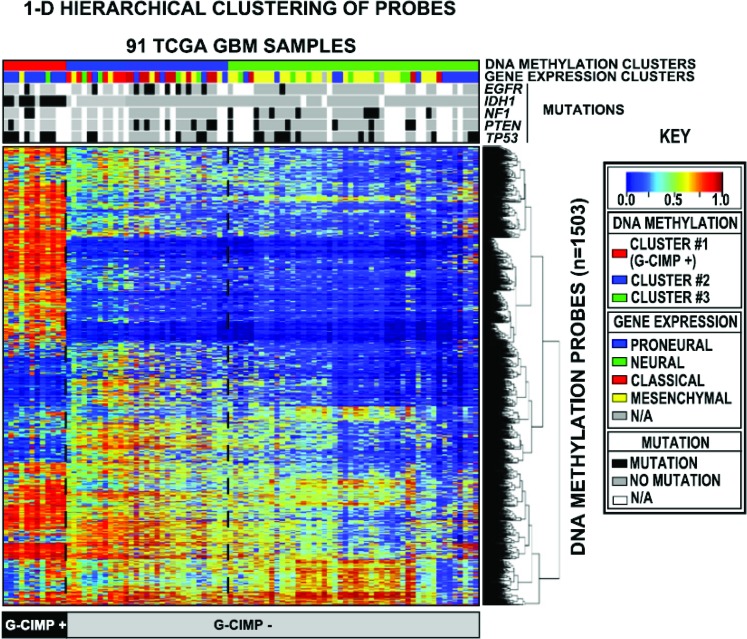
Figure 2. Identification of the CpG island methylator phenotype in glioblastoma.
DNA methylation from 91 TCGA glioblastomas was profiled and subjected to unsupervised clustering (statistical methodology, reference 57). Three distinct methylation clusters were identified, with cluster 1 (red top bar) designated as G-CIMP+ due to a high frequency of DNA methylation. The gene expression profile and mutation status of selected genes (EGFR, IDH1, NF1, PTEN, and p53) are shown in the bars below each methylation cluster. IDH1 mutations were found exclusively in G-CIMP+ tumors, and nearly all G-CIMP+ tumors had a proneural gene expression profile. Adapted from Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010;17:510–522; with permission from Elsevier. EGFR = epidermal growth factor receptor; G-CIMP = CpG hypermethylator phenotype; GBM = glioblastoma multiforme; IDH1 = isocitrate dehydrogenase 1 gene; NF1 = neurofibromatosis 1 gene; PTEN = phosphatase and tensin homologue gene; TCGA = The Cancer Genome Atlas; TP53 = p53 gene.