Abstract
Gastrointestinal stromal tumours (GISTs) are the most common mesenchymal tumours of the digestive tract. Pathogenesis is linked to activating mutations identified in two proto-oncogenes, v-kit Hardy/Zuckerman 4 feline sarcoma viral oncogene homologue KIT (KIT) and the platelet-derived growth factor α (PDGFRα). In addition, these mutations affect response to treatment with tyrosine kinase inhibitors. In the present study, we report on the molecular characterisation of GISTs in the South African population. Tumour DNA was extracted from 46 GIST samples, followed by cycle sequencing of KIT exons 11, 13 and 17 and PDGFRα exons 12, 14 and 18. Fragment length analysis was used to detect a 6-bp duplication in KIT exon 9. Wild-type duplications were analysed further by PCR and sequencing of additional KIT and PDGFRα exons was performed. Overall, 78.3% of the samples had a mutation in KIT or PDGFRα. Of these, mutations were detected in KIT exon 11 (88.9%), PDGFRα exon 18 (8.3%) and KIT exon 9 (2.8%). Mutations varied from simple substitutions and duplications to large deletions (some with nucleotide insertions) resulting in missense mutations. In addition, seven single nucleotide polymorphisms were detected in 17 patients, one of which appears novel. The incidence of mutations in KIT exon 11 and PDGFRα exon 18 is consistent with the literature, however, the low incidence of KIT exon 9 mutations detected was unexpected. In contrast to previous western and Asian studies, this mutation appears to be rare in the South African population. The present study contributes to the molecular understanding of GISTs in the South African population.
Keywords: gastrointestinal stromal tumours, KIT mutations, PDGFRα mutations, South Africa
Introduction
Gastrointestinal stromal tumours (GISTs) are rare, however they are the most common mesenchymal tumours of the digestive tract. he annual GIST incidence in the western world is estimated at 11–15 cases/million individuals (1) and the disease predominantly affects middle-aged or elderly individuals; however, the disease has been identified in a wide range of ages. GISTs are highly resistant to chemotherapy and surgical resection is the current mainstay treatment (2). Risk of recurrence or metastasis is high. Factors of malignancy prediction include tumour size and mitotic index, as well as the tumour site (1,3).
The pathogenesis of GISTs has been linked to activating mutations occurring in two proto-oncogenes, v-kit Hardy/Zuckerman 4 feline sarcoma viral oncogene homologue KIT (KIT) and the platelet derived growth factorα (PDGFRα) (4,5). The genes encode homologous type III tyrosine kinases (6). Activating mutations render the receptors constitutively activated, resulting in the dysregulation of several signalling pathways and uncontrolled cellular proliferation (7). Mutations in the juxtamembrane domain of KIT are most frequently identified and this region is encoded by exons 9 and 11, while mutations in the tyrosine kinase domain (exons 13 and 17) are rare. In PDGFRα, juxtamembrane (exon 12) mutations are rare, however, the kinase domain, encoded by exon 14 and 18, is frequently mutated (8–10).
An increased understanding of GIST pathogenesis has led to targeted treatment with tyrosine kinase inhibitors (TKIs), including imatinib and sunitinib. These drugs block the protein tyrosine kinase domain, preventing downstream signalling (1). The majority of activating mutations render the tumour more susceptible to TKIs and therefore TKI treatment is recommended as an adjuvant to surgery in patients at high risk for relapse, as well as for metastatic GIST (1,7,11). Overall, GISTs with KIT exon 11 mutations exhibit the best response to TKI therapy when compared with wild-type tumours and other mutations (12). It was previously hypothesised that the conformational changes caused by the mutations enable improved access of the drug to the tyrosine kinase domain of the protein (13). However, some primary and secondary mutations are associated with imatinib resistance. Determination of mutations in the KIT and PDGFRα genes by direct sequencing is therefore essential for individual targeted treatment of GIST patients (2).
A number of previous studies have characterised GIST samples at the molecular level, generating population-specific data for USA, Europe and Asia. Information on KIT and PDGFRα mutations associated with GIST in African individuals remains extremely scarce (14). The present study is the first to report on the molecular characterisation of 46 African GIST samples. We have developed initial data with the aim to determine the GIST mutation pattern in the South African population.
Materials and methods
Patient selection and samples
Ethics approval was granted from the Human Research Ethics Committee of the University of the Witwatersrand. Archival material was used in the present study. A total of 46 formalin-fixed paraffin-embedded (FFPE) GIST classified samples were received from all regions in South Africa. A haematoxylin and eosin slide from each block was reviewed by experienced pathologists and the tumour area selected for DNA extraction.
DNA extraction
Each FFPE tissue block was sectioned using a clean microtome blade and five 4-μm sections were cut and baked at 48.2°C for 2 h or overnight. Sections were deparaffinised in methylcyclohexane followed by rehydration in a series of 100% ethanol, 95% ethanol and distilled water washes. DNA was extracted using the Qiagen DNeasy kit (Whitehead Scientific, Brackenfell, South Africa) following the manufacturer’s instructions.
Polymerase chain reaction (PCR) amplification and DNA cycle sequencing
PCR conditions were optimised for each exon, as previously described (9,15). Primers used are listed in Table I. PCR was prioritised into two rounds, according to the most prevalent mutations. The first round was performed on all samples and amplified KIT exons 9, 11, 13 and 17 along with PDGFRα exons 12, 14 and 18. The second round PCR was only performed on the sample if the first round of sequenced results were all wild-type. The second round consisted of amplification of KIT exons 10, 12, 14, 16, 18 and 19 and PDGFRα exons 10, 13, 15, 16, 17, 19, 20 and 21. KIT exon 9 was not sequenced but resolved in a 4% MetaPhor agarose gel (Whithead Scientific). PCR fragment length analysis was used to determine the presence of a 6-bp duplication (codons 502–503) (10,16). All other amplified exons were screened on a 2% agarose gel and subsequently sequenced in both directions using the BigDye terminator kit v3.1 (Applied Biosystems, Bedford, MA, USA). Reactions were loaded on either an ABI 3170 or 3500 genetic analyser (Applied Biosystems). Sequences were analysed using Sequence Scanner v1.0 (Applied Biosystems).
Table I.
PCR primers for KIT and PDGFRα exons.
| Gene | Exon | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|---|
| KIT | 9 | GATGTGGGCAAGACTTCTG | TGGTAGACAGAGCCTAAACATCC |
| 10 | GAGTGGCTGTGGTAGAGATC | CAGTCATTAGAGCACTCTGG | |
| 11 | CCAGAGTGCTCTAATGACTG | ACTGTTATGTGTACCCAAAAAG | |
| 12 | GCACAAATGGTCCTTCAATTC | AAAGCACAACTGGCAAACTGA | |
| 13 | GCTTGACATCAGTTTGCCAGT | GGCAGCTTGGACACGGCTTTA | |
| 14 | TTATGGGAGGCAGAATTAATCTAT | CCCTTATGACCCCATGAACT | |
| 16 | ACTGTCTTTTCCTTTCCTGACC | ACACAAAACTCTTTAGAGAATCACTC | |
| 17 | GGTTTTCTTTTCTCCTCCAACC | TGCAGGACTGTCAAGCAGAG | |
| 18 | AATTTTGTTGAGCTTCTGAATTAAC | ACTTCAAGAAGATGCTCTGAGTCT | |
| 19 | GGTGTCCTGCTTCCTTGTGAT | CCTCAACATCTGGGTTTCTGTC | |
| PDGFRα | 10 | TGTTCCCGTGGCTCCACTCATTG | TTCCGCCTGGGGCAGATGC |
| 12 | TCCAGTCACTGTGCTGCTTC | GCAAGGGAAAAGGGAGTCTT | |
| 13 | GCTGGCTACGGTGCAGAAAG | TATCCCCATGCTCAAAAAATCC | |
| 14 | TGAGAACAGGAAGTTGGTAGCTCA | GATGGAGAGTGGAGGATTTAAGCCT | |
| 15 | GGACAATTCATGGCTTTTCTGTT | TTCCATTTGTGATGCCTGTAAGA | |
| 16 | CCCAGTTAGCTCCCATGCCTACCT | CATCCCTATACACTTCCCTCTAAAT | |
| 17 | TGCCTCTGCAACCTGATGATT | CTCCGTCCACACTCCACTCAC | |
| 18 | CAGCTACAGATGGCTTGATC | GGATGAGCCTGACCAGTGAG | |
| 19 | CCCTTTTCTATTTCCACTGCTGTGG | CATCCTGGGGCTTGAAAGAAC | |
| 20 | TCATGCCAAGTGTTTCAGCAAT | GCCCCTCCCTCCCCCTAGAC | |
| 21 | CCGGGGGCCTGTGTTCACAGT | TGCGGAACAGCACAGCTCAC |
Results
Patient demographics
In the present study, 46 GIST samples were obtained from 45 patients. The majority (80%) of the tumours were localised. The most common site of the tumour was the stomach (48%), followed by the small bowel (20%) and large bowel or colon (9%). The rest of the tumours (24%) occurred along various parts of the GI tract, including the liver, pancreas, retroperitoneal and intraperitoneal cavity. Patient age ranged between 17 and 86 years, with an average age of 56 years. Ethnicity was known for 35 patients and of these, 23 were African and 12 were Caucasian. There was a higher male to female ratio of 1.25:1. In addition, one patient had neurofibromatosis 1 (NF1). By assessing the tumour size, mitotic index and tumour site, the risk for malignancy was evaluated for 43 of the samples. The majority of the samples (23 GISTs) were high risk, with a mitotic count >10/50 high-powered fields and/or a tumour diameter >10 cm. Eight and 12 samples were classified as either intermediate or low risk, respectively. Follow-up data were not available for the majority of the cases.
Mutational analysis
Overall, 36 (78.3%) samples had a mutation in one of the two genes while the remaining 10 samples were considered wild-type (21.7%). Of the samples with mutations, 32 (88.9%) occurred in KIT exon 11, three (8.3%) in PDGFRα exon 18 and one (2.8%) in KIT exon 9. The wild-type samples included tumours from a juvenile boy (aged 17 years) and the known NF1 patient.
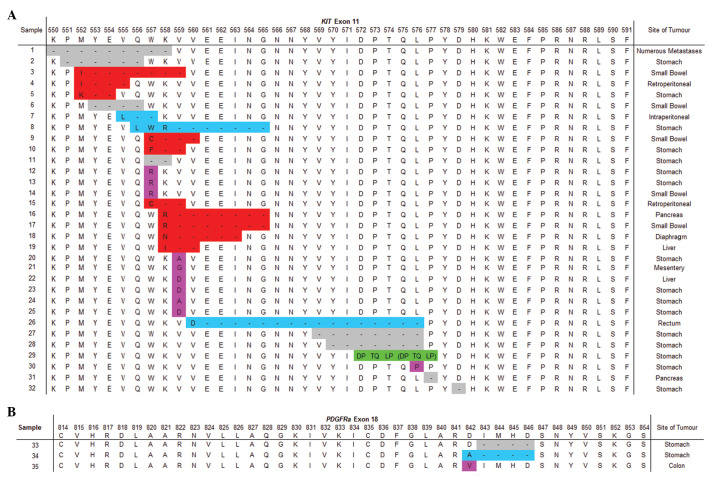
Mutations occurring in KIT exon 11 included substitutions, deletions, deletions with a missense mutation, deletions and nucleotide insertions resulting in missense mutations and a duplication (Fig. 1A) in 10, 8, 10, 3 and 1 case(s), respectively. KIT exon 11 mutations occurred in 73% of GISTs arising in the stomach, 56% in the small bowel and accounted for 85% of tumours detected in other areas of the GI tract. The most commonly (46.9%) affected KIT exon 11 codon was 559, but mutations were also identified in codons 550–579. Two of the three PDGFRα exon 18 mutations occurred in GIST arising in the stomach and the other was in the colon. PDGFRα exon 18 mutations included a substitution, deletion and deletion with nucleotide insertion resulting in a missense mutation. These affected PDGFRα exon 18 codons 842 to 846 (Fig. 1B). A single KIT exon 9 mutation was identified in a GIST arising from the liver. This patient had an additional primary GIST of the small bowel with a KIT exon 11 mutation (absent in the liver GIST).
Figure 1.
(A) KIT exon 11 mutations detected in South African GIST samples. (B) PDGFRα exon 18 mutations detected in South African GIST samples. Deletions (grey), deletions with a missense mutation (red), deletions with nucleotide insertions (blue), substitutions (pink) and duplications (green). GIST, gastrointestinal stromal tumour.
Single nucleotide polymorphisms (SNPs)
Seven different SNPs were detected in 17 patients with one or more SNPs (Table II). Seven of these patients had wild-type tumours. Thirteen were of African ethnicity, two were Caucasian and two were of unknown ethnicity. Of the seven SNPs, one in KIT exon 17 (K779K; AAA→AAG) has not been previously identified.
Table II.
SNPs detected in KIT and PDGFRα in 46 GIST samples from South Africa.
| Gene | Exon | Description | Site of tumoura | Mutation statusa | Ethnicitya |
|---|---|---|---|---|---|
| KIT | 11 | P585P (CCC>TCC) rs121913515 | Small bowel (1) | Wild-type (1) | African (1) |
| KIT | 18 | L862L (CTG>CTC) rs3733542 | Stomach (1) | Wild-type (2) | African (2) |
| Small bowel (1) | |||||
| PDGFRα | 12 | P567P (CCG>CCA) rs1873778 | Stomach (3) | K11 mutation (3) | African (4) |
| Small bowel (1) | Wild-type (1) | ||||
| PDGFRα | 13 | Intronic (AAA>A3AG) rs41279523 | Large bowel (1) | Wild-type (1) | African (1) |
| PDGFRα | 17 | K779K (AAA>AAG) Unknown | Rectum (1) | Wild-type (1) | African (1) |
| PDGFRα | 18 | V824V (GTC-GTT) rs2228230 | Stomach (9) | Wild-type (5) | African (10) |
| Small bowel (2) | K11 mutation (7) | Caucasian (2) | |||
| Large bowel (1) | P18 mutation (1) | Unknown (2) | |||
| Other (2) | |||||
| PDGFRα | 20 | R914R (CGG>CGA) rs56384252 | Rectum (1) | Wild-type (1) | African (1) |
Number of patients indicated in parentheses. SNPs, single nucleotide polymorphisms; GIST, gastrointestinal stromal tumour; K11, KIT exon 11; P18, PDGFRα exon 18.
Discussion
Molecular profiling of GIST is recommended for the determination of the best course of treatment for a patient with unresectable or metastatic disease (2). Previously, our laboratory reported KIT and PDGFRα mutational incidence in 17 samples from South African GIST patients (14). The present study expands this previous report and aimed to determine the molecular profile of 46 GIST samples. In addition, the present findings were compared with those from developing and first world countries. Overall, a mutation was identified in 78.3% of South African GISTs. The majority of these mutations occured in KIT exon 11 (88.9%) followed by PDGFRα exon 18 (8.3%). The KIT exon 9 mutation was rare and was identified in only one sample.
GIST pathology studies have been previously performed in a number of countries worldwide, including Africa and South Africa (17,18). Previously, Tran et al(19) revealed that GIST incidence was higher in African-Americans compared with Caucasians. However, information on GIST incidence and mutational profiles is scarce in Africa. The genetic profile of South African GIST patients remains poorly understood and must be determined.
The frequency of specific mutations varies worldwide, however, current results demonstrate that KIT mutations are the most common while those in PDGFRα have only been detected in KIT wild-type GISTs (Table III) (1). Variance in reported mutation rates for various populations is thought to be due to a number of factors, including ethnicity, study design and selection criteria for each study (7,20).
Table III.
KIT and PDGFRα mutational rates in various populations.
| Country | Gene | Number of studies/total sample number | Mutated exons (%)
|
Reference | |||||
|---|---|---|---|---|---|---|---|---|---|
|
KIT
|
PDGFRα
|
||||||||
| 11 | 9 | 13 | 17 | 18 | 12 | ||||
| Taiwan | KIT | 2/188 | 66.1 | 14.2 | 0.0 | 0.0 | - | - | 20, 28 |
| PDGFRα | 1/134 | - | - | - | - | 0.0 | 0.8 | ||
| Japan | KIT | 2/118 | 58.7 | 7.0 | 0.0 | NP | - | - | 30–32 |
| PDGFRα | 1/70 | - | - | - | - | 4.3 | 2.9 | ||
| China | KIT | 1/165 | 63.0 | 6.6 | 1.3 | 0.6 | - | - | 29 |
| PDGFRα | 1/165 | - | - | - | - | 3.0 | 0.0 | ||
| Brazil | KIT | 1/55 | 69.0 | 3.6 | 0.0 | 0.0 | - | - | 33 |
| PDGFRα | 1/55 | - | - | - | - | 1.8 | 5.5 | ||
| Spain | KIT | 1/166 | 47.5 | 2.4 | 0.6 | 1.6 | - | - | 27 |
| PDGFRα | 1/166 | - | - | - | - | 4.0 | 4.0 | ||
| USA | KIT | 4/818 | 56.5 | 8.9 | 0.4 | 0.6 | - | - | 5,21–23 |
| PDGFRα | 3/693 | - | - | - | - | 8.1 | 0.7 | ||
| Europe | KIT | 3/591 | 62.6 | 9.9 | 1.4 | 0.45 | - | - | 24–26 |
| PDGFRα | 3/579 | - | - | - | - | 3.9 | 0.8 | ||
| South Africa | KIT | 1/46 | 69.6 | 2.8 | 0.0 | 0.0 | - | - | Present study |
| PDGFRα | 1/46 | - | - | - | - | 6.5 | 0.0 | ||
NP, not performed.
Previous studies performed in European and North American populations (western populations) revealed that 85–90% of GISTs exhibited an activating mutation in KIT or PDGFRα(1). The most frequently observed mutation is KIT exon 11, followed by KIT exon 9 and PDGFRα exon 18 (21–25). Mutations have been detected at much lower frequencies in KIT exons 13 and 17 and PDGFRα exons 12 and 14 (21–25). These trends are comparable in other populations as well as in the present findings, despite small sample numbers. In the present study, none of the mutations identified resulted in frame shift or nonsense mutations. Point mutations in KIT exon 11 were detected in codons 557, 559 and 576 only, with codon 559 being the most frequently affected. Deletions in KIT exon 11 affected codons 558 followed by 557. Three of the five deletions, in more distal regions of this exon, involved codons 568 and/or 570. The duplication occurred following codon 570. The above findings are consistent with previous literature (7,8,26,27). Additional mutations in the remaining exons of KIT and PDGFRα that were sequenced were not identified.
Previous Taiwanese studies demonstrated that PDGFRα mutations are rare in Taiwan. Only 1/134 patients revealed a PDGFRα mutation in exon 12 and no mutations in the more prevalent exon 18 were identified (20,28). By contrast, a Chinese study was consistent with western trends described above (29). Japanese studies revealed the same trend but also demonstrated a higher prevalence of PDGFRα exon 12 mutations (30–32). In addition, this was observed in Brazilian and Hispanic populations, revealing higher mutation rates in PDGFRα exon 12 than exon 18 (27,33). In the present study, a more common pattern of PDGFRα exon 18 mutations was observed, occurring in 6.5% of GIST samples, while no exon 12 mutations were identified.
KIT exon 9 mutations are the second most prevalent mutations detected in western and Asian populations. However, the present study revealed similar findings to those of previous Brazilian and Hispanic studies, demonstrating a reduced KIT exon 9 mutation frequency than PDGFRα exons (27,33). A single sample revealed a KIT exon 9 mutation. Therefore, we assume that KIT exon 9 mutations are rare in South African GIST patients. However, a larger study is required to yield more concrete findings. The KIT exon 9 mutation was identified in a metastatic tumour of a Caucasian patient with a primary tumour in the small bowel with a KIT exon 11 deletion. However, the metastatic tumour did not possess the KIT exon 11 mutation, indicating that the metastatic tumour was an independent GIST to the primary tumour. Duplication of codons 502 and 503 of KIT exon 9 was not detected in any of the African patients.
Molecular characterisation is vital for the prediction of response, choice and dose of TKIs. Imatinib is the first choice TKI, while sunitinib is the second line of therapy in cases of resistance (12). As a general rule, mutations in KIT exon 11 are sensitive to imatinib (11). GIST cases with a KIT exon 9 mutation tend to be more aggressive and should recieve a higher daily dose of imatinib (10–12). The effect of mutations in PDGFRα have variable effects on imatinib sensitivity. However, the most common mutation identified, the D842V substitution, is resistant to imatinib treatment (10,15). In the present study, a single known resistant mutation was identified, a D842V mutant.
Genetic diversity is a common trait in African populations (34). As the majority of the patients were of African ethnicity, it was predicted that silent variations would also be detected and it was identified that 77.8% of the patients containing an SNP were of African ethnicity. A novel SNP was detected in KIT exon 17 that has not previously been described. An SNP was observed in 17/45 patients, of which 7 were identified to be wild-type for KIT and PDGFRα. A previous Brazilian study (33) analysed SNP rates and identified polymorphisms in 3/11 wild-type GIST cases. SNPs do not lead to phenotypic alterations, however, their impact on disease progression or treatment is currently unknown.
The mutation status of GISTs affects the efficacy of TKIs in treatment. Therefore, it is vital to determine the genetic profile of a GIST to provide the best possible care for patients. The genetic profile of African GIST patients, including South Africans, is currently unknown and further investigation is required in this area of research. The present study is likely to contribute to the understanding of the molecular profile of GIST in South African patients.
Acknowledgments
The authors thank Novartis Oncology and University of the Witwatersrand for their contribution towards publishing this article.
References
- 1.Bachet JP, Emile JF. Diagnostic criteria, specific mutations and genetic predisposition in gastrointestinal stromal tumors. Appl Clin Genet. 2010;3:85–101. doi: 10.2147/TACG.S7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blay JY, Reichardt P. Advanced gastrointestinal stomal tumour in Europe: a review of updated treatment recommendations. Expert Rev Anticancer Ther. 2009;9:1–8. doi: 10.1586/era.09.34. [DOI] [PubMed] [Google Scholar]
- 3.Gupta P, Tewari M, Shukla HS. Gastrointestinal stromal tumor. Surg Oncol. 2008;17:129–138. doi: 10.1016/j.suronc.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 5.Heinrich MC, Corless CL, Duensing A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 6.Stenman G, Eriksson A, Claesson-Welsh L. Human PDGFA receptor gene maps to the same region on chromosome 4 as the KIT oncogene. Genes Chromosomes Cancer. 1989;1:155–158. doi: 10.1002/gcc.2870010208. [DOI] [PubMed] [Google Scholar]
- 7.Lasota J, Miettinen M. Clinical significance of oncogenic KIT and PDGFRA mutations in gastrointestinal stromal tumours. Histopathology. 2008;53:245–266. doi: 10.1111/j.1365-2559.2008.02977.x. [DOI] [PubMed] [Google Scholar]
- 8.Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal tumors. J Clin Oncol. 2004;22:3813–3825. doi: 10.1200/JCO.2004.05.140. [DOI] [PubMed] [Google Scholar]
- 9.Corless C, Schroeder A, Griffith D, et al. PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol. 2005;23:5357–5364. doi: 10.1200/JCO.2005.14.068. [DOI] [PubMed] [Google Scholar]
- 10.Cassier PA, Blay JY. Molecular response prediction in gastrointestinal stromal tumors. Target Oncol. 2010;5:29–37. doi: 10.1007/s11523-010-0134-9. [DOI] [PubMed] [Google Scholar]
- 11.Blay JY. A decade of tyrosine kinase inhibitor therapy: Historical and current perspectives on targeted therapy for GIST. Cancer Treat Rev. 2011;37:373–384. doi: 10.1016/j.ctrv.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Demetri GD, von Mehren M, Antonescu CR, et al. NCCN Task force report: Update on the management of patients with gastrointestinal stromal tumours. J Natl Compr Canc Netw. 2010;8:S1–41. doi: 10.6004/jnccn.2010.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mol CD, Dougan DR, Schneider TR, et al. Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase. J Biol Chem. 2004;279:31655–31663. doi: 10.1074/jbc.M403319200. [DOI] [PubMed] [Google Scholar]
- 14.Babb C, Schnugh D, Louw M, et al. Getting the gist: what is the importance of molecular genetics in gastro-intestinal stromal tumours (GIST) S Afr Gastroenterol Rev. 2010;8:4–5. [Google Scholar]
- 15.Heinrich MC, Owzar K, Corless CL, et al. Correlation of kinase genotype and clinical outcome in the North American Intergroup Phase III Trial of imatinib mesylate for treatment of advanced gastrointestinal stromal tumor: CALGB 150105 Study by Cancer and Leukemia Group B and Southwest Oncology Group. J Clin Oncol. 2008;26:5360–5367. doi: 10.1200/JCO.2008.17.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 17.Abdulkareem FB, Rotimi O, Elesha SO, Banjo AA. Immunophenotyping of gastrointestinal mesenchymal tumours in Lagos, Nigeria. West Afr J Med. 2009;28:358–362. doi: 10.4314/wajm.v28i6.55022. [DOI] [PubMed] [Google Scholar]
- 18.Hartley RJ, Becker JHR, Van Der Walt H, Luyhengo T. Gastro-intestinal stromal tumours (GISTs) - the Pretoria experience and a literature review. S Afr J Surg. 2011;49:128–131. [PubMed] [Google Scholar]
- 19.Tran T, Davila JA, El-Serag HB. The epidemiology of malignant gastrointestinal stromal tumors: an analysis of 1,458 cases from 1992 to 2000. Am J Gastroenterol. 2005;100:162–168. doi: 10.1111/j.1572-0241.2005.40709.x. [DOI] [PubMed] [Google Scholar]
- 20.Tzen CY, Wang MN, Mau BL. Spectrum and prognostication of KIT and PDGFRA mutation in gastrointestinal stromal tumors. Eur J Surg Oncol. 2008;34:563–568. doi: 10.1016/j.ejso.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Agaram NP, Baren A, Arkun K, Dematteo NP, Besmer P, Antonescu CR. Comparative ultrastructural analysis and KIT/PDGFRA genotype in 125 gastrointestinal stromal tumors. Ultrastruct Pathol. 2006;30:443–452. doi: 10.1080/01913120600854186. [DOI] [PubMed] [Google Scholar]
- 22.Miettinen M, Sobin LH, Lasota J. Gastrointestinal stromal tumors of the stomach: a clinicopathologic, immunohistochemical and molecular genetic study of 1765 cases with long-term follow-up. Am J Surg Pathol. 2005;29:52–68. doi: 10.1097/01.pas.0000146010.92933.de. [DOI] [PubMed] [Google Scholar]
- 23.Miettinen M, Makhlouf H, Sobin LH, Lasota J. Gastrointestinal stromal tumors of the jejunum and ileum: a clinicopathologic, immunohistochemical and molecular genetic study of 906 cases before imatinib with long-term follow-up. Am J Surg Pathol. 2006;30:477–489. doi: 10.1097/00000478-200604000-00008. [DOI] [PubMed] [Google Scholar]
- 24.Debiec-Rychter M, Dumez H, Judson I, et al. Use of c-KIT/PDGFRA mutational analysis to predict the clinical response to imatinib in patients with advanced gastrointestinal stromal tumours entered on phase I and II studies of the EORTC Soft Tissue and Bone Sarcoma Group. Eur J Cancer. 2004;40:689–689. doi: 10.1016/j.ejca.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 25.Debiec-Rychter M, Sciot R, Le Cesne A, et al. KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur J Cancer. 2006;42:1093–1103. doi: 10.1016/j.ejca.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 26.Andersson J, Bumming P, Meis-Kindblom JM, et al. Gastrointestinal stromal tumors with KIT exon 11 deletions are associated with poor prognosis. Gastroenterology. 2006;130:1573–1581. doi: 10.1053/j.gastro.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 27.Martín J, Poveda A, Llombart-Bosch A, et al. Deletions affecting codons 557–558 of the c-KIT gene indicate a poor prognosis in patients with completely resected gastrointestinal stromal tumors: a study by the Spanish group for Sarcoma Research (GEIS) J Clin Oncol. 2005;23:6190–6198. doi: 10.1200/JCO.2005.19.554. [DOI] [PubMed] [Google Scholar]
- 28.Yeh CN, Chen TW, Lee HL, et al. Kinase mutations and imatinib mesylate response for 64 Taiwanese with advanced GIST: preliminary experience from Chang Gung Memorial Hospital. Ann Surg Oncol. 2007;14:1123–1128. doi: 10.1245/s10434-006-9288-1. [DOI] [PubMed] [Google Scholar]
- 29.He HY, Fang WG, Zhong HH, et al. Status and clinical implication of c-KIT and PDGFRA mutations in 165 cases of gastrointestinal stromal tumor (GIST) Zhonghua Bing Li Xue Za Zhi. 2006;35:262–266. (In Chinese). [PubMed] [Google Scholar]
- 30.Sakurai S, Fukasawa T, Chong JM, Tonaka A, Fukayama M. C-kit gene abnormalities in gastrointestinal stromal tumors (tumors of interstitial cells of Cajal) Jpn J Cancer Res. 1999;90:1321–1328. doi: 10.1111/j.1349-7006.1999.tb00715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakurai S, Oguni O, Hironaka M, Fukayama M, Morinaga S, Saito K. Mutations in c-kit gene exons 9 and 13 in gastrointestinal stromal tumors among Japanese. Jpn J Cancer Res. 2001;92:494–498. doi: 10.1111/j.1349-7006.2001.tb01121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirota S, Ohashi A, Nishida T, Isozaki K, Kinoshita K, Shinomura Y, Kitamura O. Gain-of-function mutations of platelet-derived growth factor receptor α gene in gastrointestinal stromal tumors. Gastroenterology. 2003;125:660–667. doi: 10.1016/s0016-5085(03)01046-1. [DOI] [PubMed] [Google Scholar]
- 33.Braggio DA, Braggio E, Small IA, et al. The profile of platelet-derived growth factor receptor alpha (PDGFRA) gene alterations in GIST patients (pts) from Brazil. J Clin Oncol. 2008;26:10561. [Google Scholar]
- 34.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins and complex disease mapping. Annu Rev Genomics Hum Genet. 2008;9:303–333. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]