Abstract
Subsequent to the two rounds of whole-genome duplication that occurred in the common ancestor of vertebrates, a third genome duplication occurred in the stem lineage of teleost fishes. This teleost-specific genome duplication (TGD) is thought to have provided genetic raw materials for the physiological, morphological, and behavioral diversification of this highly speciose group. The extreme physiological versatility of teleost fish is manifest in their diversity of blood–gas transport traits, which reflects the myriad solutions that have evolved to maintain tissue O2 delivery in the face of changing metabolic demands and environmental O2 availability during different ontogenetic stages. During the course of development, regulatory changes in blood–O2 transport are mediated by the expression of multiple, functionally distinct hemoglobin (Hb) isoforms that meet the particular O2-transport challenges encountered by the developing embryo or fetus (in viviparous or oviparous species) and in free-swimming larvae and adults. The main objective of the present study was to assess the relative contributions of whole-genome duplication, large-scale segmental duplication, and small-scale gene duplication in producing the extraordinary functional diversity of teleost Hbs. To accomplish this, we integrated phylogenetic reconstructions with analyses of conserved synteny to characterize the genomic organization and evolutionary history of the globin gene clusters of teleosts. These results were then integrated with available experimental data on functional properties and developmental patterns of stage-specific gene expression. Our results indicate that multiple α- and β-globin genes were present in the common ancestor of gars (order Lepisoteiformes) and teleosts. The comparative genomic analysis revealed that teleosts possess a dual set of TGD-derived globin gene clusters, each of which has undergone lineage-specific changes in gene content via repeated duplication and deletion events. Phylogenetic reconstructions revealed that paralogous genes convergently evolved similar functional properties in different teleost lineages. Consistent with other recent studies of globin gene family evolution in vertebrates, our results revealed evidence for repeated evolutionary transitions in the developmental regulation of Hb synthesis.
Keywords: gene duplication, genome duplication, gene family evolution, convergent evolution
Introduction
Evidence suggests that two successive rounds of whole-genome duplication that occurred early in vertebrate evolution may have played an important role in the evolution of vertebrate-specific innovations (Holland et al. 1994; Meyer 1998; Meyer and Schartl 1999; Shimeld and Holland 2000; Wada 2001; Hoegg and Meyer 2005; Wada and Makabe 2006; Zhang and Cohn 2008; Van de Peer et al. 2009; Hoffmann, Opazo, and Storz 2012). Roughly 320–400 Ma, a third genome duplication occurred in the stem lineage of teleost fish (infraclass Teleostei) following divergence from nonteleost ray-finned fish (Amores et al. 1998, 2011; Postlethwait et al. 2000; Taylor et al. 2001, 2003; Van de Peer et al. 2003; Hoegg et al. 2004; Jaillon et al. 2004; Meyer and Van de Peer 2005; Kasahara et al. 2007; Sato and Nishida 2010). The teleost-specific genome duplication (TGD) is thought to have provided raw materials for the physiological, morphological, and behavioral diversification of teleost fish, perhaps facilitating the radiation of this speciose group into diverse marine and freshwater environments across the planet. Evidence in support of a causal connection between the TGD and phenotypic innovation is provided by studies of TGD-derived gene duplicates that evolved distinct physiological or developmental functions in various teleost lineages (Meyer and Málaga-Trillo 1999; Lister et al. 2001; Mulley et al. 2006; Braasch et al. 2006, 2007; Hashiguchi and Nishida 2007; Hoegg and Meyer 2007; Sato and Nishida 2007; Siegel et al. 2007; Yu et al. 2007; Douard et al. 2008; Braasch, Brunet, et al. 2009; Braasch, Volff, et al. 2009; Sato et al. 2009a, 2009b; Arnegard et al. 2010).
The extreme physiological versatility of teleost fishes is manifest in their diversity of blood–gas transport traits (Wells 2009). This diversity reflects the myriad solutions that have evolved to maintain tissue O2 delivery in the face of changing metabolic demands and environmental O2 availability during different ontogenetic stages. Relative to air-breathing vertebrates, fish generally contend with far greater vicissitudes of environmental O2 availability, which is largely because O2 solubility (and hence, the availability of dissolved O2 for respiration) varies as a function of water temperature. During ontogeny, regulatory changes in blood-O2 transport are mediated by the expression of multiple, functionally distinct hemoglobin (Hb) isoforms that are adapted to the particular O2-transport challenges encountered by the developing embryo or fetus (in viviparous or oviparous species) and in free-swimming larvae and adults (reviewed by Ingermann 1997; Jensen et al. 1998). As in other vertebrates, the developmental regulation of Hb synthesis in fish involves differential expression of duplicated genes that encode the α- and β-chain subunits of distinct tetrameric α2β2 Hb isoforms (Chan et al. 1997; Brownlie et al. 2003; Maruyama, Yasumasu, and Iuchi 2004; Maruyama, Yasumasu, Naruse, et al. 2004; Tiedke et al. 2011).
Most teleost fish also coexpress functionally distinct Hb isoforms during posthatching life, and these isoforms can be broadly classified (based on electrophoretic mobility at pH > 8.0) as “anodic” or “cathodic.” The anodic Hbs have relatively low O2 affinities and a pronounced Bohr effect (decreased Hb-O2 affinity at low pH), whereas the cathodic Hbs have relatively high O2 affinities, an enhanced responsiveness to allosteric regulation by organic phosphates, and a reversed Bohr effect (increased Hb-O2 affinity at low pH) in the absence of organic phosphates (Weber and Jensen 1988; Weber 1990, 2000; Jensen et al. 1998; Weber et al. 2000; Wells 2009). Experimental evidence for some species suggests that regulatory changes in intraerythrocytic Hb isoform composition may play a role in the acclimatization response to environmental hypoxia (e.g., Rutjes et al. 2007), but it has not been possible to formulate any broadly consistent empirical generalizations (Weber and Jensen 1988; Weber, 1990, 2000; Ingermann 1997; Wells 2009). A remarkable feature of nearly all anodic Hb isoforms of teleost fish is the Root effect, an extreme reduction in Hb-O2 binding capacity at low pH, even when blood O2 tension remains high. The Root effect is considered a key evolutionary innovation in teleost fish, as it plays a critical role in secreting O2 into the swim bladder for buoyancy control and in supplying O2 to the avascular retina (Pelster and Weber 1991; Berenbrink et al. 2005; Berenbrink 2007; Wells 2009).
The proto α- and β-globin genes of jawed vertebrates (Gnathostomata) represent the product of an ancient gene duplication event that occurred roughly 450–500 Ma in the Ordovician, before the divergence between cartilaginous fish (Chondrichthyes) and the common ancestor of ray-finned fish (Actinopterygii) and lobe-finned fish + tetrapods (Sarcopterygii; Goodman et al. 1987; Storz et al. 2011, 2012; Hoffmann, Opazo, and Storz 2012). Subsequent rounds of duplication and divergence gave rise to diverse repertoires of α- and β-like globin genes that are developmentally regulated in different ways in different vertebrate lineages (Hardison 2001; Hoffmann, Storz, et al. 2010). The ancestral linkage arrangement of the α- and β-globin genes is still retained in at least some cartilaginous fish (Marino et al. 2007), teleosts (Chan et al. 1997; Miyata and Aoki 1997; Gillemans et al. 2003; Pisano et al. 2003), and amphibians (Hentschel et al. 1979; Jeffreys et al. 1980; Kay et al. 1980; Hosbach et al. 1983; Fuchs et al. 2006). In amniote vertebrates, by contrast, the α- and β-globin gene clusters are located on different chromosomes due to transposition of the proto β-globin gene to a new genomic location sometime after the stem lineage of amniotes split from the line leading to amphibians (Hardison 2008; Patel et al. 2008, 2010; Hoffmann, Opazo, and Storz 2012).
The main objective of the present study was to assess the relative contributions of whole-genome duplication, large-scale segmental duplication, and small-scale gene duplication in producing the extraordinary functional diversity of teleost Hbs. To accomplish this, we integrated phylogenetic reconstructions with analyses of conserved synteny to characterize the genomic organization and evolutionary history of the globin gene clusters of teleost fish. These results were then integrated with available experimental data on functional properties and developmental patterns of stage-specific gene expression. Results of the phylogenetic and comparative genomic analyses revealed repeated evolutionary transitions in stage-specific expression in different teleost lineages. Our analyses also revealed that functionally distinct anodic and cathodic adult Hbs evolved independently in different teleost lineages, providing evidence for convergence in the physiological division of labor between coexpressed Hb isoforms.
Materials and Methods
Data Collection
We used bioinformatic techniques to manually annotate the full complement of globin genes in the genomes of six teleost fish available in release 67 of the ensembl database (fugu, Takifugu rubripes; medaka, Oryzias latipes; green spotted puffer, Tetraodon nigroviridis; tilapia, Oreochromis niloticus; three-spined stickleback, Gasterosteus aculeatus; and zebrafish, Danio rerio). We also annotated the globin genes from a live-bearing teleost (platyfish, Xiphophorus maculatus) and a nonteleost ray-finned fish (spotted gar, Lepisosteus oculatus), both available from the Pre!ensembl database. We compared the ensembl data with previous reports on the genomic organization of the globin gene clusters in fugu (Flint et al. 2001), medaka (Maruyama, Yasumasu, and Iuchi 2004; Maruyama, Yasumasu, Naruse, et al. 2004), and zebrafish (Brownlie et al. 2003). We also included coding sequences from the full complement of globin genes from Atlantic cod (Gadus morhua; Borza et al. 2009, Wetten et al. 2010) and Atlantic salmon (Salmo salar; Quinn et al. 2010). However, the fragmentary state of the cod and salmon genome assemblies precluded a detailed comparative analysis of the globin gene clusters in these two species. Finally, we included additional records from tetrapod vertebrates and cartilaginous fish as outgroup sequences for phylogenetic analyses, and we included genomic contigs from representative tetrapods for the purpose of making synteny comparisons. When possible, the annotated genomic sequences were validated by comparison with the relevant expressed sequence tag (EST) databases.
Assessments of Conserved Synteny
To examine patterns of conserved synteny, we annotated the genes found upstream and downstream of the globin gene clusters of seven teleost species (fugu, medaka, platyfish, green-spotted puffer, stickleback, tilapia, and zebrafish) and one nonteleost ray-finned fish (spotted gar). Initial ortholog predictions were derived from the EnsemblCompara database (Vilella et al. 2009) and were visualized using the program Genomicus (Muffato et al. 2010). In addition, we also used the program Genscan (Burge and Karlin 1997) to identify additional unannotated genes lying upstream and downstream of the annotated globin genes. The unannotated genes were compared with the nonredundant protein database using Basic Local Alignment Search Tool (BLAST) (Altschul et al. 1990). Partial sequences for genes of interest (representing pseudogenes or artifacts related to incomplete sequence coverage) were identified and annotated with BLAST. To examine large-scale patterns of sequence conservation, we conducted pairwise comparisons of sequence similarity between globin gene clusters using the Pipmaker and Multipipmaker programs (Schwartz et al. 2000, 2003). To facilitate comparisons, genes have been labeled following the Zebrafish Model Organism Database nomenclature guidelines. Finally, we conducted an analysis of conserved synteny between the globin gene clusters of medaka and the reconstructed protokaryotype of the pre-TGD teleost common ancestor provided by Kasahara et al. (2007) and Nakatani et al. (2007).
Sequence Alignment
Separate alignments of the α- and β-globin coding sequences were based on conceptual translations of nucleotide sequences. Alignments were performed using Muscle v 3.8 (Edgar 2004) and the E-INS-i, G-INS-I, and L-INS-i strategies from Mafft v6.8 (Katoh et al. 2009). We employed MUMSA (Lassmann and Sonnhammer 2005, 2006) to select the best-scoring multiple alignment, and we then used the selected alignment to estimate phylogenetic relationships. These sequence manipulations were carried out in the Mobyle platform server (Néron et al. 2009) hosted by the Institut Pasteur (http://mobyle.pasteur.fr, last accessed September 2012). All sequence alignments are provided in supplementary data file S1, Supplementary Material online.
Phylogenetic Analyses
We reconstructed separate phylogenies for the α- and β-globin genes using Bayesian and maximum likelihood approaches. We performed maximum likelihood analyses in Treefinder, version March 2011 (Jobb et al. 2004), and we evaluated support for the nodes with 1,000 bootstrap pseudoreplicates. We used the “propose model” tool of Treefinder to select the best-fitting models of amino acid and nucleotide substitution, with an independent model for each codon position in analyses based on nucleotide sequences. Model selection was based on the Akaike information criterion with correction for small sample size. We estimated Bayesian phylogenies in MrBayes v.3.1.2 (Ronquist and Huelsenbeck 2003), running six simultaneous chains for 2 × 107 generations, sampling every 2.5 × 103 generations, and using default priors. A given run was considered to have reached convergence once the likelihood scores reached an asymptotic value and the average standard deviation of split frequencies remained <0.01. We discarded all trees that were sampled before convergence, and we evaluated support for the nodes and parameter estimates from a majority rule consensus of the last 2,500 trees.
Results and Discussion
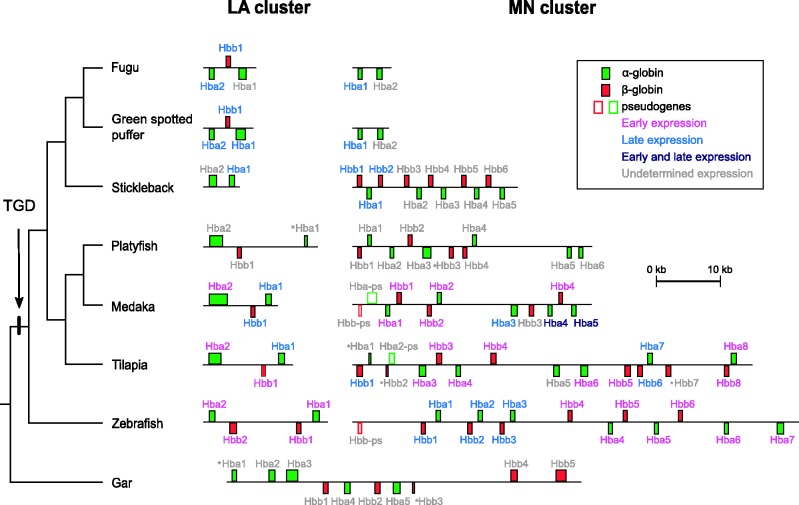
The comparative genomic analysis revealed that the Hb genes of teleost fish are located in two separate chromosomal regions that are clearly delineated by distinct sets of flanking loci (fig. 1). In contrast, the Hb genes of the nonteleost gar are located in a single chromosomal region. Following Hardison (2008), the teleost globin gene cluster flanked by the mpg and nprl3 genes was labeled the “MN” cluster, and the teleost globin cluster flanked by the lcmt1 and aqp8 genes was labeled the “LA” cluster. In the platyfish assembly, we identified two separate scaffolds containing the MN and LA clusters (fig. 1) and a third scaffold (JH559524) that contained a single, putatively functional β-globin gene. We excluded this latter scaffold from all subsequent analyses because it likely represents an assembly artifact. The MN and LA clusters correspond to the medaka E1 and A1 clusters, respectively, that were described by Maruyama et al. (Maruyama, Yasumasu, and Iuchi 2004; Maruyama, Yasumasu, Naruse, et al. 2004). To facilitate comparison, we report the order of genes in the same orientation as they appear in the zebrafish genome assembly, regardless of how they are found in the ensembl database. Since the MN and LA clusters of most teleosts harbor globin genes in both forward and reverse orientations, we use the terms left and right to describe linear gene order. The individual α- and β-globin genes in the MN cluster were numbered from left to right, such that the functional globin gene in the leftmost position of the MN cluster of zebrafish is labeled MN Hbb1, the next gene to the right is MN Hba1, and so forth, whereas the genes in the LA cluster were numbered from right to left, starting with the gene closest to aqp8 (fig. 2). In the case of cod and salmon globin genes, we retained the labels from the original studies (Borza et al. 2009; Quinn et al. 2010). Sequence sources for the globin gene clusters used in this study are provided in table 1, and the annotations for each cluster are provided in supplementary table S1, Supplementary Material online. To facilitate comparisons with previous studies, we compiled a list of previously used names for each of the annotated globin genes (supplementary table S2, Supplementary Material online).
Fig. 1.
Unscaled depiction of the genomic organization of the MN and LA globin gene clusters from representative teleost fishes, with the human α-globin gene cluster provided as reference. To facilitate comparisons, all clusters are presented in the same orientation as the zebrafish. Genes in the forward orientation are shown on top of the chromosome, whereas genes in the reverse orientation are shown below.
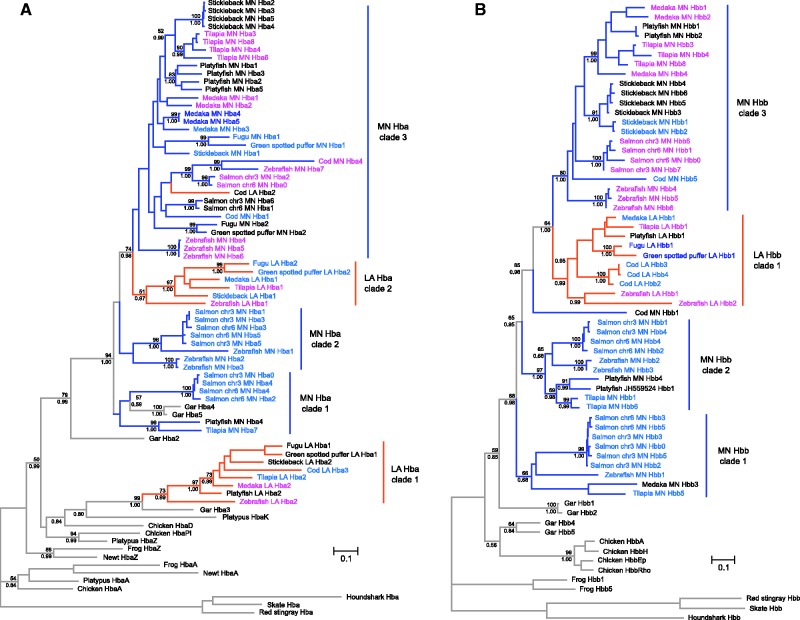
Fig. 2.
Genomic structure of the MN and LA globin gene clusters of teleost fish. To facilitate comparisons, all clusters are presented in the same orientation as the zebrafish. Genes in the forward orientation are shown on top of the chromosome, whereas genes in the reverse orientation are shown below. The green-spotted puffer globin genes are assumed to have the same stage-specific expression profiles as their orthologous counterparts in fugu. The Hbb pseudogene in the zebrafish MN cluster is not drawn to scale. Gene labels are color coded based on the timing of their expression. Genes marked with an asterisk were not included in the phylogenetic analyses.
Table 1.
Data Sources, Genomic Coordinates and Orientations of the Globin Gene Clusters in Fugu, Green-Spotted Puffer, Gar, Medaka, Platyfish, Tilapia, Stickleback, and Zebrafish.
| Species | Release | Cluster | Location | Orientation | Start | End |
|---|---|---|---|---|---|---|
| Fugu (T. rubripes) | Fugu 4.0 | LA | Sc_ 3 | lcmt1 → rhbdf1b | 2,511,982 | 2,517,737 |
| MN | Sc_15 | kank → nprl3 | 417,195 | 420,598 | ||
| Green-spotted puffer (Tet. nigroviridis) | 46 | LA | Chr 2 | rhbdf1b → aqp8 | 5,887,638 | 5,893,221 |
| MN | Chr 3 | kank → nprl3 | 12,162,093 | 12,165,924 | ||
| Gar (Lepisosteus oculatus) | LepOcu1 | Hb | LG13 | nprl3 → luc7l | 2,809 | 54,885 |
| Medaka (O. latipes) | Medaka 1.0 | LA | Chr 19 | → aqp8 | 1,478,030 | 1,487,664 |
| MN | Chr 8 | nprl3 → kank | 8,378,078 | 8,412,019 | ||
| Platyfish (X. maculatus) | Xipmac4.4.2 | LA | JH557783 | rhbdf1b → aqp8 | 22,438 | 37,618 |
| MN | JH556906 | nprl3 → kank | 106,543 | 141,798 | ||
| Unassigned | JH559524 | 5,235 | 8,503 | |||
| Tilapia (Ore. niloticus) | Orenil1.0 | LA | GL831136 | rhbdf1b → aqp8 | 111,303 | 122,995 |
| MN | GL831149 | nprl3 → kank | 110,462 | 169,554 | ||
| Stickleback (G. aculeatus) | BROADS1 | LA | Sc 112 | c17orf28 → aqp8 | 339,530 | 343,463 |
| MN | Gr XI | kank → nprl3 | 13,640,461 | 13,663,356 | ||
| Zebrafish (D. rerio) | Zv9 | LA | Chr 12 | rhbdf1b → aqp8 | 21,688,806 | 21,705,956 |
| MN | Chr 3 | nprl3 → kank | 55,938,147 | 55,999,373 |
Note.—In all cases, data were obtained from Ensembl. The start and end points correspond to the most distant edges from the two genes on either end of the cluster.
Genomic Structure of the MN and LA Globin Gene Clusters in Teleosts
Patterns of Conserved Synteny
The genomic context of the teleost globin gene clusters is relatively well conserved, especially in the case of the MN cluster. In all teleost species analyzed, there is perfect conservation for the five genes to the left of the MN cluster: aanat, mgrn1, rhbdf1a, mpg, and nprl3 (fig. 1). The two genes to the right, kank2 and dock6, are also conserved in all species. The genomic organization of the LA globin gene cluster is not as strongly conserved. Four of the seven teleost species possess a single copy of rhbdf1b to the left of the LA cluster, which is paralogous to the rhbdf1a gene found adjacent to the MN cluster. On the right side of the LA cluster, all teleost species possess copies of lcmt1 and arhgap17. Each of the teleost species possess one or two copies of aqp8, with the exception of the two tetraodontid species (fugu and green spotted puffer) that have secondarily lost this gene (fig. 1).
Hb Gene Repertoires of Teleost Fish
There were several cases where our manual annotations of the globin gene clusters differed from annotations provided in the most recent releases of the various teleost genome assemblies. For example, no MN-linked globin genes were annotated in the most recent release of the fugu genome in the ensembl database. However, BLAST comparisons with an independent record of the fugu MN globin cluster (AY016024) revealed the presence of two unannotated α-globin genes between nprl3 and kank2, as reported by Flint et al. (2001). In addition, the only annotated β-globin gene in the green-spotted puffer genome (green-spotted puffer LA Hbb1) contained a 4 bp insertion in the second exon that would render it nonfunctional. Comparisons with cDNA-derived sequence databases revealed several putatively functional transcripts that lacked the inactivating 4 bp insertion but were otherwise identical in sequence. We assumed that the insertion was either a sequencing or assembly artifact, and we therefore used the cDNA-derived sequence for all further analyses.
The MN and LA clusters of the different species exhibited substantial variation in both physical extent and gene content (fig. 2). From the start codon of the first globin gene to the stop codon of the last globin gene, the MN cluster ranged from 3.4 kb in fugu to 68.5 kb in zebrafish, and the LA cluster ranged from 3.4 kb in stickleback to 17.2 kb in zebrafish. With respect to gene content, the number of globin genes in these clusters ranged from 2 in the MN clusters of fugu and green-spotted puffer and the LA cluster of stickleback, to 13 in the MN clusters of tilapia and zebrafish (not including two genes with partial sequence coverage in the tilapia assembly; fig. 2). Interspecific comparisons revealed a higher rate of globin gene turnover in the MN cluster than in the LA cluster. The MN clusters of fugu and green-spotted puffer possess only two α-globin genes in the reverse orientation, whereas the MN clusters of all other teleosts contain interspersed α- and β-globin genes in both head-to-head and head-to-tail orientations (fig. 2). In the case of stickleback, all of the α-globin genes are found in the reverse orientation, and all of the β-globin genes are in the forward orientation (fig. 2). In all other teleosts, in contrast, multiple α- and β-globin genes are found in both forward and reverse orientations. In all species examined, the LA cluster harbors two tandemly duplicated α-globin genes, and when present, the β-globin genes are sandwiched in between the α-globin genes but in the opposite orientation. The comparative genomic analysis revealed that the β-globin genes of stickleback are only present in the MN cluster, whereas the single β-globin genes of fugu and green-spotted puffer are only present in the LA cluster. Thus, the β-globin genes of stickleback and the two tetraodontid species are not 1:1 orthologs. Furthermore, the set of three globin genes in the LA clusters of fugu and green-spotted puffer appear to have been inverted relative to those of medaka, stickleback, and zebrafish. This inversion hypothesis predicts that the LA Hba1 genes from fugu and green-spotted puffer should be most closely related to the LA Hba2 genes of medaka, platyfish, stickleback, tilapia, and zebrafish.
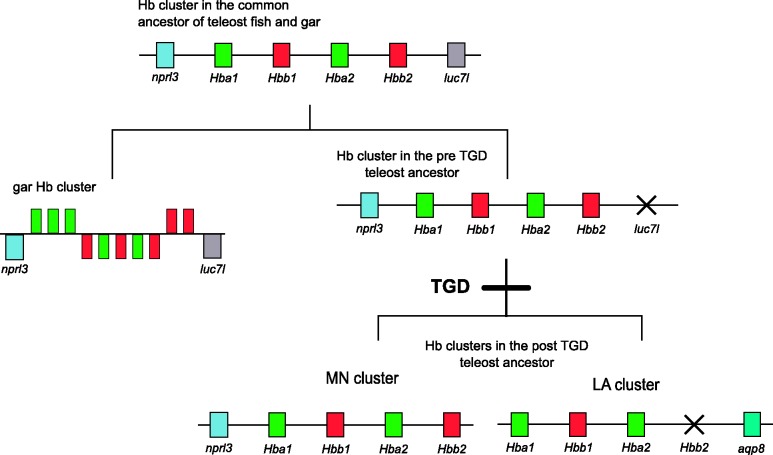
The Globin Gene Cluster in Gar and the Origin of the MN and LA Clusters of Teleosts
In most cases, orthologs of genes flanking the MN and LA globin clusters in teleost fish are located in the vicinity of the α-globin gene cluster in human and chicken, which appears to represent the ancestral location of the proto-Hb gene in jawed vertebrates (Hoffmann, Opazo, and Storz 2012). The 2:1 pattern of conserved synteny between teleost fish and tetrapods suggests that the MN and LA globin clusters of teleost fish derive from the TGD, as suggested by Quinn et al. (2010). This inference is also supported by the presence of duplicate copies of rhbdf1 in teleosts, which are co-orthologs of the single-copy rhbdf1 in tetrapods. Additional bioinformatic searches in the vicinity of the globin gene clusters revealed that most teleosts also possess duplicate copies of shisa9 and mlk2, one on the LA cluster and one on the MN cluster, that are co-orthologous to single-copy genes on the same chromosome as the α-globin gene cluster in human and chicken.
Two additional lines of evidence support the hypothesis that the LA and MN clusters represent paralogous products of the TGD. First, we tested the prediction that the spotted gar (a nonteleost ray-finned fish) would possess a single globin gene cluster, since the gar and teleost lineages diverged before TGD. Consistent with this prediction, our comparative genomic analysis revealed that the spotted gar does indeed possess a single globin gene cluster, ∼52 kb in length, that contains 5 α- and 5 β-globin genes in both forward and reverse orientations (fig. 2). The cluster is flanked by copies of c16or33, polr3k, mgrn1, fox1j, aanat, rhbdf1, mpg, and nprl3 on the left, and by copies of luc7l and itfg3 on the right (fig. 1). Second, we tested the prediction that the LA and MN gene clusters of teleosts descend from the same linkage group in the reconstructed protokaryotype of the pre-TGD teleost ancestor. Consistent with this prediction, an analysis of conserved synteny revealed that the MN and LA clusters of medaka are embedded in paralogous chromosomal segments that trace their duplicative origin to chromosome “e” in the pre-TGD teleost protokaryotype inferred by Kasahara et al. (2007) and Nakatani et al. (2007).
Phylogenetic Relationships among Teleost α- and β-Globin Genes
After characterizing the genomic organization of the globin gene clusters in spotted gar and the seven teleost fish, we performed phylogenetic analyses to reconstruct the duplicative history of the α- and β-globin genes. For this analysis, we added the globin gene repertoires of cod and salmon to those of fugu, green-spotted puffer, medaka, platyfish, stickleback, tilapia, and zebrafish, and we also included sequences from representative tetrapods and cartilaginous fish for comparative purposes. All of the different alignment strategies produced very similar results for the α- and β-globin data sets, and in both cases, we selected the L-INS-i alignment for use in the phylogenetic reconstructions because it had the highest MUMSA score. Before estimating phylogenies, we selected the best-fitting models of amino acid and nucleotide substitution based on the Akaike information criterion with correction for small sample size. In analyses based on nucleotide sequences, we selected an independent model for each codon position. Results of the model estimation procedure can be found in supplementary table S3, Supplementary Material online.
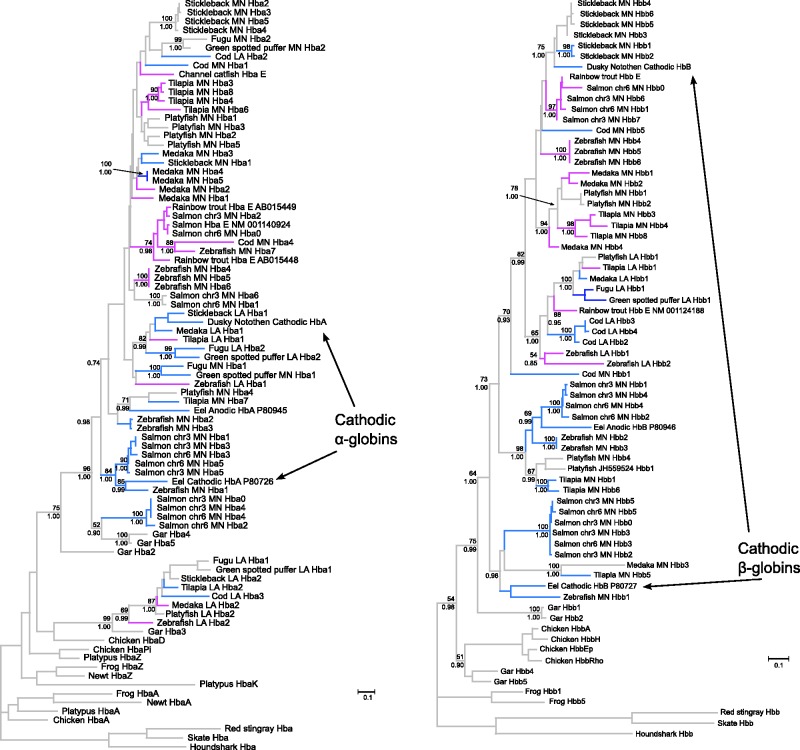
The estimated phylogenies of vertebrate globin sequences suggested that neither α- or β-globin genes of ray-finned fish are monophyletic relative to their tetrapod counterparts (fig. 3A and B). In the case of α-globin genes, a clade of fish sequences that included a subset of genes derived from the teleost LA cluster (LA Hba clade 1 + gar Hba3) were placed sister to the chicken αD-globin gene, whereas all other fish α-globins were placed in a second monophyletic group (fig. 3A). In the case of the β-like globin genes, a clade of two gar sequences, including gar Hbb4 and Hbb5, was placed sister to the chicken β-globins. These arrangements suggest that multiple α- and β-globin genes were present in the common ancestor of Actinopterygii + Sarcopterygii.
Fig. 3.
Maximum likelihood phylogram depicting relationships among the globin sequences of seven representative teleost fishes. Phylogenetic reconstructions were based on the coding sequences of α- and β-globin genes (panels A and B, respectively). Cartilaginous fish globins were used as outgroup sequences, and tetrapod sequences were included for comparative purposes. Values on the nodes denote bootstrap support values (above) and Bayesian posterior probabilities (below). Branches are color coded according to the location of the genes: MN-linked genes are shown in blue, LA-linked genes are shown in orange, and the gar genes are in green. Labels are color coded based on the timing of their expression. The substitution models selected are listed in supplementary table S3, Supplementary Material online.
The phylogeny shown in figure 3A revealed that fish α-globins can be arranged into two distinct clades, defined by the presence of gar Hba2 and gar Hba3, respectively. In turn, teleost α-globins were arranged into five clades that (with the exception of the cod LA Hba2 sequence) reflect their cluster of origin. The discordant position of the cod LA Hba2 sequence probably represents an assembly artifact. Aside from this cod sequence, all α-globin genes derived from the LA cluster were grouped into two strongly supported clades: LA Hba clade 1 is sister to gar Hba3, and LA Hba clade 2 is embedded in a strongly supported clade that includes all MN α-globin sequences in addition to LA Hba2 from cod and Hba2, Hba4, and Hba5 from spotted gar (fig. 3A). Genealogical relationships within these two clades of LA α-globins are largely congruent with the known organismal relationships, and in both cases, the deepest split separated the zebrafish genes from those of the remaining euteleost taxa. As expected under the cluster-inversion hypothesis, the leftmost α-globin genes in the LA cluster of fugu and green-spotted puffer are most closely related to the rightmost α-globin genes of medaka, platyfish, stickleback, tilapia, and zebrafish, and vice versa (fig. 3A). Relationships among the α-globin sequences in the MN cluster are more complex and are not easily reconciled with the organismal phylogeny. The MN-linked genes are organized into three weakly supported clades (fig. 3A). MN Hba clade 1 contains salmon and platyfish sequences in addition to two gar sequences, whereas MN Hba clade 2 contains zebrafish and salmon sequences. MN Hba clade 3 was placed sister to LA Hba clade 2 and includes sequences from all teleosts in addition to cod LA Hba2. All species examined possess an α-globin gene repertoire that includes representatives of at least three of the five clades, and zebrafish possesses α-globin genes that are represented in four of the five clades.
In contrast to the α-globin genes, all teleost β-globin genes were placed in a moderately well-supported clade, which was placed sister to a clade of two gar Hbb sequences (Hbb1 and Hbb2). The other two gar Hbb sequences were placed sister to chicken Hbbs (fig. 3B). The β-globin genes could be arranged into four separate clades, three of which were strongly supported, with sequences from the MN cluster forming a paraphyletic group relative to those from the LA cluster. The β-globins from the LA cluster were placed in a monophyletic group, while those from the MN cluster can be grouped into three separate clades, with the exception of Cod MN Hbb1, which is distantly related to the rest. MN Hbb clade 1 contains sequences from medaka, salmon, tilapia, and zebrafish; MN Hbb clade 2 contains sequences from platyfish, salmon, tilapia, and zebrafish; and MN Hbb clade 3 contains sequences from cod, medaka, platyfish, salmon, stickleback, tilapia, and zebrafish (fig. 3B). Within each of these clades, paralogs from the same species almost invariably formed monophyletic groups, which likely reflects a history of lineage-specific duplication, as with the Hba genes of the MN cluster. This is particularly clear in the case of MN Hbb clade 3, where relationships among the different paralogs are congruent with the known organismal phylogeny after accounting for lineage-specific duplications.
With the exception of β-globin genes from the LA cluster, globin genes of the same subunit type from the MN or LA clusters did not form monophyletic groups. Taken together, the analyses of conserved synteny (figs. 1 and 2) and the phylogenetic reconstructions (fig. 3) indicate that the pre-TGD globin gene cluster of teleost fish contained at least two α-globin genes and 2 β-globin genes. Further, the positions of the gar sequences in the phylogenies of α- and β-like globins indicate that multiple globins of each subunit type were present in the common ancestor of gar and teleosts. If further analyses confirm the paraphyly of ray-finned fish α- and β-globins relative to their tetrapod homologs, it would indicate that multiple α- and β-globin genes were present in the common ancestor of Actinopterygii and Sarcopterygii. As for teleosts, after the TGD but before divergence between zebrafish and the remaining euteleost species, one of the two ancestral β-globin paralogs in the LA cluster was secondarily lost such that the post-TGD globin repertoire was reduced from 8 to 7 genes (fig. 4). Similar lineage-specific patterns of gene turnover have been documented in the α- and β-globin gene clusters of mammals and other vertebrates (Hoffmann et al. 2008a, 2008b; Opazo et al. 2008a, 2008b, 2009; Hoffmann, Storz, et al. 2010). On a deeper evolutionary timescale, lineage-specific duplications and deletions have produced extensive variation in the size and membership composition of the globin gene superfamily among different vertebrate classes and among different deuterostome phyla and subphyla (Ebner et al. 2010; Hoffmann, Storz, et al. 2010; Hoffmann et al. 2011; Storz et al. 2011; Hoffmann, Opazo, Hoogewijs, et al. 2012; Hoogewijs et al. 2012).
Fig. 4.
Evolutionary model describing the duplicative origins of the LA and MN globin gene clusters of teleost fish and the inferred globin gene repertoire in the common ancestor of teleosts and gar, a nonteleost ray finned fish. All clusters depicted are hypothetical with the exception of the gar cluster. The order of the α- and β-globin genes on the hypothetical clusters is arbitrary.
The phylogenies in figure 3 indicate that all salmon α- and β-globin genes are exclusively found in association with MN-linked globin genes from other species. This reflects the fact that salmonid fish have experienced an additional lineage-specific genome-duplication and that all globin genes were deleted from the duplicated LA clusters and were retained exclusively in the duplicated MN clusters (Quinn et al. 2010). With the exception of fugu and green-spotted puffer, which possess identical globin gene repertoires, all other species in our study show evidence of lineage-specific duplications, which are much more frequent in the MN cluster. In fact, aside from fugu and green-spotted puffer, all other species have expanded the repertoire of α- and β-globin genes via lineage-specific duplications. The most striking contrast is between the MN α-globins from platyfish, tilapia, and zebrafish, and the MN β-globins from stickleback. The stickleback β-globins in the MN cluster derive from a recent set of duplications, whereas the α-globins from the MN clusters of platyfish, tilapia, and zebrafish derive from a combination of recent, lineage-specific duplications of genes deriving from more ancient duplications that likely occurred before the TGD.
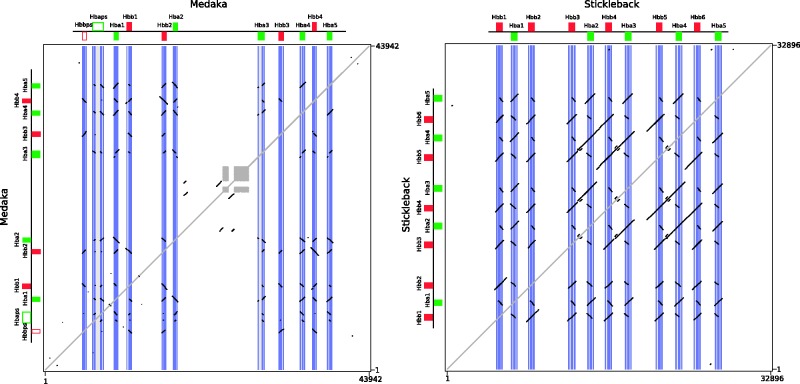
In addition to the differences in timing, these lineage-specific duplications also appear to involve different mechanisms. In many instances, the expansions derive from single gene duplications, such as the one giving rise to the duplicate Hbb paralogs in the zebrafish LA cluster. On the other hand, the structure of the MN clusters of medaka, stickleback, and zebrafish suggest that en bloc duplications are partly responsible for their lineage-specific expansions in gene family size. In the case of the stickleback, the presence of extensive internal colinearity within the MN cluster suggests that it expanded by en bloc duplications involving either the Hbb–Hba pair or an Hbb–Hba–Hbb–Hba four-gene set (fig. 5). The same can be said for the zebrafish MN Hba2–Hbb2 gene pair and the MN Hba3–Hbb3 gene pairs in zebrafish (supplementary fig. S1, Supplementary Material online). However, comparisons of zebrafish MN Hba4–6 and MN Hbb4–6 gene pairs revealed low levels of sequence similarity in flanking regions (supplementary fig. S1, Supplementary Material online).
Fig. 5.
Dot plots of intrachromosomal sequence similarity in the MN globin gene clusters of medaka and stickleback. The fragment includes all genes in the clusters in addition to 5 kb of flanking sequence. The diagonal self-identity plot is shown in gray, as are the low-complexity areas in the medaka cluster. Note that the intragenomic dot plot for the stickleback gene cluster shows longer tracts of internal similarity off the self-identity diagonal relative to that for the medaka gene cluster, shown in black.
Repeated Evolutionary Transitions in Functional Properties and Stage-Specific Expression
In light of evidence that the developmental regulation of Hb synthesis has evolved independently in multiple tetrapod lineages (Hoffmann, Storz, et al. 2010; Storz et al. 2011, 2012), we tested for evidence of a similar phenomenon in teleosts by reconstructing phylogenetic relationships among α- and β-like globin genes that are differentially expressed during development. For the purposes of this analysis, globin genes were classified as “early-expressed” if they are preferentially expressed during embryonic or larval developmental stages, whereas genes were classified as “late-expressed” if they are preferentially expressed in juveniles or adults (supplementary table S4, Supplementary Material online, fig. 2). Since fugu and green-spotted puffer possess a single β-globin gene, we assumed that this gene is expressed during all ontogenetic stages. For comparative purposes, we included additional teleost globins that are known to be preferentially expressed during embryogenesis in channel catfish (Ictalurus punctatus; Chen et al. 2010), rainbow trout (Oncorhynchus mykiss; Maruyama et al. 1999), and salmon (Leong et al. 2010). We also analyzed late-expressed α- and β-globin genes whose products are incorporated into tetrameric Hbs with highly distinct functional properties, such as the well-characterized anodic and cathodic Hbs of the European eel (Anguilla anguilla; Fago et al. 1995, 1997) and dusky notothen (Trematomus newnesi; Mazzarella et al. 1999). Expression data for cod, medaka, and zebrafish were obtained from the literature. The cod sequences were classified following Wetten et al. (2010), the medaka sequences were classified following Maruyama, Yasumasu and Iuchi (2004), and the zebrafish sequences were classified following Tiedke et al. (2011). For globin genes in fugu, gar, green-spotted puffer, platyfish, salmon, and tilapia, we inferred the timing of expression by identifying matches with sequences in EST databases (supplementary table S4, Supplementary Material online). In the cases of sequences with no matches from the same species as the query sequence or lack of developmental information for the EST matches, the sequences were left as unclassified.
Intriguingly, results of our analyses revealed repeated evolutionary transitions in stage-specific expression during development. In some cases, paralogous genes in different species evolved convergent expression patterns, and in other cases, orthologous genes evolved divergent expression patterns. In the case of the α-like globin genes, LA Hba clades 1 and 2 provide clear examples of probable 1:1 orthologs that evolved differences in stage-specific expression (e.g., the early-expressed zebrafish LA Hba1 and the late-expressed medaka LA Hba1; fig. 3A). In the case of β-like globin genes, LA Hbb clade 1 illustrates a similar pattern of replicated expression divergence (e.g., the early expressed zebrafish LA Hbb1 and Hbb2 genes are clearly co-orthologous to the adult-expressed medaka LA Hbb1; fig. 3B). These results demonstrate that the developmental timing of globin gene expression is evolutionarily labile.
In the α- and β-globin gene clusters of most amniotes, the linear order of the genes reflects their temporal order of expression during development, with early-expressed genes at the 5′ end of the cluster and late-expressed genes at the 3′ end of the cluster (Hardison 2001). In the globin gene clusters of teleosts, in contrast, linear gene order is not as strong a predictor of stage-specific expression. In the case of the zebrafish MN cluster, all late-expressed genes are on the left and all early-expressed genes are on the right, whereas in medaka, all genes on the left side are early-expressed and the genes on the right are variable with respect to the developmental timing of expression, and in tilapia, the early- and late-expressed genes are interspersed. Our results indicate that the genes in the LA cluster provide the clearest evidence of lineage-specific changes in gene expression.
Since embryonic/fetal Hbs and adult-expressed Hbs exhibit consistent differences in O2-affinity and sensitivity to allosteric regulators (Ingermann 1997), convergence in stage-specific expression also likely entailed convergence in functional properties. Similarly, adult α- and β-globin genes that encode the subunits of cathodic Hbs of European eel and dusky notothen are clearly not 1:1 orthologs (fig. 6), indicating that specialized Hbs with similar functional properties evolved independently in different teleost lineages. In fact, the dusky notothen cathodic Hba is closely related to sequences in the LA cluster, whereas the eel cathodic Hba is closely related to sequences to the MN cluster, suggesting they trace their duplicative origin at least to the TGD. Consistent with other studies of vertebrate globins (Berenbrink et al. 2005; Hoffmann et al. 2010), these results demonstrate that similar expression patterns and functional properties in the Hbs of distinct lineages may sometimes represent products of convergent evolution. Although tandemly duplicated globin genes often evolve in concert due to interparalog gene conversion (Hoffmann et al. 2008a, 2008b; Opazo et al. 2009; Runck et al. 2009; Storz et al. 2011), paralogous genes that are products of genome duplications (also known as “ohnologs”) can escape the homogenizing effects of gene conversion because they are located on different chromosomes. This is one possible reason why paralogous gene copies derived from genome duplications may be more likely to diverge in function than tandem gene duplicates.
Fig. 6.
Maximum likelihood phylogram depicting relationships among the globin genes of the seven fish species for which full genome sequence data were available, plus sequences of functionally annotated globins from other teleost species. The phylogenetic reconstructions were based on the amino acid sequences of α- and β-globins (panels A and B, respectively). Cartilaginous fish globins were used as outgroup sequences, and tetrapod sequences were included for comparative purposes. Values on the nodes denote bootstrap support values (above) and Bayesian posterior probabilities (below). Genes are color coded according to their time of expression. Genes expressed at the embryonic/larval stages are shown in magenta, genes expressed at the juvenile and/or adult stages are shown light blue, and genes expressed across all ontogenetic stages are shown in dark blue. Genes with no record of expression and genes from nonactinopterygian vertebrates are shown in gray. The substitution models selected are listed on supplementary table S3, Supplementary Material online.
Conclusion
Results of our combined phylogenetic and comparative genomic analyses indicate that some of the teleost α- and β-like globins are representatives of ancient gene lineages, with duplicative origins that trace back at least to the common ancestor of gar and teleost fish, and potentially back to the common ancestor of Actinopterygii and Sarcopterygii (superclass Osteichthyes). Such a scenario is consistent with the fact that Hb multiplicity has also been documented in cartilaginous fish (Fyhn and Sullivan 1975; Mumm et al. 1978; Weber et al. 1983; Galderisi et al. 1996; Dafre and Reischl 1997). Our results indicate that the common ancestor of ray-finned fish possessed a fairly diverse globin gene repertoire, and in teleosts, this inherited repertoire was further augmented by the TGD, which produced dual sets of α- and β-like globin genes on two paralogous chromosomes. These TGD-derived gene clusters underwent lineage-specific changes in size and membership composition, and the MN gene cluster underwent an especially high rate of gene turnover. The phylogenetic analyses of teleost globins revealed repeated transitions in stage-specific expression patterns, demonstrating a surprising fluidity in the genetic regulatory control of Hb synthesis during development.
Supplementary Material
Supplementary tables S1–S4, figure S1, and data file S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Andi Kautt, Axel Meyer, David A. Ray, and two anonymous reviewers for critical reviews of this manuscript. M.F.N. holds a CONICYT doctoral fellowship. This work was supported by National Science Foundation (EPS-0903787) grant to F.G.H., National Institutes of Health/NHLBI (R01 HL087216 and HL087216-S1) and the National Science Foundation (IOS-0949931) grant to J.F.S., and Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT 1120032) grant to J.C.O.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics. 2011;188:799–808. doi: 10.1534/genetics.111.127324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, et al. (13 co-authors) Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Arnegard ME, Zwickl DJ, Lu Y, Zakon HH. Old gene duplication facilitates origin and diversification of an innovative communication system—twice. Proc Natl Acad Sci U S A. 2010;51:22172–221777. doi: 10.1073/pnas.1011803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenbrink M. Historical reconstructions of evolving physiological complexity: O2 secretion in the eye and swimbladder of fishes. J Exp Biol. 2007;209:1641–1652. doi: 10.1242/jeb.003319. [DOI] [PubMed] [Google Scholar]
- Berenbrink M, Koldkjaer P, Kepp O, Cossins AR. Evolution of oxygen secretion in fishes and the emergence of a complex physiological system. Science. 2005;307:1752–1757. doi: 10.1126/science.1107793. [DOI] [PubMed] [Google Scholar]
- Borza T, Stone C, Gamperl AK, Bowman S. Atlantic cod (Gadus morhua) hemoglobin genes: multiplicity and polymorphism. BMC Genet. 2009;10:51. doi: 10.1186/1471-2156-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Brunet F, Volff J, Schartl M. Pigmentation pathway evolution after whole-genome duplication in fish. Genome Biol Evol. 2009;1:479–493. doi: 10.1093/gbe/evp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Salzburger W, Meyer A. Asymmetric evolution in two fish-specifically duplicated receptor tyrosine kinase paralogons involved in teleost coloration. Mol Biol Evol. 2006;23:1192–1202. doi: 10.1093/molbev/msk003. [DOI] [PubMed] [Google Scholar]
- Braasch I, Schartl M, Volff J. Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol Biol. 2007;7:74. doi: 10.1186/1471-2148-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Volff J, Schartl M. The endothelin system: evolution of vertebrate-specific ligand-receptor interactions by three rounds of genome duplication. Mol Biol Evol. 2009;26:783–799. doi: 10.1093/molbev/msp015. [DOI] [PubMed] [Google Scholar]
- Brownlie A, Hersey C, Oates AC, et al. (13 co-authors) Characterization of embryonic globin genes of the zebrafish. Dev Biol. 2003;255:48–61. doi: 10.1016/s0012-1606(02)00041-6. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Chan FY, Robinson J, Brownlie A, Shivdasani RA, Donovan A, Brugnara C, Kim J, Lau BC, Witkowska HE, Zon LI. Characterization of adult alpha- and beta-globin genes in the zebrafish. Blood. 1997;89:688–700. [PubMed] [Google Scholar]
- Chen F, Lee Y, Jiang Y, et al. (11 co-authors) Identification and characterization of full-length cDNAs in channel catfish (Ictalurus punctatus) and blue catfish (Ictalurus furcatus) PLoS One. 2010;5:e11546. doi: 10.1371/journal.pone.0011546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafre AL, Reischl E. Asymmetric hemoglobins, their thiol content, and blood glutathione of the scalloped hammerhead shark, Sphyrna lewini. Comp Biochem Physiol B. 1997;116:323–331. doi: 10.1016/s0305-0491(96)00215-5. [DOI] [PubMed] [Google Scholar]
- Douard V, Brunet F, Boussau B, Ahrens I, Vlaeminck-Guillem V, Haendler B, Laudet V, Guiguen Y. The fate of the duplicated androgen receptor in fishes: a late neofunctionalization event? BMC Evol Biol. 2008;8:336. doi: 10.1186/1471-2148-8-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner B, Panopoulou G, Vinogradov SN, Kiger L, Marden MC, Burmester T, Hankeln T. The globin gene family of the cephalochordate amphioxus: implications for chordate globin evolution. BMC Evol Biol. 2010;10:370. doi: 10.1186/1471-2148-10-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fago A, Bendixen E, Malte H, Weber RE. The anodic hemoglobin of Anguilla anguilla. Molecular basis for allosteric effects in a root-effect hemoglobin. J Biol Chem. 1997;272:15628–15635. doi: 10.1074/jbc.272.25.15628. [DOI] [PubMed] [Google Scholar]
- Fago A, Carratore V, di Prisco G, Feuerlein RJ, Sottrup-Jensen L, Weber RE. The cathodic hemoglobin of Anguilla anguilla. Amino acid sequence and oxygen equilibria of a reverse Bohr effect hemoglobin with high oxygen affinity and high phosphate sensitivity. J Biol Chem. 1995;270:18897–18902. doi: 10.1074/jbc.270.32.18897. [DOI] [PubMed] [Google Scholar]
- Flint J, Tufarelli C, Peden J, et al. (14 co-authors) Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the alpha globin cluster. Hum Mol Genet. 2001;10:371–382. doi: 10.1093/hmg/10.4.371. [DOI] [PubMed] [Google Scholar]
- Fuchs C, Burmester T, Hankeln T. The amphibian globin gene repertoire as revealed by the Xenopus genome. Cytogenet Genome Res. 2006;112:296–306. doi: 10.1159/000089884. [DOI] [PubMed] [Google Scholar]
- Fyhn UEH, Sullivan B. Elasmobranch hemoglobins: dimerization and polymerization in various species. Comp Biochem Physiol. 1975;50:119–129. doi: 10.1016/0305-0491(75)90311-9. [DOI] [PubMed] [Google Scholar]
- Galderisi U, Fucci L, Garaci G. Multiple hemoglobins in the electric ray: Torpedo marmorata. Comp Biochem Physiol B. 1996;113:645–651. [Google Scholar]
- Gillemans N, McMorrow T, Tewari R, et al. (12 co-authors) Functional and comparative analysis of globin loci in pufferfish and humans. Blood. 2003;101:2842–2849. doi: 10.1182/blood-2002-09-2850. [DOI] [PubMed] [Google Scholar]
- Goodman M, Miyamoto M, Czelusniak J. Pattern and process in vertebrate phylogeny revealed by coevolution of molecules and phylogenies. In: Patterson C, editor. Molecules and morphology in evolution: conflict or compromise? Cambridge (United Kingdom): Cambridge University Press; 1987. pp. 140–176. [Google Scholar]
- Hardison RC. Organization, evolution, and regulation of the globin genes. In: Steinberg MH, Forget BG, Higgs DR, Nagel RL, editors. Disorders of hemoglobin: genetics, pathophysiology, and clinical management. Cambridge (United Kingdom): Cambridge University Press; 2001. pp. 95–115. [Google Scholar]
- Hardison RC. Globin genes on the move. J Biol. 2008;7:35. doi: 10.1186/jbiol92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi Y, Nishida M. Evolution of trace amine associated receptor (TAAR) gene family in vertebrates: lineage-specific expansions and degradations of a second class of vertebrate chemosensory receptors expressed in the olfactory epithelium. Mol Biol Evol. 2007;24:2099–2107. doi: 10.1093/molbev/msm140. [DOI] [PubMed] [Google Scholar]
- Hentschel CC, Kay RM, Williams JG. Analysis of Xenopus laevis globins during development and erythroid cell maturation and the construction of recombinant plasmids containing sequences derived from adult globin mRNA. Dev Biol. 1979;72:350–363. doi: 10.1016/0012-1606(79)90124-6. [DOI] [PubMed] [Google Scholar]
- Hoegg S, Brinkman H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol. 2004;5:190–203. doi: 10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- Hoegg S, Meyer A. Hox clusters as models for vertebrate genome evolution. Trends Genet. 2005;21:421–424. doi: 10.1016/j.tig.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Hoegg S, Meyer A. Phylogenomic analyses of KCNA clusters in vertebrates: why do some clusters stay intact? BMC Evol Biol. 2007;7:139. doi: 10.1186/1471-2148-7-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Hoogewijs D, Hankeln T, Ebner B, Vinogradov S, Bailly X, Storz JF. Evolution of the globin gene family in deuterostomes: lineage-specific patterns of diversification and attrition. Mol Biol Evol. 2012;29:1735–1745. doi: 10.1093/molbev/mss018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF. Rapid rates of lineage-specific gene duplication and deletion in the α-globin gene family. Mol Biol Evol. 2008a;25:591–602. doi: 10.1093/molbev/msn004. [DOI] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF. New genes originated via multiple recombinational pathways in the β-globin gene family of rodents. Mol Biol Evol. 2008b;25:2589–2600. doi: 10.1093/molbev/msn200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF. Gene cooption and convergent evolution of oxygen transport hemoglobins in jawed and jawless vertebrates. Proc Natl Acad Sci U S A. 2010;107:14274–14279. doi: 10.1073/pnas.1006756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF. Differential loss and retention of myoglobin, cytoglobin, and globin-E during the radiation of vertebrates. Genome Biol Evol. 2011;3:588–600. doi: 10.1093/gbe/evr055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF. Whole-genome duplications spurred the functional diversification of the globin gene superfamily in vertebrates. Mol Biol Evol. 2012;29:303–312. doi: 10.1093/molbev/msr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann FG, Storz JF, Gorr TA, Opazo JC. Lineage-specific patterns of functional diversification in the α- and β-globin gene families of tetrapod vertebrates. Mol Biol Evol. 2010;27:1126–1138. doi: 10.1093/molbev/msp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PW, Garcia-Fernàndez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994;1994:125–133. [PubMed] [Google Scholar]
- Hoogewijs D, Ebner B, Germani F, et al. (11 co-authors) Androglobin: a chimeric globin in metazoans that is preferentially expressed in mammalian testes. Mol Biol Evol. 2012;29:1105–1114. doi: 10.1093/molbev/msr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosbach HA, Wyler T, Weber R. The Xenopus laevis globin gene family: chromosomal arrangement and gene structure. Cell. 1983;32:45–53. doi: 10.1016/0092-8674(83)90495-6. [DOI] [PubMed] [Google Scholar]
- Ingermann RI. In: Handbook of physiology. Bethesda (MD): American Physiological Society; 1997. Vertebrate hemoglobins. pp. 357–408. [Google Scholar]
- Jaillon O, Aury J, Brunet F, et al. (61 co-authors) Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Wood D, Simons JP, Kay RM, Williams JG. Linkage of adult alpha- and beta-globin genes in X. laevis and gene duplication by tetraploidization. Cell. 1980;21:555–564. doi: 10.1016/0092-8674(80)90493-6. [DOI] [PubMed] [Google Scholar]
- Jensen FB, Fago A, Weber RE. Hemoglobin structure and function. In: Perry SF, Tufts BL, editors. Fish physiology, Vol. 17: Fish respiration. San Diego (CA): Academic Press; 1998. pp. 1–40. [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. Treefinder: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kasahara M, Naruse K, Sasaki S, et al. (37 co-authors) The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- Kay RM, Harris R, Patient RK, Williams JG. Molecular cloning of cDNA sequences coding for the major alpha- and beta-globin polypeptides of adult Xenopus laevis. Nucleic Acids Res. 1980;8:2691–2707. doi: 10.1093/nar/8.12.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer ELL. Automatic assessment of alignment quality. Nucleic Acids Res. 2005;33:7120–7128. doi: 10.1093/nar/gki1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer ELL. Kalign, Kalignvu and Mumsa: web servers for multiple sequence alignment. Nucleic Acids Res. 2006;34:W596–W599. doi: 10.1093/nar/gkl191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong JS, Jantzen SG, von Schalburg KR, et al. (12 co-authors) Salmo salar and Esox lucius full-length cDNA sequences reveal changes in evolutionary pressures on a post-tetraploidization genome. BMC Genomics. 2010;11:279. doi: 10.1186/1471-2164-11-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister JA, Close J, Raible DW. Duplicate mitf genes in zebrafish: complementary expression and conservation of melanogenic potential. Dev Biol. 2001;237:333–344. doi: 10.1006/dbio.2001.0379. [DOI] [PubMed] [Google Scholar]
- Marino K, Boschetto L, de Pascale D, Cocca E. Organization of the hb1 genes of the antarctic skate Bathyraja eatonii: new insights into the evolution of globin genes. Gene. 2007;406:199–208. doi: 10.1016/j.gene.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Yasumasu S, Iuchi I. Characterization and expression of embryonic globin in the rainbow trout, Oncorhynchus mykiss: intra-embryonic initiation of erythropoiesis. Dev Growth Differ. 1999;41:589–599. doi: 10.1046/j.1440-169x.1999.00460.x. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Yasumasu S, Iuchi I. Evolution of globin genes of the medaka Oryzias latipes (Euteleostei; Beloniformes; Oryziinae) Mech Dev. 2004;121:753–769. doi: 10.1016/j.mod.2004.03.035. [DOI] [PubMed] [Google Scholar]
- Maruyama K, Yasumasu S, Naruse K, Mitani H, Shima A, Iuchi I. Genomic organization and developmental expression of globin genes in the teleost Oryzias latipes. Gene. 2004;335:89–100. doi: 10.1016/j.gene.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Mazzarella L, D'Avino R, di Prisco G, Savino C, Vitagliano L, Moody PC, Zagari A. Crystal structure of Trematomus newnesi haemoglobin re-opens the root effect question. J Mol Biol. 1999;287:897–906. doi: 10.1006/jmbi.1999.2632. [DOI] [PubMed] [Google Scholar]
- Meyer A. Hox gene variation and evolution. Nature. 1998;391:225, 227–228. doi: 10.1038/34530. [DOI] [PubMed] [Google Scholar]
- Meyer A, Málaga-Trillo E. Vertebrate genomics: more fishy tales about Hox genes. Curr Biol. 1999;9:R210–R213. doi: 10.1016/s0960-9822(99)80131-6. [DOI] [PubMed] [Google Scholar]
- Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;11:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Miyata M, Aoki T. Head-to-head linkage of carp α- and β-globin genes. Biochim Biophys Acta. 1997;1354:127–133. doi: 10.1016/s0167-4781(97)00111-5. [DOI] [PubMed] [Google Scholar]
- Mulley JF, Chiu CH, Holland PW. Breakup of a homeobox cluster after genome duplication in teleosts. Proc Natl Acad Sci U S A. 2006;103:10369–10372. doi: 10.1073/pnas.0600341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffato M, Louis A, Poisnel C, Crollius HR. Genomicus: a database and a browser to study gene synteny in modern and ancestral genomes. Bioinformatics. 2010;26:1119–1121. doi: 10.1093/bioinformatics/btq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm DP, Atha DH, Riggs A. The hemoglobin of the common sting-ray, Dasyatis sabina: structural and functional properties. Comp Biochem Physiol B. 1978;60:189–193. doi: 10.1016/0305-0491(78)90129-3. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Takeda H, Kohara Y, Morishita S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007;17:1254–1265. doi: 10.1101/gr.6316407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Néron B, Ménager H, Maufrais C, Joly N, Maupetit J, Letort S, Carrere S, Tuffery P, Letondal C. Mobyle: a new full web bioinformatics framework. Bioinformatics. 2009;25:3005–3011. doi: 10.1093/bioinformatics/btp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF. Genomic evidence for independent origins of β-like globin genes in monotremes and therian mammals. Proc Natl Acad Sci U S A. 2008a;105:1590–1595. doi: 10.1073/pnas.0710531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Hoffmann FG, Storz JF. Differential loss of embryonic globin genes during the radiation of placental mammals. Proc Natl Acad Sci U S A. 2008b;105:12950–12955. doi: 10.1073/pnas.0804392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo JC, Sloan AM, Campbell KL, Storz JF. Origin and ascendancy of a chimeric fusion gene: the β/δ-globin gene of paenungulate mammals. Mol Biol Evol. 2009;26:1469–1478. doi: 10.1093/molbev/msp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VS, Cooper SJB, Deakin JE, Fulton B, Graves T, Warren WC, Wilson RK, Graves JAM. Platypus globin genes and flanking loci suggest a new insertional model for β-globin evolution in birds and mammals. BMC Biol. 2008;6:34. doi: 10.1186/1741-7007-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VS, Ezaz T, Deakin JE, Marshall Graves JA. Globin gene structure in a reptile supports the transpositional model for amniote α- and β-globin gene evolution. Chromosome Res. 2010;18:897–907. doi: 10.1007/s10577-010-9164-5. [DOI] [PubMed] [Google Scholar]
- Pelster B, Weber RE. The physiology of the root effect. Adv Comp Environ Physiol. 1991;8:51–77. [Google Scholar]
- Pisano E, Cocca E, Mazzei F, Ghigliotti L, di Prisco G, Detrich HW, 3rd, Ozouf-Costaz C. Mapping of α- and β-globin genes on antarctic fish chromosomes by fluorescence in-situ hybridization. Chromosome Res. 2003;11:633–640. doi: 10.1023/a:1024961103663. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- Quinn NL, Boroevich KA, Lubieniecki KP, Chow W, Davidson EA, Phillips RB, Koop BF, Davidson WS. Genomic organization and evolution of the Atlantic salmon hemoglobin repertoire. BMC Genomics. 2010;11:539. doi: 10.1186/1471-2164-11-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Runck AM, Moriyama H, Storz JF. Evolution of duplicated β-globin genes and the structural basis of hemoglobin isoform differentiation in Mus. Mol Biol Evol. 2009;26:2521–2532. doi: 10.1093/molbev/msp165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutjes HA, Nieveen MC, Weber RE, Witte F, Van den Thillart GEE. Multiple strategies of Lake Victoria cichlids to cope with lifelong hypoxia include hemoglobin switching. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1376–R1383. doi: 10.1152/ajpregu.00536.2006. [DOI] [PubMed] [Google Scholar]
- Sato Y, Hashiguchi Y, Nishida M. Temporal pattern of loss/persistence of duplicate genes involved in signal transduction and metabolic pathways after teleost-specific genome duplication. BMC Evol Biol. 2009a;9:127. doi: 10.1186/1471-2148-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Hashiguchi Y, Nishida M. Evolution of multiple phosphodiesterase isoforms in stickleback involved in cAMP signal transduction pathway. BMC Syst Biol. 2009b;3:23. doi: 10.1186/1752-0509-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Nishida M. Post-duplication charge evolution of phosphoglucose isomerases in teleost fishes through weak selection on many amino acid sites. BMC Evol Biol. 2007;7:204. doi: 10.1186/1471-2148-7-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Nishida M. Teleost fish with specific genome duplication as unique models of vertebrate evolution. Environ Biol Fish. 2010;88:169–188. [Google Scholar]
- Schwartz S, Elnitski L, Li M, Weirauch M, Riemer C, Smit A, Green ED, Hardison RC, Miller W. Multipipmaker and supporting tools: alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 2003;31:3518–3524. doi: 10.1093/nar/gkg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. Pipmaker—a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimeld SM, Holland PW. Vertebrate innovations. Proc Natl Acad Sci U S A. 2000;97:4449–4452. doi: 10.1073/pnas.97.9.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel N, Hoegg S, Salzburger W, Braasch I, Meyer A. Comparative genomics of ParaHox clusters of teleost fishes: gene cluster breakup and the retention of gene sets following whole genome duplications. BMC Genomics. 2007;8:312. doi: 10.1186/1471-2164-8-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J F, Opazo JC, Hoffmann FG. Phylogenetic diversification of the globin gene superfamily in chordates. IUBMB Life. 2011;63:313–322. doi: 10.1002/iub.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz J F, Opazo JC, Hoffmann FG. Gene duplication, genome duplication, and the functional diversification of vertebrate globins. Mol Phyl Evol. Forthcoming 2012 doi: 10.1016/j.ympev.2012.07.013. http://dx.doi.org/10.1016/j.ympev.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Van de Peer Y, Braasch I, Meyer A. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos Trans R Soc Lond B Biol Sci. 2001;356:1661–1679. doi: 10.1098/rstb.2001.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedke J, Gerlach F, Mitz SA, Hankeln T, Burmester T. Ontogeny of globin expression in zebrafish (Danio rerio) J Comp Physiol B. 2011;181:1011–1021. doi: 10.1007/s00360-011-0588-9. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Taylor JS, Meyer A. Are all fishes ancient polyploids? J Struct Funct Genomics. 2003;3:65–73. [PubMed] [Google Scholar]
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. Ensemblcompara genetrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H. Origin and evolution of the neural crest: a hypothetical reconstruction of its evolutionary history. Dev Growth Differ. 2001;43:509–520. doi: 10.1046/j.1440-169x.2001.00600.x. [DOI] [PubMed] [Google Scholar]
- Wada H, Makabe K. Genome duplications of early vertebrates as a possible chronicle of the evolutionary history of the neural crest. Int J Biol Sci. 2006;2:133–141. doi: 10.7150/ijbs.2.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber RE. Functional significance and structural basis of multiple hemoglobins with special reference to ectothermic vertebrates. In: Truchot JP, Lahlou B, editors. Animal nutrition and transport processes. 2. Transport, respiration and excretion: comparative and environmental aspects. Basel (Switzerland): Karger; 1990. pp. 58–75. [Google Scholar]
- Weber RE. Adaptations for oxygen transport: lessons from fish hemoglobins. In: Di Prisco G, Giardina B, Weber RE, editors. Hemoglobin function in vertebrates: molecular adaptation in extreme and temperate environments. New York: Springer; 2000. pp. 23–37. [Google Scholar]
- Weber RE, Fago A, Val AL, Bang A, van Hauwert ML, DeWilde S, Zal F, Moens L. Isohemoglobin differentiation in the bimodal-breathing Amazon catfish Hoplosternum littorale. J Biol Chem. 2000;275:17297–17305. doi: 10.1074/jbc.M001209200. [DOI] [PubMed] [Google Scholar]
- Weber RE, Jensen FB. Functional adaptations in hemoglobins from ectothermic vertebrates. Ann Rev Physiol. 1988;50:161–179. doi: 10.1146/annurev.ph.50.030188.001113. [DOI] [PubMed] [Google Scholar]
- Weber RE, Wells RMG, Rossetti JE. Allosteric interactions governing oxygen equilibria in the haemoglobin system of the spiny dogfish. Squalus acanthias. J Exp Biol. 1983;103:109–120. doi: 10.1242/jeb.103.1.109. [DOI] [PubMed] [Google Scholar]
- Wells RMG. Blood-gas transport and hemoglobin function: adaptations for functional and environmental hypoxia. In: Richards JG, Farrell AP, Brauner CJ, editors. Fish physiology. Vol. 27. New York: Elsevier; 2009. pp. 255–299. [Google Scholar]
- Wetten OF, Nederbragt AJ, Wilson RC, Jakobsen KS, Edvardsen RB, Andersen Ø. Genomic organization and gene expression of the multiple globins in atlantic cod: conservation of globin-flanking genes in chordates infers the origin of the vertebrate globin clusters. BMC Evol Biol. 2010;10:315. doi: 10.1186/1471-2148-10-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WP, Yew K, Rajasegaran V, Venkatesh B. Sequencing and comparative analysis of fugu protocadherin clusters reveal diversity of protocadherin genes among teleosts. BMC Evol Biol. 2007;7:49. doi: 10.1186/1471-2148-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Cohn MJ. Genome duplication and the origin of the vertebrate skeleton. Curr Opin Genet Dev. 2008;18:387–393. doi: 10.1016/j.gde.2008.07.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.