Abstract
Annotation of the human genome is inching forward. Seven human chromosomes have now been fully annotated, covering 17 per cent of the genome, and at least one chromosome has been re-annotated. The enormity of the task forces a dependence on automated tools for detecting and assembling the genes, followed by hand curation to correct errors and polish the gene models. The accuracy of gene prediction algorithms is very good for internal exons from intact genes, but these programs do peculiar and exasperating things to pseudogenes. These programs can actually resurrect pseudogenes from the dead, making them into viable gene models for intact proteins, albeit science-fictional proteins. This process is demonstrated for four human pseudogenes from the cytochrome P450 family and one putatively functional P450 gene, CYP2U1, having a non-consensus intron boundary. These examples are offered as a call-to-arms to improve pseudogene prediction as an art in itself, and not as a by-product of gene annotation. Failure to do so will flood the databases with thousands of false-positive predictions. Indeed, they are already there.
Keywords: pseudogenes, pseudogene prediction, genome annotation, cytochrome P450 (CYP) genes, GNOMON
Introduction
The assembly of the human genome from whole-genome-shotgun sequence reads was once deemed impossible, and the probable outcome was dubbed the 'MAD Magazine version of the human genome' [1]. This may have referred to MAD's fold-in back cover, where a drawing and the caption under it are folded twice to reveal a new picture and a new caption to go with it. This fold-in back cover is rather like alternative splicing, or, more likely, the intent was to imply production of a crazy-quilt genome structure caused by the incorrect joining of the millions of repeat sequences in the human genome. In spite of these concerns, this did not happen. Paired-end sequencing of three sizes of clones largely avoided the misas-sembly problem and resulted in a faithful rendition of the genome, now with only 498 contigs in Build 34 version 2 [2]. The spectre of the MAD Magazine version of the genome is coming back in a new guise, however: the corrupt annotation of pseudogenes.
Programs designed to scan genomic DNA for genes are trained to look for GT and AG boundaries and some consensus regions around these boundaries. They also evaluate statistical properties of exons to try to assemble a gene. When they encounter a pseudogene that is nearly intact, they try mightily to make it code for an intact protein. The outcome is a Frankenstein gene that never existed in reality, but was cobbled together from spare parts, beginning with the pseudogene carcass. Although it is thought that the number of pseudogenes in the human genome is 20,000 or more, [3] these genome features are not annotated as completely as intact genes. Locus Link has 21,382 protein coding genes, but only 2,592 pseudogenes are listed (as of 13th February, 2004) [4]. Complete annotation of the human genome must also include identification and naming of all the pseudogenes.
Pseudogenes live
Several examples are given from the human cytochrome P450 (CYP) pseudogenes. The human and mouse cytochromes P450 have been annotated in detail [5]. There are 57 intact and putatively functional huma CYP genes, as well as 58 pseudogenes [6]. These examples illustrate the trouble that the pseudogenes present for gene prediction algorithms. In the first example, CYP2G2P encodes a nearly intact P450 (Figure 1A) with two in-frame stop codons, one in exon 1 and one in exon 3. The intron-exon boundaries are normal. Frequently, pseudogenes are predicted to be short on the ends. A frameshift or stop codon in exon 1 may trick the program into accepting the next downstream Met as the starting Met; this happened in CYP2G2P, with a Met immediately after the exon 1 stop codon being assigned as the start codon. A stop codon or frameshift in an internal exon can have multiple effects. If the phase of the two intron-exon junctions bracketing the defective exon is the same, then the exon may just be skipped. This happened in CYP2G2P. The result is a shortened protein (Figure 1B), missing part of exon 1 and all of exon 3. The Ensembl prediction for this same protein is even shorter; it is missing all of exons 1, 3, 8 and 9. Figure 1C shows the alignment of the lower-case portions of the pseudogene with the functional mouse CYP2G1 gene, to show what was missed by the prediction algorithm.
Figure 1.

(A) The human CYP2G2P pseudogene. The nine exons are shown, one per line, with the intron phase indicated in parenthese. The * are stop codons. Lower-case letters are the parts of the pseudogene missed in the Genscan prediction shown in panel B. (B) Genscan prediction of the CYP2G2P pseudogene taken from the University of California Santa Cruz Genome browser. The gene is shown in the same format as in Panel A, with blank space for the missed regions. (C) Alignments of the two regions left out of the Genscan prediction against the mouse CYP2G1 functional gene.
More subtle effects can come from internal exon defects. A frameshift or stop codon in an internal exon may trigger several possible solutions to be created by the program. Figure 2 illustrates a second possibility not seen in Figure 1. In the CYP2AB1P pseudogene, a bad GT boundary at the end of exon 2 and a stop codon in exon 3 are both solved by creating a new cryptic exon. This new exon bypasses both defects in the protein-coding region. The result shortens exon 2 by 13 amino acids and replaces the first 25 amino acids of exon 3 with 17 amino acids derived from an alternative frame translation from the first part of exon 3. Later in the pseudogene, a defective exon boundary between exons 6 and 7 and a frameshift in exon 7 are avoided by creation of a second cryptic exon. This new exon replaces exon 6 and about half of exon 7 and bypasses both genetic defects. There is a missing stop codon at the end of this pseudogene. This is handled by extending downstream to the next available stop. This pseudopseudogene is now taking on a new artificial look, like a car that has been in a wreck and now has a different coloured door.
Figure 2.

(A) The human CYP2AB1P pseudogene, using the same conventions as in Figure 1. (B) Genscan prediction of the CYP2AB1P pseudogene. New cryptic exons are underlined. (C) Alignments of the two regions left out of the Genscan prediction against the mous CYP2AB1 putatively functional gene.
The CYP51 pseudogenes pose an even greater challenge for gene prediction. The CYP51P1 pseudogene has only 130 of 508 amino acids correctly predicted (Figure 3A, upper case). Curiously, the Genscan prediction adds one bogus exon 72,022 base pairs (bp) upstream; this distant exon has no significant BLAST hits other than itself in the nr section, of Genbank, and no hits in the dbEST section. It is hard to imagine why this sequence is included as part of the CYP51P1 prediction.
Figure 3.

(A) The human CYP51P1 pseudogene. Upper-case sequence (130/508 amino acids) is correctly predicted by Genscan. (B) The underlined sequence is a new first exon, plus an upstream extension from the start codon, to find a phase 2 junction. Most of the pseudogene is not predicted, although the pseudogene is 92 per cent identical to CYP51 over the entire length of the protein sequence.
Gene fusions
The CYP51 errors continue with CYP51P2. The prediction for this pseudogene is 937 amino acids long, about twice the size of any P450 protein (Figure 4). Investigation of the sequence shows that, after 211 P450 residues (about 42 per cent), most of the transporter ADD1 gene (major facilitator superfamily) NM_181785 has been added. There are 387 more amino acids after this, so another fusion was suspected. The University of California Santa Cruz genome browser [7] shows another gene in this region with multiple mRNAs, but it is on the opposite strand. Therefore, the Genscan prediction for CYP51P2 fuses less than half of the true P450 pseudogene with another downstream gene and additional exon predictions that cannot be real since they overlap a third gene on the opposite strand. This problem of gene fusions is not rare. While annotating P450 genes in the Drosophila pseudoobscura genome, a predicted triple-gene fusion was discovered which was 2,182 amino acids long (Contig3323_Contig6140.179) [8]. This sequence sandwiched a P450 gene (CYP9F2 orthologue, minus the last seven amino acids) between a DNA ligase III homologue and an angiotensin-converting enzyme (ACER) homologue. Erroneous gene fusions of multiple CYP genes in gene clusters have been observed in Caenorhabditis elegans, Arabidopsis thaliana and Takifugu rubripes (pufferfish).
Figure 4.
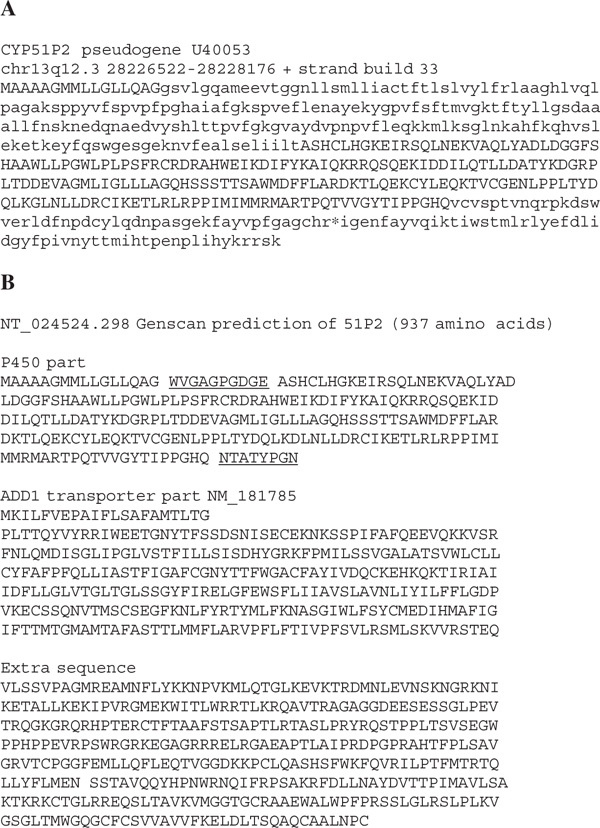
(A) The human CYP51P2 pseudogene. Upper-case sequence (211/508 amino acids) is correctly predicted. (B) Genscan prediction for CYP51P2. This is a fusion of part of CYP51P2 with the ADD/ transporter gene and an additional sequence derived from the opposite strand of another gene.
Stumbling on a GC boundary
Real genes with no defects may still be misassembled by gene prediction programs. CYP2U1 , a human P450 gene, has a verified GC boundary at the end of exon 3 (Figure 5A). The joint at this position is confirmed by cDNA sequences. The Genscan prediction misses the correct intron 1 GT boundary and jumps to the next one of the same phase. This adds 17 amino acids to the first exon. Genscan skips the GC boundary at exon 3 and extends exon 3 by 11 amino acids. A stop codon then truncates the protein prematurely. The last two real exons are missed. This gene is correctly annotated in Genbank because the mRNA sequence is known; however, there are genes with no known mRNA sequences that can be used to correct errors like this.
Figure 5.

(A) The human CYP2U1 gene known from full-length cDNA has a GC intron boundary at the end of exon 3. (B) The Genscan prediction of CYP2U1. The underlined sequence is not found in the true protein and the last two exons are missed after the GC boundary is read through.
Taking a lesson from the re-annotation of Drosophila
A recent re-annotation effort on the Drosophila melanogaster genome has predicted approximately 2,000 more genes than is the case for the current Berkeley Drosophila Genome Project (BDGP) annotations [9,10]. This represents an increase of almost 15 per cent. This result was obtained by relaxing the stringency of the FGENESH prediction algorithm; 7,464 gene models were predicted above and beyond the BDGP annotations. Many of these gene models will be pseudogenes or false-positive predictions. Validation by spotting the gene model exon sequences in a microarray and probing for mRNA sequences that bound to the array did give support for a large percentage of these predictions. Further analysis by reverse transcriptase polymerase chain reaction (RT-PCR) of a subset of the microarray-positive models confirmed that many of these genes are expressed and need to be added to the official gene count [9].
These findings are important to human genome annotators for two reasons. First, the number of weakly supported or unsupported gene models was large, but a significant fraction of these weak predictions were verified by microarray and RT-PCR experiments. Some of these genes had no BLAST hits in Genbank and so represent new protein families; this argues that the conservative estimates (approximately 25,000 genes) used by some human annotators [11] are missing many real genes. Secondly, it is perilous not to identify and properly annotate the pseudogenes. There are an estimated 20,000 pseudogenes in the human genome and this number may be much higher [3,5]. The collection of these defective genes may be called the human pseudogenome. As discussed above, what comes out of this set -- after Genscan or Ensembl gets through with it -- is pretty frightening. These are vivisected genes raised from the dead, as sure as Shelley's monster.
Conclusions
The human genome is large and it contains large genes. The recent annotation of chromosome 6 illustrates this point [12,13]. The BPAG1 gene contains 101 exons. Another chromosome 6 gene, TCBA1 , contains an intron that is 479 kilobases long. ZNF451 , a zinc finger protein, contains a single exon of 9,114 bp. These are all potential challenges for gene prediction programs. Unfortunately, prediction algorithms are not written to cope with pseudogenes, sequence errors or GC boundaries, giving rise to the results shown in Figures 1-5. Even the detailed manual annotation of chromosome 6 found only 633 pseudogenes, as compared with 1,557 genes. Other evidence suggests that the number of human pseudogenes should be closer to the number of intact genes. Part of this problem lies in the definition of pseudogenes. Perhaps the working definition for the chromosome 6 annotation did not include some small solo exons or detritus exons scattered near documented genes [5]. It is possible, however, that there might still be about 900 undocumented pseudogenes on chromosome 6.
The Genscan program is quite successful in predicting genes, especially the internal exons of real genes [14]. It has been optimised for different G + C compositions and different species, but not for pseudogenes. In order to be able to annotate all of the human genome and other genomes, there is a need for a second program, which is optimised for detecting pseudogenes. These pseudogenes need to be named and documented on the sequence records and genome browsers in order to avoid the aforementioned problem of creating Frankenstein genes.
One program that may fulfil some of these criteria is GNOMON, the National Center for Biotechnology Information's Hidden Markov Model ab initio prediction program [15]. BLAST searches against the GNOMON predictions can be performed from MapViewer [16]. This program assumes that very close exon predictions (less than 50 bp apart) are separated by a frameshift, because very short introns are rare (at least in vertebrates). A frameshift is introduced to merge the two exons into one. The Hidden Markov Model used by GNOMON allows non-consensus splice sites such as the GC boundary in CYP2U1. The CYP2U1 gene is correctly predicted by GNOMON. Stop codons in the middle of an otherwise strong alignment are disregarded and the stops are included in the model, which is treated as a pseudogene rather than as a gene. These features ought to allow the correct prediction of the pseudogenes in Figures 1-4. In practice, even GNOMON makes a few errors. For example, all amino acids of CYP2G2P are correctly predicted, but 20 extra amino acids are added between exons 2 and 3. The CYP2AB1P sequence predicted by GNOMON has 69 more correctly predicted amino acids than Genscan, and includes two of the in-frame stops, but it misses exon 7 and adds the same cryptic exon as Genscan did after exon 5. GNOMON finds most of the C-terminal part of CYP51P1 , but misses the first two-thirds. CYP51P2 coverage is improved, but both N- and C-terminal regions are missed, as well as two internal segments. In short, GNOMON is better than Genscan for these five genes but still misses the gold standard of hand curation.
Finally, once pseudogenes are correctly assembled, nomenclature groups must devise a suitable nomenclature for them. This will not be easy. Pseudogenes are a heterogeneous set. Our own efforts at CYP pseudogene nomenclature identify at least four types of pseudogenes [5]. This nomenclature could be applied to other gene families, or other proposals could be made, but this problem needs to be addressed. In the future, progress in genome annotation should make it increasingly hard to find the misleading and erroneous peptide predictions that currently abound, as these become replaced by accurate annotated and named pseudogene models.
References
- Anon. USA Today. Arlington; 1998. 'The book of life: Twin efforts will attempt to write it'; p. 06D. [Google Scholar]
- National Center for Biotechnology Information. 'Statistics: Genome annotation'. 2003. http://www.ncbi.nlm.nih.gov/mapview/stats/BuildStats.cgi?taxid = 9606&build = 34&ve = 2 Human genome Build 34 version 2, from data freeze of 19th November, (cited 8th March, 2004)
- Harrison PM, Hegyi H, Balasubramanian S. et al. 'Molecular fossils in the human genome: Identification and analysis of the pseudogenes in chromosomes 21 and 22'. Genome Res. 2002;12:272–280. doi: 10.1101/gr.207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Biotechnology Information (NCBI) LocusLink/Statistics. 2004. http://www.ncbi.nlm.nih.gov/LocusLink/statistics.html (cited 18th February, 2004).
- Nelson DR, Zeldin DC, Hoffman SMG. et al. 'Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes, and alternative-splice variants'. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- Nelson DR. 'Human P450s in FASTA format'. 2003. http://drnelson.utmem.edu/human.blast.file.html (cited 8th March, 2004).
- Kent WJ. UCSC Genome Browser, July, Human genome assembly. 2003. http://genome.ucsc.edu/ (cited 18th February, 2004).
- Nelson DR. ' Drosophila pseudoobscura P450s'. 2004. http://drnelson.utmem.edu/pseudoobscura.html (cited 18th February, 2004).
- Hild M, Beckmann B, Haas SA. et al. 'An integrated gene annotation and transcriptional profiling approach towards the full gene content of the Drosophila genome'. Genome Biol. 2003;5:R3. doi: 10.1186/gb-2003-5-1-r3. http://genomebiology.com/2003/5/I/R3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver B, Leblanc B. 'How many genes in a genome?'. Genome Biol. 2003;5:204. doi: 10.1186/gb-2003-5-1-204. http://genomebiology.com/mkt/5114/2003/5/1/204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Keibler E, Hu P. 'Leveraging the mouse genome for gene prediction in human: from whole-genome shotgun reads to a global synteny map'. Genome Res. 2003;13:46–54. doi: 10.1101/gr.830003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall AJ, Palmer SA, Sims SK. et al. 'The DNA sequence and analysis of human chromosome 6'. Natur. 2003;425:805–811. doi: 10.1038/nature02055. [DOI] [PubMed] [Google Scholar]
- Grimwood J, Schmutz J. 'Six is seventh'. Natur. 2003;425:775–776. doi: 10.1038/425775a. [DOI] [PubMed] [Google Scholar]
- Burge C. The New GENSCAN We b Server at MIT, modified. 2003. http://genes.mit.edu/GENSCAN.html (cited 18th February, 2004).
- National Center for Biotechnology Information. Gnomon description. 2003. http://www.ncbi.nlm.nih.gov/genome/guide/gnomon.html (cited 8 March, 2004)
- http://www.ncbi.nlm.nih.gov/genome/seq/page.cgi?F = HsBlast.html&&ORG = Hs


