. Specifically, the log of the intensity ratio (C) in Figure 1b is equal to the difference between the target cell line and the normal reference average using log intensity. The log intensity value of the target cancer cell line on SNP j with genotype g is denoted as and the corresponding reference average is denoted as . The difference between the two is used to substitute for the log of the intensity ratio (C) in the formula shown in Figure 1b, giving the copy number estimation its final form:
Figure 1.
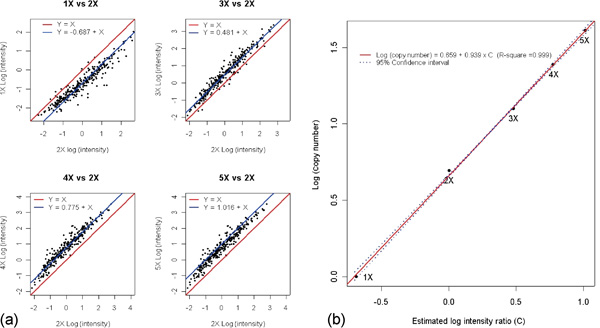
Plot of the standardised log intensity of 1X, 3X, 4X and 5X against 2X. The signal intensities are based on the average of two replicates across 302 single nucleotide polymorphisms that map to the X chromosome using National Center for Biotechnology Information Build 33. Figure 1b plots log (copy number) as a function of estimated log (intensity ratio) (C). The black dots indicate different samples (1X to 5X). The red line is the linear regression result using log (copy number) as the response and estimated log intensity ratio as the predictor. The blue lines indicate the 95 per cent confidence interval for the response, ie the natural log of the copy number.
An independent quantitative PCR (qPCR) method for measuring DNA copy number changes was used to verify observed regions of chromosomal gains and losses. PCR reactions on a set of 99 autosomal SNPs were carried out using genomic DNA templates from SK-BR-3 and normal individuals. This set of SNPs was not completely random, and contained both previously known as well as putative novel gains and losses identified in the cancer cell line. Figure 2 shows the relationships between ΔCt (Ct difference between the normal DNA sample and the cancer sample) derived from quantitative PCR, the calculated WGSA copy number and the calculated WGSA significance level (p -value). Figure 2a shows that the estimated copy number using WGSA is approximately an exponential function of ΔCt and falls near the theoretical estimating function 2ΔCt+1. The trend is tight when ΔCt values are low but becomes more scattered with increasing ΔCt. Figure 2b shows a strong positive correlation between ΔCt and the significance level calculated using the SPA algorithm. Except for a few points, the majority of the SNPs with a large ΔCt difference show very strong significance, while SNPs with a small ΔCt difference show moderate to low statistical significance. This figure also illustrates the importance of the discrimination ratio as a supplementary metric to PM intensity. For the data point circled in blue, the ΔCt value is less than -5, suggesting a homozygous deletion. The significance based on PM intensity is only moderate. This SNP shows increased significance, however, with a p -value of less than 10-6 when DR is applied (data not shown), allowing the deletion to be correctly identified. Figure 2c shows the relationship between the estimated copy number and the statistical significance. As expected, when the copy number approaches 0 (indicating a homozygous deletion), or approaches a large positive number (indicating high level amplification), the significance becomes very strong. These combined results using qPCR as an independent measure indicate that WGSA can detect chromosomal copy number changes in a quantitative manner. This result is also consistent with reports that the SNP array detects similar patterns of copy number changes when compared with bacterial artificial chromosome (BAC)-array CGH. [26,27]
Figure 2.
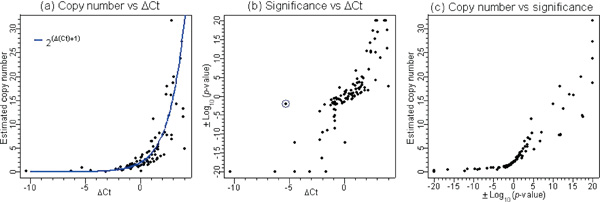
The results for 99 autosomal single nucleotide polymorphisms using the SK-BR-3 breast cancer cell line. The pairwise scatter-plots are based on three measurements: copy number, significance and the change in threshold cycle (Δ Ct). The significance measure is represented by the log 10 transformed p-value derived from the algorithm. To distinguish between deletions and amplifications, the -log10 (p -value) is used when the target value is higher than the reference mean, ie denoting amplification, and the log10 (p -value) is used when the target value is lower than the reference mean, ie denoting deletion. Copy number is estimated using the following formula: . ΔCt denotes the difference between the normal DNA sample versus SK-BR-3. The Ct is the cycle number at which the reporter fluorescence passes a fixed threshold above baseline. Positive ΔCt suggests amplification, while negative ΔCt suggests deletion.
The breast cancer cell line panel was surveyed for copy number changes in two well-characterised regions, namely chromosome 8q and chromosome 9p. CGH analysis of 38 breast cancer cell lines showed gains of 8q in 75 per cent of the samples [28] and loss of chromosome 9p has been reported in breast cancer [29]. Specifically, the c-MYC oncogene at chromosome 8q24 has been shown to be commonly amplified in breast cancer,[30,31] while the p16/INK4 tumour suppressor on chromosome 9p21 has been shown to be deleted in a variety of tumour types [32,33]. Figure 3a shows a comparison across four samples for a region of chromosome 8 from 50 to 140 Mb. The genomic region near c-MYC appears to be amplified in three cancer cell lines with moderate to very strong significance and does not appear to be amplified in the normal control (Hs-578Bst). This is consistent with published CGH results that show that all three cell lines contain gains in 8q23-q24 [34]. Quantitative PCR was carried out with a c-MYC primer pair and confirmed the copy number increase. The estimated c-MYC copy number by qPCR for SK-BR-3, MCF-7, ZR-75-30 and Hs-578Bst was 21.0, 7.5, 10.6 and 3.0, respectively. While the array does not contain SNPs from the c-MYC gene itself, the two nearest SNPs are SNP 55150, which is located 300 kb proximal to c-MYC, and SNP 511315, which is located 196 kb distal to c-MYC. WGSA and qPCR results for these SNPs are summarised in Table 1 and confirm that the region surrounding c-MYC is amplified in three of the four cell lines.
Figure 3.
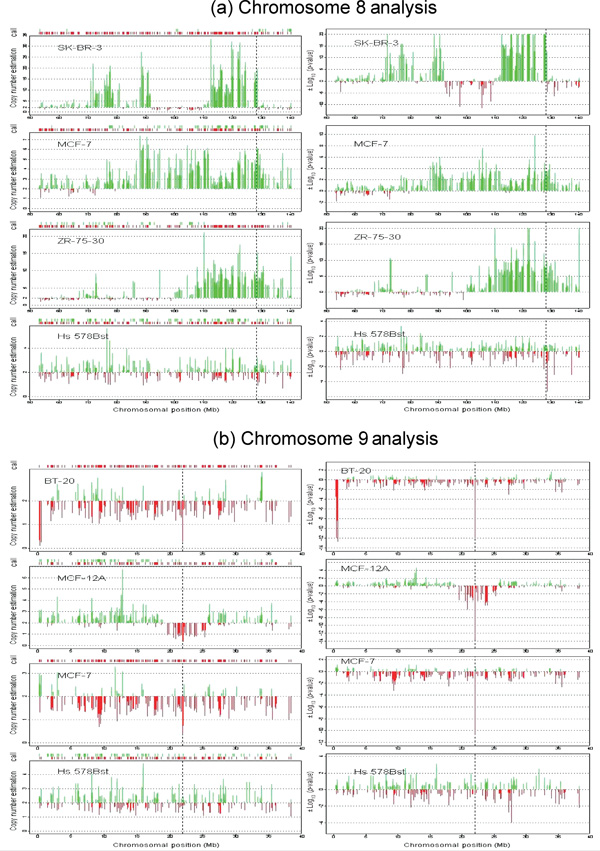
(see facing page). Chromosome 8 (panel a) and chromosome 9 (panel b) analysis. The graphs on the left-hand side of panels (a) and (b) represent copy number estimation and genotype information. The x-axis is the chromosomal position (National Center for Biotechnology Information (NCBI) Build 33). For each sample, the genotype information is presented on top of each panel. The downward red line indicates a homozygous genotype, while the upward green line indicates a heterozygous genotype. Each panel shows the copy number estimation on the y-axis. The vertical green and red lines are individual single nucleotide polymorphism copy number estimates. The upward green lines represent an estimate that is larger than the baseline value of 2, while the downward red lines represent an estimate that is lower than 2. The black dotted lines indicate the relative location of the c-MYC and p-16 genes on chromosomes 8 and 9, respectively. The panels on the right-hand side represent the significance results. The x-axis is the chromosomal position (NCBI Build 33) and the black vertical lines represent the location of the c-MYC (panel a) and p-16 (panel b) genes. The y-axis is the log 10 transformed p-value of each given SNP. To distinguish deletions from amplifications, the log10 ( p-value) (upward green lines) is used when the target value is higher than the reference mean (amplifications) and the log10 (p-value) (downward red lines) is used when the target value is lower than the reference mean (deletions).
Table 1.
qPCR and WGSA results on c-MYC and p16 genes
| c-Myc region on chromosome 8 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Marker/sample | SNP 55150 (300 kb distal) | SNP 511315 (196 kb distal) | |||||||
| 12ΔCt+1 | 2WGSA | 3Sig | 2ΔCt+1 | WGSA | Sig | ||||
| Sk-BR-3 | 32.00 | 15.87 | < - 20 | 22.63 | 21.12 | - 11.89 | |||
| MCF-7 | 9.19 | 4.54 | - 3.47 | 7.46 | 6.25 | - 1.89 | |||
| ZR-75-30 | 13.00 | 7.64 | -7.67 | 11.31 | 16.31 | -9.95 | |||
| Hs578 Bst | 2.60 | 2.54 | -0.86 | 2.64 | 3.21 | -0.77 | |||
| p 16 region on chromosome 9 | |||||||||
| Marker/sample | SNP 827951 (235 kb proximal) | SNP 139369 (inside p 16) | SNP 87445 (21 kb distal) | ||||||
| 2ΔCt+1 | WGSA | Sig | 2ΔCt+1 | WGSA | Sig | 2ΔCt+1 | WGSA | Sig | |
| BT-20 | 1.82 | 1.92 | -0.31 | 0.008 | 0.23 | -12.06 | 1.32 | 1.57 | -0.71 |
| MCF-12A | 1.29 | 1.02 | -1.46 | 0.014 | 0.27 | -10.44 | 0.08 | 0.57 | -8.12 |
| MCF-7 | 1.33 | 1.82 | -0.37 | 0.002 | 0.25 | -10.83 | 1.00 | 0.95 | -2.68 |
| Hs578 Bst | 2.28 | 1.87 | -0.35 | 1.073 | 1.61 | -0.60 | 1.23 | 1.75 | -0.56 |
12Δct+1: Copy number estimated by quantitative polymerase chain reaction.
2WGSA: Copy number estimated by whole genome sampling analysis assay.
3Sig: Log10 (p-value). p -value is derived from the presented algorithm by comparing the target sample to a reference set consisting of normal people. SNP, single nucleotide polymorphism.
Figure 3b also shows a comparison across four cell lines for a region of chromosome 9 from 0 to 40 Mb harbouring p16. WGSA results show that three of these cell lines have a significant deletion in the region of p16, as determined by SNP 139369, which is located within the p16 structural gene. This SNP, as well as two flanking SNPs, were further analysed by quantitative PCR, and the results are summarised in Table 1. The PCR results independently confirm the p16 deletion. In summary, PCR and the copy number algorithm show highly correlated results for two genomic regions with known alterations, namely c-MYC and p-16, and suggest that the identification of novel regions with copy number alterations should be feasible.
The SK-BR-3 chromosome 8 plot and the BT-20 chromosome 9 plot also illustrate the high resolution capabilities of the WGSA algorithm. SK-BR-3 shows two adjacent amplified segments (119 to 125.4 Mb and 127.5 to 127.7 Mb) near c-MYC. Twelve representative SNPs from the first and second segments were analysed by PCR and confirmed the WGSA copy number increase. There is a single SNP (719292) disrupting these two segments, which is scored as unamplified using both quantitative PCR (ΔCt = -0.3) and the copy number algorithm (p value = 0.43). BT-20 contains a single-point homozygous deletion (p16) flanked by SNPs that show no copy number alterations (Table 1). These two examples suggest that the algorithm is capable of single point resolution, which can result in improvements to the boundary delineations of gains and losses and result in highly refined genomic structures.
CPA
As described in the previous sections, the algorithm is able to detect homozygous deletions and amplifications with large copy number increases; however, the detection rate of regions with small copy number changes is relatively low. At a 1 per cent false-positive rate, the detection rate for X chromosome SNPs using the 1X, 3X,[35] 4X and 5X samples is 22.0 per cent, 12.4 per cent, 31.3 per cent and 54.9 per cent, respectively, as shown in Figure 4 (panels a and c). This moderate detection rate is due to dispersion of the reference set distribution in some SNPs rather than the lack of dosage response [36] CPA assumes that the greater the number of consecutive SNPs that display the same type of alteration (gain or loss), the greater the confidence in the significance of the changes [37] and is therefore applied to improve the detection rate. Figure 4 summarises the comparison between SPA and CPA. CPA results in a substantial shift of the receiver operating characteristic (ROC) curves toward the upper left-hand corner, indicating highly improved sensitivity and specificity. Panels c and d in Figure 4 are detailed views of panels a and b for the sub-region with a < 1 per cent false-positive rate. These graphs show that with a < 0.2 per cent false-positive rate, the true-positive (detection) rates for the 1X, 4X and 5X samples are 91.1 per cent, 91.4 per cent and 98.3 per cent, respectively. The true-positive rate for the 3X sample is improved to more than 50 per cent by using a false-positive rate of < 1 per cent. CPA shows much stronger power than SPA in these X chromosome examples because the span of the changes is continuous and large and the majority of the SNPs consistently show the same trend towards gain or loss.
Figure 4.
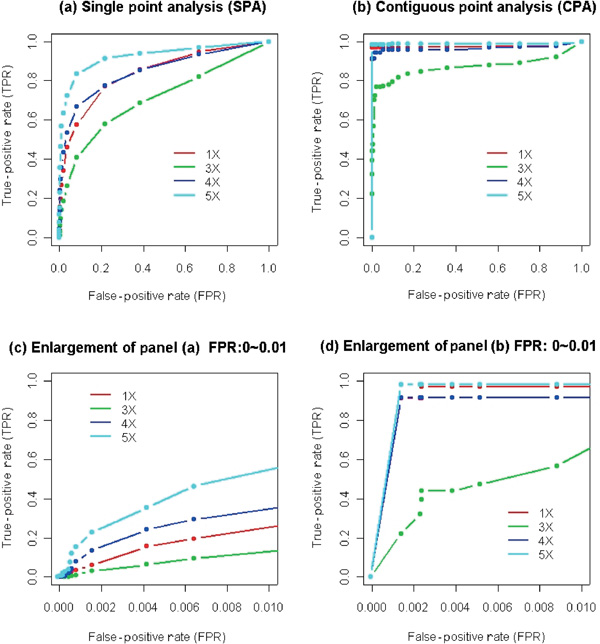
Receiver operating characteristic (ROC) curves for contiguous point analysis and single point analysis. In each panel, the false-positive rate is estimated by the average of leave-one-out cross-validation on 62 normal females (2X). The true-positive rate is estimated using 1X, 3X, 4X and 5X samples. With a range of p-value thresholds, a series of false-positive rates and true-positive rates can be calculated which form the basis of the ROC curves. Panels (c) and (d) are enlargements of (a) and (b), respectively, with the false-positive rate extending only to 1 per cent rather than 100 per cent.
LOH and copy number analysis
The matched Hs578 samples were used to compare traditional LOH identification (comparison of WGSA SNP genotypes between matched samples) with the application of a probability model for LOH identification. This application may be particularly useful when there is no matched normal control sample available for analysis. The model uses the allele frequency information of the reference set and calculates the probability that any given stretch of homozygous genotypes may occur due to random chance. The significance increases as the number of homozygous SNPs in the covered region increases. Thus, the use of a stringent significance cut-off may allow genomic regions with many consecutive homozygous calls to serve as a surrogate for conventionally-defined regions of LOH.
Using the matched Hs578 pair, the method was evaluated in terms of how well it captured traditionally-defined LOH markers. The comparative results are summarised in Table 2. There are, in total, 1,293 autosomal SNPs defined by traditional LOH analysis. These SNPs are heterozygous in the normal control and homozygous in the tumour sample. Among these SNPs, more than 80 per cent have a significance of less than 10-6 using the probability model. Yet, approximately 10 per cent of the SNPs have non-significant p -values (> 0.01). The stretches with significance < 10-6 have a mean span of 31.32 Mb, while the stretches with significance > 0.01 have a mean span of 1.11 Mb. This indicates that the majority of the traditionally defined LOH SNPs are located in long stretches of homozygous calls, while ~ 10 per cent of the SNPs reside in short stretches. By contrast, for all of the 11,205 autosomal SNPs in the normal control sample, there are no SNPs which belong to stretches with p -values lower than 10-6. Thus, for this particular sample pair, a p-value threshold of 10-6 captures more than 80 per cent of the traditionally-defined LOH, while the normal sample contains no regions at this level of significance. This result shows that the probability model can identify genomic regions that have undergone LOH in the paired cell lines and may serve as an alternative approach to LOH identification, especially when normal matched samples are not available.
Table 2.
Comparison between probability model and traditional loss of heterozygosity on Hs578 matched pair
| p-value | Normal match (%) | Tumour sample (%) |
|---|---|---|
| < 1 × 10-8 | 0 (0.00%) | 955 (73.78%) |
| < 1 × 10-6 | 0 (0.00%) | 1,037 (80.12%) |
| < 1 × 10-4 | 81 (0.72%) | 1,086 (83.91%) |
| < 1 × 10-2 | 1,179 (10.52%) | 1,158 (89.48%) |
| Total | 11,205 (100.00%) | 1,293 (100.00%) |
Copy number analysis of SNPs undergoing LOH in this tumour cell line reveals that approximately 32 per cent have one copy, 51 per cent have two copies, 17 per cent show moderate amplification (copy number less than eight) and less than 0.2 per cent show homozygous deletions or large-fold amplifications. Interestingly, the matched pair identifies regions of LOH where no obvious copy number alterations occur. By comparing the tumour and normal genotype calls, the entire length of chromosome 12 and chromosome 17, as well a ~ 90 to 170 Mb on chromosome 5, can be defined as LOH, yet there are no significant copy number alterations. This pattern is also observed in MCF-7 (Figure 3a), where a putative stretch of LOH containing 77 SNPs defined with the probability model from 57 to 77 Mb (p -value 7.2 ×10-16) shows no copy number reduction. Additionally, SK-BR-3 and ZR-75-30 both show a region of putative LOH from 110 to 125-135 Mb with respective p-values of 3.8 × 10-18 (80 SNPs) and 1.8 × 10-24 (120 SNPs), but show significant copy number increases. These examples of LOH with either no copy number reduction or copy number increase are not readily identified by many currently used single molecular approaches, and underscore the power in coupling LOH measurements with genome-wide copy number profiling.
Mixing experiment
Tumour samples can often be contaminated by normal cells of either stromal or lymphocytic origin. While methods such as laser capture micro-dissection or flow cytometry have been successfully used to enrich for tumour cells, the resulting populations are rarely completely pure and thus molecular methods that are used for genome-wide DNA copy number profiling must be sufficiently robust to accommodate heterogeneous samples. The matched pair Hs-578 was used to assess the tolerance of the WGSA assay and copy number algorithm to mixed DNA samples by testing the effect of increasing amounts of normal DNA (Hs-578Bst) mixed into the cancer sample (Hs-578T). Mixed samples were analysed for changes in LOH and for changes in the detection of copy number alterations. DNA derived from the cancer cell line was mixed prior to the WGSA assay with the normal matched DNA at increasing percentages of 0 per cent (pure cancer sample), 10 per cent, 20 per cent, 30 per cent, 40 per cent, 50 per cent, 60 per cent, 70 per cent, 80 per cent, 90 per cent and 100 per cent (pure normal samples). The modal chromosome number of Hs-578Bst and Hs-578T is 46 (diploid) and 59 (hypo-triploid), respectively, thus mixing by DNA mass approximates mixing by cell number. Figure 5 summarises the changes seen as a result of mixing on the identification of traditional LOH SNPs, as well as putative LOH regions, using the probability model. As the contribution of normal DNA increases, the number of traditionally defined LOH SNPs (red line) decreases. Following the same trend, the total length (green line) and total number (blue line) of LOH regions defined by the probability model also decrease. Overall, when the percentage of normal DNA is less than or equal to 30 per cent, more than 70 per cent of the LOH changes are retained. A significant shift occurs when the mixed normal DNA reaches 30 to 50 per cent of the total, resulting in nearly 60 per cent loss of detection of LOH. When normal DNA is present at 60 per cent or greater, most SNPs (> 98 per cent) undergoing LOH are undetectable. We also examined the relationship between the transition points of LOH detection and the copy number of these SNPs. This comparison involved three groups of LOH SNPs with different copy numbers, which comprised 99.8 per cent of the total: one-copy (407 SNPs), two-copy (663 SNPs) and moderate copy number (three to eight) increases (221 SNPs). On average, as the percentage of normal DNA increased in the mixed sample, the ability to detect a heterozygous call occured first for SNPs with one copy, followed next by those with two copies and lastly with those of moderate copy number. The difference between the three groups was statistically significant, with a p-value of 3.3 × 10-5 using the Kruskal-Wallis test. The Wilcoxon rank sum test was used to compare each pair. The following p-values for the differences between groups were found: 0.00742 (one-copy and two-copy), 0.00487 (two-copy and moderate copy) and 1.35 × 10-5 (one-copy and moderate copy). All comparisons are significant at a 0.05 level with Bonferroni correction, with the difference between the one-copy and the moderate copy number groups being the most significant.
Figure 5.
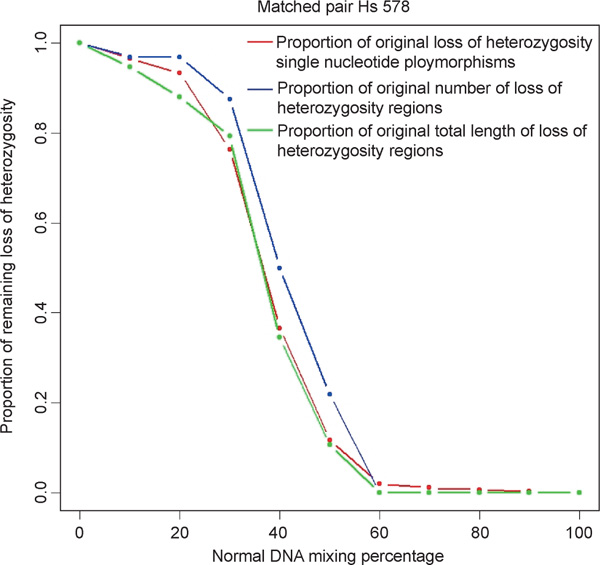
Loss of heterozygosity (LOH) analysis on mixed samples. The x-axis is the percentage of mixing of the normal DNA sample. The y-axis is the proportion of LOH signal remaining using three measurements: LOH single nucleotide polymorphisms (red dots and line), total length of LOH (blue dots and line) and total number of LOH regions (green dots and line). The definition of LOH regions and length is described in detail in the methods section.
The effect of mixed samples on detection of gains and losses was examined as well. The relative percentage of copy number alterations that are detected in mixed samples with CPA is greater than with SPA. At mixing levels of 10 per cent, 20 per cent and 30 per cent normal DNA, the detectable signals remaining from the original total were, respectively, 89.0 per cent, 85.7 per cent and 57.6 per cent (CPA) and 50 per cent, 25 per cent and 21.43 per cent (SPA). Once the proportion of normal DNA reaches 40 per cent of the total sample, there is a significant reduction in the detection of these amplified and deleted SNPs. This trend is true for both CPA and SPA. These results indicate that detection of LOH and copy number alterations using the WGSA assay and algorithm can tolerate a mixed sample containing up to 20 to 30 per cent normal DNA.
Discussion
We have developed an algorithm for genome-wide copy number estimation using high-density DNA oligonucleotide arrays in conjunction with target DNA preparations using WGSA. A comparison of experimental samples with a reference set consisting of more than 100 normal individuals allows p-values to be computed and statistically significant gains and losses to be identified. SNP-specific reference distributions are used to account for the inherent variability in normalised signal intensities across SNPs. Although the specific selection of probe sequences is constrained by the requirement for SNP genotyping by allele-specific hybridisation, and thus may not necessarily be optimised with regard to sensitivity and specificity for detection of copy number alterations, more than 96 per cent of the X chromosome SNPs have a correlation greater than 0.85 between log (signal intensity) and log (copy number). Copy number changes identified by the algorithm were well correlated with quantitative PCR results and could also be detected in samples containing mixtures of normal and tumour DNA. Lastly, the identification of genomic intervals with statistically significant stretches of homozygous markers can potentially allow detection of regions of LOH without the need for a matched normal control sample.
We have used SPA as an initial approach. An alternative to this is CPA, where consecutive SNPs displaying a consistent trend towards gains or losses are given additional weight and significance. CPA improves the sensitivity in the example of the X chromosome copy number alterations. CPA may require caution due to a bias towards long regions of copy number change, however, and may underestimate complex structures which do not span large distances. Also, CPA may have an impact on regions near the boundary of copy number changes in which moderate yet consistent signals are detected and therefore can lead to an overestimation of the absolute length of the alteration. Thus, the absolute false-positive rate for a give p-value threshold using SPA is lower than that using CPA for X chromosome SNPs. CPA could conceivably serve as a screening tool when the identification of all putative moderate alterations (high true-positive rate) is needed, while SPA may be more appropriate as a diagnostic tool due to the high specificity it displays. Since gene amplifications can be relatively simple continuous regions ranging from one to several hundred kb, such as in neuroblastomas, [38] rather than complex, irregular regions up to 20 Mb, as seen in breast cancers [39,40] SPA is essential in order to capture local alterations when marker density is not high. For both SPA and CPA, with more than 10,000 markers, an inevitable issue that arises is the multiple hypothesis testing problem. As a partial solution, the p -value threshold is stringently set so as to ensure high specificity (low false-positive rate) with concomitant lower sensitivity (higher false-negative rate) with regard to gains and losses. There are several alternative statistical methods that could be used to analyse the array data, such as kernel smoothing to average neighbouring points [41] change point method [42,43] and hidden Markov chain models [27,44]. The development of these approaches, while beyond the scope of this paper, would benefit from a training set of true-positive control samples containing a range of defined alterations with respect to length and copy number.
The identification of regions that may have undergone LOH using a probability-based model, in lieu of conventional methods using paired samples, offers analysis of unmatched cancer samples. This approach calculates the likelihood of a stretch of homozygous genotype calls by using allele frequencies derived from the normal reference set. This model-based approach can therefore serve as a guideline to regions of LOH in cases where a normal control sample is not available. Since regions of linkage disequilibrium can vary across the genome,[45] the probability model may tend to overestimate the significance of regions of LOH by treating each SNP independently. Once a significant stretch of homozygosity is identified, the interpretation of whether it truly represents LOH may be difficult due to the presence of homozygous segments in the human genome [46] Using 8,000 short tandem-repeat polymorphisms, several CEPH families showed homozygous segments greater than 10 centimorgans [47].
In conclusion, we have developed an algorithm which uses the Affymetrix GeneChip® Mapping 10K assay (Xba_131 array) to identify genome-wide copy number gains and losses. While copy number estimations across the genome can be made independently of SNP genotype calls (LOH analysis), linking the two datasets offers insights into complex genomic structures which few alternative single methods are capable of. The integration of transcriptional profiles of samples to the copy number profiles should further reveal functional roles for genomic regions with allelic imbalances. As the information content on the high density array increases with decreasing feature size, the WGSA assay is easily scalable beyond 100,000 SNPs. This will result in unprecedented resolution across the genome and should prove to be useful in elucidating genomic changes underlying the complex chromosomal make-up of tumour cells.
Acknowledgements
We thank Weiwei Liu, Nike Beaubier and Julia Yeh for technical assistance, and Kyle Cole for critical reading of the manuscript.
References
- Albertson DG, Collins C, McCormick F. et al. 'Chromosome aberrations in solid tumors'. Nat Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. 'Genetic instabilities in human cancers'. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Cavenee WK, Dryja TP, Phillips RA. et al. 'Expression of recessive alleles by chromosomal mechanisms in retinoblastoma'. Nature. 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D. et al. 'Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors'. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- Schrock E, du Manoir S, Veldman T. et al. 'Multicolor spectral karyotyping of human chromosomes'. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Landegent J, Collins C. et al. 'Fluorescence in situ hybridization with human chromosome-specific libraries: Detection of trisomy 21 and translocations of chromosome 4'. Proc Natl Acad Sci USA. 1988;85:9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsyn NA, Lisitsina NM, Dalbagni G. et al. 'Comparative genomic analysis of tumors: Detection of DNA losses and amplification'. Proc Natl Acad Sci USA. 1995;92:151–155. doi: 10.1073/pnas.92.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucito R, Nakimura M, West JA. et al. 'Genetic analysis using genomic representations'. Proc Natl Acad Sci USA. 1998;95:4487–4492. doi: 10.1073/pnas.95.8.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TL, Maierhofer C, Speicher MR. et al. 'Digital kar-yotyping'. Proc Natl Acad Sci USA. 2002;99:16156–16161. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucito RJ, Healy J, Alexander A. et al. 'Representational digonucleotide microarray analysis: A high-resolution method to detect genome copy number variation'. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor SP, Read JL, Pirrung MC. et al. 'Light-directed, spatially addressable parallel chemical synthesis'. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Fodor SP, Rava RP, Huang XC. et al. 'Multiplexed biochemical assays with biological chips'. Nature. 1993;364:555–556. doi: 10.1038/364555a0. [DOI] [PubMed] [Google Scholar]
- Pease AC, Solas D, Sullivan EJ. et al. 'Light-generated oligo-nucleotide arrays for rapid DNA sequence analysis'. Proc Natl Acad Sci USA. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Tanenbaum DM, Daly MJ. et al. 'Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleo-tide polymorphism arrays'. Nat Biotechnol. 2000;18:1001–1005. doi: 10.1038/79269. [DOI] [PubMed] [Google Scholar]
- Mei R, Galipeau PC, Prass C. et al. 'Genome-wide detection of allelic imbalance using human SNPs and high-density DNA arrays'. Genome Res. 2000;10:1126–1137. doi: 10.1101/gr.10.8.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert EL, Hsu L, Cousens LA. et al. 'Single nucleotide polymorphism array analysis of flow-sorted epithelial cells from frozen versus fixed tissues for whole genome analysis of allelic loss in breast cancer'. Am J Pathol. 2002;160:73–79. doi: 10.1016/S0002-9440(10)64351-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumur CI, Dechsukhum C, Ware JL. et al. 'Genome-wide detection of LOH in prostate cancer using human SNP microarray technology'. Genomics. 2003;81:260–269. doi: 10.1016/S0888-7543(03)00020-X. [DOI] [PubMed] [Google Scholar]
- Kennedy GC, Matsuzaki H, Dong S. et al. 'Large-scale gen-otyping of complex DNA'. Nat Biotechnol. 2003;21:1233–1237. doi: 10.1038/nbt869. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Loi H, Dong S. et al. 'Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high density oligonucleotide array'. Genome Res. 2004;14:414–425. doi: 10.1101/gr.2014904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett AJ, Smith HS, Springer EL. et al. 'Two syngeneic cell lines from human breast tissue: the aneuploid mammary epithelial (Hs578T) and the diploid myoepithelial (Hs578Bst) cell lines'. J Natl Cancer Inst. 1977;58:1795–1806. doi: 10.1093/jnci/58.6.1795. [DOI] [PubMed] [Google Scholar]
- All references to the function log default to e as the base (natural log) unless stated otherwise (such as log10)
- Liu WM, Di X, Yang G. et al. 'Algorithms for large-scale genotyping microarrays'. Bioinformatics. 2003;19:2397–2403. doi: 10.1093/bioinformatics/btg332. [DOI] [PubMed] [Google Scholar]
- If the genotype of the target cell line is missing (no call), or the number of reference samples with that particular genotype is small (less than 10), all 110 reference samples are used to estimate the distribution
- Based on Shapiro-Wilk's W test for normality, only 3.3 per cent of the reference distributions have p-values of less than 0.001, which is further reduced to 0.7 per cent when a more stringent cut-off of 0.0001 is used
- The total number of outliers that are removed is low: 90.38 per cent of these distributions have no outliers removed; 9.23 per cent of the distributions have one outlier removed; 0.38 per cent of the distributions have two outliers removed; 0.01 per cent of the distributions have three outliers removed; and in no cases have more than three outliers been removed
- Bignell GR, Huang J, Greshock J. et al. 'High-resolution analysis of DNA copy number using oligonucleotide microarrays'. Genome Res. 2004;14:287–295. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li C, Paez JG, 'An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays'. Cancer Res. 2004. in press . [DOI] [PubMed]
- Forozan F, Mahlamaki EH, Monni O. et al. 'Comparative genomic hybridization analysis of 38 breast cancer cell lines: A basis for interpreting complementary DNA microarray data'. Cancer Res. 2000;60:4519–4525. [PubMed] [Google Scholar]
- Struski S, Doco-Fenzy M, Cornillet-Lefebvre P. 'Compilation of published comparative genomic hybridization studies'. Cancer Genet Cytogenet. 2002;135:63–90. doi: 10.1016/S0165-4608(01)00624-0. [DOI] [PubMed] [Google Scholar]
- Escot C, Theillet C, Lidereau R. et al. 'Genetic alteration of the c-myc protooncogene (MYC) in human primary breast carcinomas'. Proc Natl Acad Sci USA. 1986;83:4834–4838. doi: 10.1073/pnas.83.13.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rummukainen J, Kytola S, Karhu R. et al. 'Aberrations of chromosome 8 in 16 breast cancer cell lines by comparative genomic hybridization, fluorescence in situ hybridization, and spectral karyotyping'. Cancer Genet Cytogenet. 2001;126:1–7. doi: 10.1016/S0165-4608(00)00387-3. [DOI] [PubMed] [Google Scholar]
- Kamb A, Gruis NA, Weaver-Feldhaus J. et al. 'A cell cycle regulator potentially involved in genesis of many tumor types'. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- Cairns P, Polascik TJ, Eby Y. et al. 'Frequency of homozygous deletion at p16/CDKN2 in primary human tumours'. Nat Genet. 1995;11:210–212. doi: 10.1038/ng1095-210. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Piper J. et al. 'Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization'. Proc Natl Acad Sci USA. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Since log (intensity) has a strong correlation with log (copy number), the difference in log (intensity) should be similar for 1X versus 2X and 4X versus 2X, and smaller for 3X versus 2X
- The overall dosage response is strong, with a correlation greater than 0.72 between log (intensity) and log (copy number) for all 302 X chromosome SNPs. Furthermore, 292 SNPs (96.7 per cent) among this group have a correlation greater than 0.85
- Salamon H, Kato-Maeda M, Small PM. et al. 'Detection of deleted genomic DNA using a semiautomated computational analysis of GeneChip data'. Genome Res. 2000;10:2044–2054. doi: 10.1101/gr.GR-1529R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amler LC, Schwab M. 'Amplified N-myc in human neuroblastoma cells is often arranged as clustered tandem repeats of differently recombined DNA'. Mol Cell Biol. 1989;9:4903–4913. doi: 10.1128/mcb.9.11.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan XY, Meltzer PS, Dalton WS. et al. 'Identification of cryptic sites of DNA sequence amplificationin human breast cancer by chromosome microdissection'. Nat Genet. 1994;8:155–161. doi: 10.1038/ng1094-155. [DOI] [PubMed] [Google Scholar]
- Szepetowski P, Perucca-Lostanlen D, Gaudray P. 'Mapping genes according to their amplification status in tumor cells: Contribution to the map of 11q13'. Genomics. 1993;16:745–750. doi: 10.1006/geno.1993.1257. [DOI] [PubMed] [Google Scholar]
- Wand MC, Jones MC. 'Kernel Smoothing'. Chapman and Hall, London, UK; 1995. [Google Scholar]
- Sen A, Srivastava MS. 'On tests for detecting a change in mean'. Annals of Statistics. 1975;3:98–108. doi: 10.1214/aos/1176343001. [DOI] [Google Scholar]
- Olshen AB, Venkatraman ES. 'Change-point analysis of array-based comparative genomic hybridization data', in: 'Proceedings of the Joint Statistical Meetings'. American Statistical Association, Alexandria, VA; 2002. [Google Scholar]
- Rabiner LR. 'A tutorial on hidden Markov models and selected applications in speech recognition'. Proc IEEE. 1989;77:257–285. doi: 10.1109/5.18626. [DOI] [Google Scholar]
- Ardlie KG, Kruglyak L, Seielstad M. 'Patterns of linkage disequilibrium in the human genome'. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- Clark AG. 'The size distribution of homozygous segments in the human genome'. Am J Hum Genet. 1999;65:1489–1492. doi: 10.1086/302668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Weber JL. 'Long homozygous chromosomal segments in reference families from the centre d'Etude du polymorphisme humain'. Am J Hum Genet. 1999;65:1493–1500. doi: 10.1086/302661. [DOI] [PMC free article] [PubMed] [Google Scholar]


