Abstract
Gram-negative bacteria produce outer membrane vesicles (OMVs) that contain biologically active proteins and perform diverse biological processes. Unlike other secretion mechanisms, OMVs enable bacteria to secrete insoluble molecules in addition to and in complex with soluble material. OMVs allow enzymes to reach distant targets in a concentrated, protected, and targeted form. OMVs also play roles in bacterial survival: Their production is a bacterial stress response and important for nutrient acquisition, biofilm development, and pathogenesis. Key characteristics of OMV biogenesis include outward bulging of areas lacking membrane-peptidoglycan bonds, the capacity to upregulate vesicle production without also losing outer membrane integrity, enrichment or exclusion of certain proteins and lipids, and membrane fission without direct energy from ATP/GTP hydrolysis. Comparisons of similar budding mechanisms from diverse biological domains have provided new insight into evaluating mechanisms for outer membrane vesiculation.
Keywords: gram-negative bacteria, bacterial envelope, vesiculation, secretion, blebs, membrane budding, stress response
INTRODUCTION TO SECRETED VESICLES AND THE ENVELOPE
Gram-negative bacteria, like all other cells, interact with the environment. These interactions are often accomplished by the distal effects of secreted molecules. Secretion is an important aspect of gram-negative bacteria that offers distinct advantages over cell-associated material. Secretion allows cells to interact with a wide area of their environment without expending energy in moving themselves. Secreted material is smaller and nonviable, so it can influence an environment that is inaccessible to the whole bacterium, either due to size or growth restriction. Secreted material is typically considered to be soluble; however, cells in all domains of life (eukaryote, archaea, and prokaryote) have specific mechanisms to secrete insoluble material as well. In this review we focus on the function and biogenesis of gram-negative bacterial outer membrane vesicles (OMVs), an insoluble secretion pathway.
To discuss OMV secretion, it is first necessary to be familiar with the unique architecture of the gram-negative envelope, which consists of two membranes, called the inner and outer membranes, the peptidoglycan layer, and the periplasm (10). The membranes differ in lipid and protein composition. In most gram-negative bacteria, the outer leaflet of the outer membrane (OM) is composed mainly of lipopolysaccharide (LPS), while the inner leaflet and both leaflets of the inner membrane are composed of phospholipids. A viscous, ~13-nm periplasmic space between the two membranes makes up 7–40% of the total cell volume (13, 42, 74). The periplasm is an oxidizing environment devoid of any known energy source such as ATP or NADPH. Within the periplasm is a thin, rigid peptidoglycan layer attached to both membranes by membrane-anchored proteins such as Braun’s lipoprotein (Lpp) and OmpA.
The gram-negative envelope contains a large number of proteins and is the location for several important functions, such as nutrient acquisition, adherence, secretion, signaling, and protection from the environment (12, 76). The importance of the envelope is underlined by the fact that disruptions to any of its layers can be lethal to the cell. To protect the environment, gram-negative bacteria have evolved multiple mechanisms to monitor and repair damage caused by envelope stressors (64).
Gram-negative bacteria release OM and periplasm through the production of OMVs, small spherical structures 20–250 nm in diameter. OMVs are produced when small portions of OM bulge away from the cell, pinch off, and release. Soluble proteins are associated with OMVs as entrapped periplasm and as externally adherent material. Secreted OMVs can disseminate far from the cell and impart biological functions on the environment and on other cells, including playing a role in pathogenesis, quorum signaling, nutrient acquisition, and horizontal gene transfer (25, 43, 54, 55, 65, 84). OMV production is a common feature of all gram-negative bacteria, and because of the energy cost implicit in replacing the shed lipids and proteins, OMV formation has presumably evolved for a reason.
Many studies of OMVs have been directed at determining OMV composition, OMV functions, conditions that affect OMV production, and the design of OMV-based vaccines. Although several mechanisms for vesiculation have been proposed, there is little concerted evidence supporting these ideas. It is possible that vesiculation occurs by a well-conserved mechanism; however, different types of OMVs may exist and originate by different mechanisms. Understanding how bacteria produce OMVs can help us comprehend how other domains of life produce similar secreted structures, and vice versa. Such comparisons will yield insight into the general principles involved in forming a vesicle from a biological membrane.
Here we review the functions of OMVs in relation to bacterial physiology and the conditions and genes involved in regulating OMV production. Then, based on an aggregate consideration of compositional, regulatory, and genetic data, as well as analysis of analogous biological systems, we review the principles of OMV biogenesis.
KEY DISCOVERIES AND ANALYSES OF OUTER MEMBRANE VESICLES
OMVs were first detected when it was observed that the cell-free supernatant of Escherichia coli cultures grown under lysine-limiting growth conditions contained soluble LPS (9). Electron microscopy of these cultures revealed that the OM of the cells was actually shed in the form of small spherical structures with a single membrane surrounding an electron-dense center (41, 83). The authors hypothesized that the lysine-limiting growth conditions inhibited peptidoglycan synthesis without affecting OM synthesis, resulting in an excess of OM that could not remain bound to the cell. However, subsequent studies showed that similar structures were also formed under normal growth conditions in vitro as well as in infected host tissues and serum, and that OMVs are produced by a large variety of gram-negative bacteria, indicating that this process was more widely relevant for gram-negative bacteria (21, 29, 31, 67).
Studies of the density and LPS composition of purified OMVs confirmed their origin (31). By protein analyses, OMVs lacked, or were at least highly depleted in, inner membrane and cytoplasmic components (38). Thus, OMVs were not simply a result of cell lysis, which would create vesicles with a mixture of proteins and lipids from all fractions of the cell. Biochemical comparisons of the protein profiles of OMVs and bacterial OM fractions and comparisons of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) banding patterns showed that OMVs consisted mainly of the typical, abundant OM proteins (31, 37, 52). Because OMVs were found to contain periplasmic proteins by activity assays (34, 35, 48), extra bands representing soluble periplasmic proteins were expected in the OMV lanes. Further differences in protein profiles were also observed. Some OM proteins were disproportionately enriched in OMVs, whereas others were excluded (32, 37, 52, 80). Similar observations were also made for the lipid composition of OMVs (32, 35, 37, 48). OMVs from Pseudomonas aeruginosa were nearly exclusively composed of B-band LPS, which is only present in small proportions in the OM (35, 48). Not only did these studies further support the origin of OMVs, but they were also critical in elucidating key features about OMVs: Enrichment and exclusion of proteins in OMVs meant that the OM does not simply randomly pinch off, and the inclusion of periplasmic content in OMVs meant that the OM is not merely fragmenting, sloughing off, and resealing. Instead, OMVs are the product of a biological mechanism.
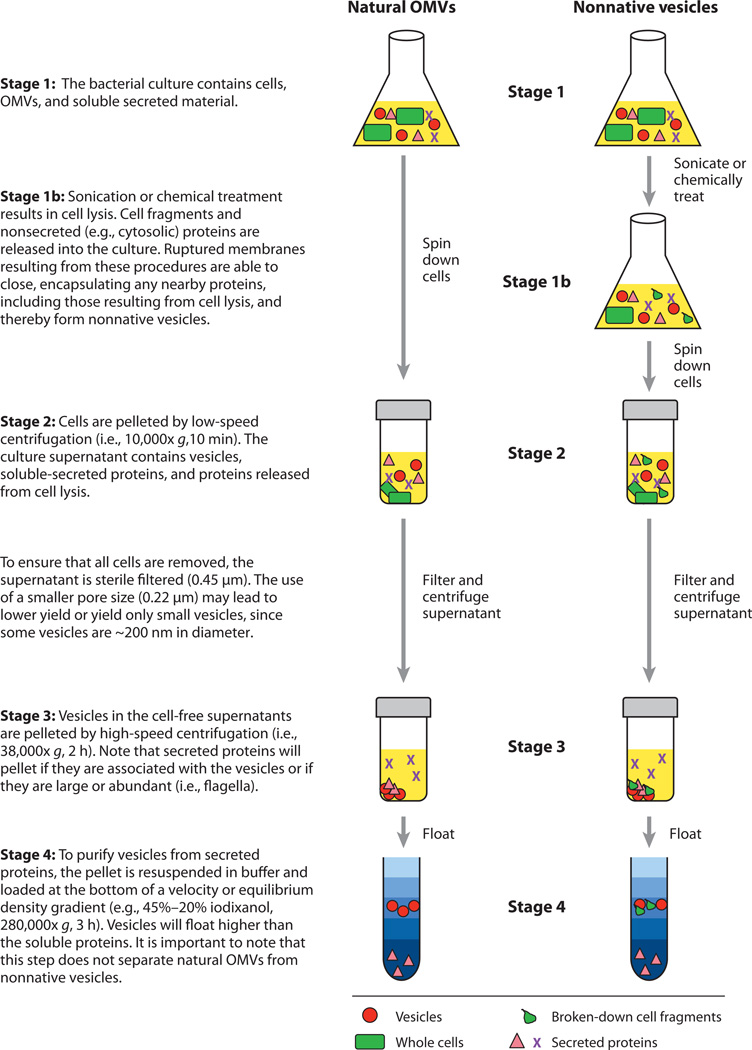
In light of these studies on the composition of OMVs, it should be emphasized that the OMV purification method is crucial in studying their properties (outlined in Figure 1). OMVs are typically separated from cell-free culture supernatants by high-speed (≥40,000× g) centrifugation. However, sedimentation of cell-free supernatants can also cause larger macro-molecules such as fimbriae and flagella to be included in the OMV pellet (5). To characterize a pure population, OMVs must be separated from other supernatant material. This is most readily accomplished by density gradient centrifugation. It should be noted that concentration by ammonium sulfate precipitation prior to density gradient separation can artificially induce nonspecific binding of extracellular proteins to OMVs. Unfortunately, many studies have utilized mechanical shearing, detergent, or chemical treatments to produce greater quantities of material also termed “outer membrane vesicles” (see Figure 1). These studies have confused the literature because such vesicles have different properties than OMVs produced naturally. These vesicles consist of proteins and lipids from the entire bacterium that were randomly captured by membrane fragments that sealed in solution. Thus, results of these studies should be interpreted carefully.
Figure 1.
General schemes for purifying native (left) and nonnative (right) vesicles. Detailed explanations for each stage are provided along the left side of the figure. OMVs, outer membrane vesicles.
FUNCTIONAL ROLES FOR OUTER MEMBRANE VESICLES
As secreted complexes of insoluble and soluble bacterial envelope components, OMVs are poised to play myriad biological roles. An important feature of OMVs is that the proteins associated with them exhibit biological activities (25, 35, 48, 55, 76). OMVs act as delivery vehicles, as nucleators in the formation of bacterial communities (biofilms), and as contributors to bacterial survival and virulence (22, 23, 57, 69).
A Secretion and Delivery System
OMVs are a secretion and delivery system in that they can disseminate bacterial products and interact with the environment (Figure 2). Like other secretion systems, OMV-mediated secretion can be regulated temporally and spatially. However, OMV secretion is distinct from the better-studied soluble protein secretion systems. In OMV secretion, soluble material is released in a complex with and/or surrounded by insoluble material. By contrast, soluble secretory pathways export specific monomeric or small protein complexes, and secretion is not accompanied by the release of other cellular material. Characteristics unique to OMV secretion and delivery are discussed below.
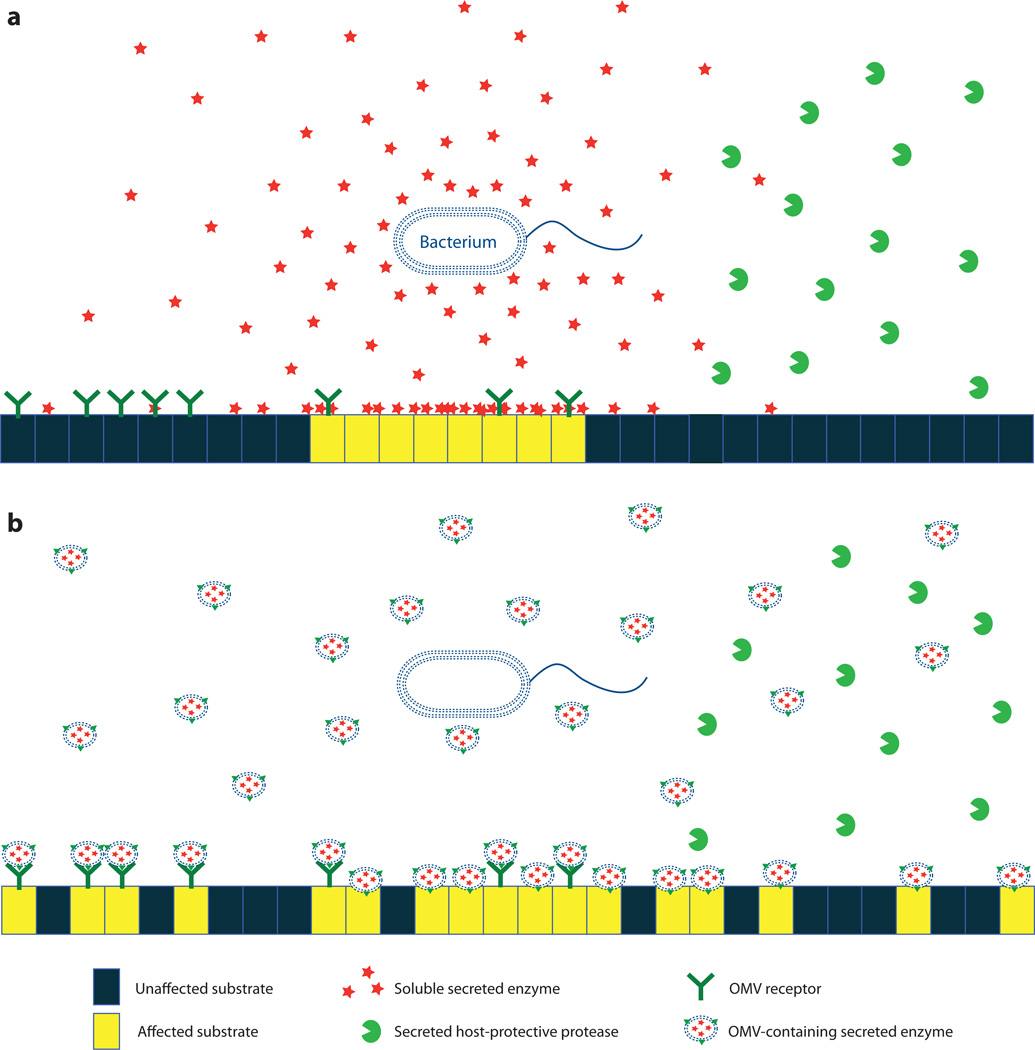
Figure 2.
Comparison of soluble and outer membrane vesicle (OMV)-based secretion and delivery. Soluble secretion pathways (a) result in general diffusion of secreted enzymes. Soluble secretion is most effective over short distances, especially if a threshold concentration is required for activity. Also, if the secreted enzymes are sensitive to proteases and other degradative factors in the environment, fewer enzymes reach target cells. OMV-mediated secretion pathways (b) generate OMVs that contain sufficient concentrations of enzymes to affect the substrate. Further, other molecules on the surface of OMVs can bind to host receptors, thereby allowing the enzyme to be targeted to specific cells or distal sites. OMVs protect the associated enzymes from extracellular degradation, allowing the enzymes to have a greater effective range.
OMV-mediated secretion
One characteristic unique to OMV secretion is that it allows the secretion of bacterial lipids, membrane proteins, and other insoluble compounds. For pathogenic bacteria, integral OM proteins called adhesins are particularly important in colonization of host tissues because they mediate coaggregation. Coadherence and coaggregation of bacteria require functionally displayed, multivalent adhesins. Because OMVs present multivalent complexes of membrane adhesins, OMVs can satisfy this role. Indeed, Porphyromonas gingivalis OMVs cause cellular aggregation and aid in the development of dental plaque biofilms (25, 33).
The highly hydrophobic P. aeruginosa quorum-signaling molecule, PQS, is another type of insoluble compound secreted by OMVs (17, 54). It has recently been shown that PQS cosediments with OMVs and contributes to OMV generation by intercalating into the OM and inducing membrane curvature (53, 54). OMV association allows dispersion of this very hydrophobic molecule in an aqueous environment.
A second characteristic unique to OMV secretion is that it is a means by which soluble proteins can be released in a protective complex (Figure 2). Soluble proteins are included in the OMV lumen as well as bound to the OMV surface. There is expected to be some compensation to the bacteria for this seemingly energetically unfavorable co-release of bystander insoluble material. One possible advantage is the protection of the soluble material within OMVs. In the bacterial environment, particularly in biological niches, extracellular proteases often degrade secreted proteins. Both periplasmic molecules inside the OMV lumen as well as soluble proteins associated with the external surface of OMVs are highly resistant to proteases (39). Thus, OMV-mediated transport can allow less stable molecules, such as protease-susceptible toxins, to reach further destinations or behave like time-release capsules, which provide a beneficial activity at a later time.
A third characteristic unique to OMV secretion is that it allows proteins to be delivered at high concentrations and in close proximity to other factors (Figure 2). If bacteria are far from a target and the secreted molecules are effective only at high concentrations, these soluble effectors will have to be secreted at high levels. By using OMVs, bacteria can deliver effectors at a higher concentration over greater distances. Furthermore, if multiple enzymes are necessary for a particular distal activity, OMVs can co-transport proteins so that they reach a remote site simultaneously.
Finally, OMVs can not only reach a distant site in a hostile environment with an active, specific, and/or highly concentrated cohort of molecules, but they can also be specifically targeted to a particular distal site through the binding specificity between surface-exposed bacterial adhesins and environmental ligands or receptors (Figure 2). The combination of these characteristics makes OMV secretion particularly effective, albeit energy consuming.
OMV-mediated delivery
Soluble OMV content is delivered to a target via two proposed mechanisms. In the first mechanism, OMVs spontaneously lyse, allowing the contents to subsequently diffuse. In the second delivery mechanism, OMVs attach to the target and deliver content by proximal lysis, internalization, or fusion (35).
Little evidence exists for spontaneous OMV lysis. One study did show that exogenously added DNA associated with OMVs in a DNase-resistant manner, suggesting DNA could enter OMVs through OMV lysis and resealing (70). However, this study did not demonstrate the release of lumenal content, and other studies demonstrated that exogenous DNA does not always associate with OMVs (15). Further, many studies have demonstrated that OMVs are stable (36, 63), and large amounts of environmental content have not typically been observed in OMVs. As an alternative explanation for the data regarding DNase-resistant DNA, DNA attached to the surface of OMVs could be protected, or DNA could enter OMVs by the mechanism used in natural bacterial transformation.
Although spontaneous lysis of OMVs is rare, triggered OMV lysis near a target site has been demonstrated. OMVs from over a dozen species lysed near both gram-positive and gram-negative bacteria, indicating that OMV release of autolysins may enable bacterial competition for an environmental niche (35, 49). It was proposed that the OMV membrane was disrupted by interactions with the positively charged peptidoglycan of gram-positive bacteria, whereas OM fusion of OMVs was proposed in the delivery of autolysins to gram-negative cells.
OMV attachment followed by membrane fusion can deliver OMV content directly into target cells (35, 36). In studies by the Beveridge lab, immunogold-labeled LPS was used to track OMVs (36). After coincubation, OMV lipids were detected in the OM of the target bacteria and host cells, and OMV content (notably autolysins and gentamicin) was con-comitantly deposited inside the target cells (35). Although this was clear evidence of OMV fusion and delivery of lumenal content, it is not clear why OMVs are able to fuse with target cells whereas bacterial cells do not readily fuse with each other or with eukaryotic cells. Fusion-inhibiting factors might be present on the surface of the cells but excluded from OMVs, or OMV curvature might facilitate membrane fusion. Restricted membrane fluidity due to the peptidoglycan layer could also prevent fusion of two bacterial membranes.
The combination of OMV-mediated membrane fusion and the ability of OMVs to carry DNA implicates OMVs in evolutionary and population dynamics. Indeed, OMVs have been reported to facilitate horizontal gene transfer between different strains of gram-negative bacteria and even across species (16, 84). In other studies, although DNA was observed in OMVs, transformation could not be achieved (65). In order to transform a recipient cell, OMV-associated DNA would presumably enter into the periplasm along with the other lumenal content during OMV-cell fusion and then transverse the inner membrane. However, the data could also be explained if the DNA was actually on the OMV surface and if DNA brought in proximity to a recipient cell by OMV adherence resulted in gene transfer and transformation by a low level of natural bacterial competence. The latter mechanism is supported by the extremely low frequency (3 × 10−10 with 0.165 ng of DNA) of OMV-mediated transformation (84).
A final mechanism of OMV delivery to consider is endocytosis into eukaryotic cells. OMVs enter host cells by multiple endocytic pathways (22, 40). Endocytosis-mediated delivery bypasses the need for heterotypic fusion of a bacterial OM bilayer into a eukaryotic plasma membrane and results in the entry of the entire OMV into the interior of the cell. The consequences of OMV endocytosis are particularly evident for bacterial pathogens. For example, endocytic delivery of OMVs into antigen-presenting cells results in the display of a cohort of bacterial epitopes to the immune system (1). Also, caveolae-mediated attachment and internalization of enterotoxigenic E. coli–derived OMVs result in both enterotoxic and proinflammatory responses (40).
OMV-Enabled Bacterial Survival
Many qualities of OMVs may aid in bacterial survival. For instance, OMVs can titrate a variety of damaging agents, either intrinsic or extrinsic, away from the bacterial population (51, 57, 69). OMVs can also contribute to nutrient acquisition either by carrying nutrient-generating enzymes or by transporting key nutrients that can be recovered by the cells (76). Furthermore, OMVs can nucleate and maintain cohesion of biofilm communities (69, 86).
Defense and resistance
Under conditions of environmental stress that affect the envelope, such as the accumulation of protein aggregates or exposure to surface-damaging agents, OMVs are a means to quickly relieve the cell of misfolded or toxic material (57). Although induction of OMV secretion requires a significant energy cost, the benefit in life-and-death situations must have been sufficient to allow OMV secretion to evolve.
OMV production can quickly remove a surface-attacking agent from bacteria. For example, OMV production increases the survival of bacteria treated with lytic phage (A. Manning & M.J. Kuehn, unpublished data). Vesiculation increases during the onset of phage adhesion, which helps the bacteria avoid infection by immediately releasing the phage before the phage DNA is injected and by creating an abundance of decoy membranes in the environment (51). OMVs also absorb other molecules, such as complement and antibiotics (12, 75). As with phage binding, OM-targeting antimicrobial treatments can induce OMV production (A. Manning & M.J. Kuehn, unpublished data). Not only do OMVs bind some antibacterial molecules, OMV association can lead to their inactivation (12, 76). OMVs tightly associated with bacterial communities (biofilms) are likely to contribute significantly to the antibiotic resistance of biofilm bacteria. In contrast, planktonic cells must continuously produce OMVs to maintain a sufficient concentration to provide protection. Because this would be expensive in terms of energy, it suggests that OMV-mediated protection by planktonic cells is only an immediate, short-term solution to lethal doses of antibacterial agents. In other words, OMV production can be used to allow the bacteria to survive long enough to establish a more permanent resistance.
OMVs are an envelope stress response pathway. Hypervesiculating mutant strains of E. coli were better able to survive conditions of envelope stress, using both external (ethanol and polymyxin treatment) and internal (overexpression of a toxic periplasmic protein) stressors (57). In contrast, hypovesiculating mutants of E. coli were typically significantly more sensitive than wild type in these conditions. Stressors can influence vesiculation levels as well as OMV content. Overexpression of a periplasmic fusion protein designed to mimic a misfolded OM protein in E. coli causes hypervesiculation and OMV enrichment of the misfolded mimic at least tenfold over a housekeeping protein (57). This enrichment could demonstrate a key feature of the OMV stress response pathway whereby misfolded proteins are selectively removed from the cell by OMVs. Aggregates of misfolded proteins could become resistant to proteolytic cleavage and interfere with essential processes. Therefore, OMVs could be used to remove aggregated proteins from the cell before they become toxic. OMVs seem a particularly good mechanism for secreting large aggregates because the size of the aggregates is not restricted by an OM pore.
Nutrient acquisition
OMVs carry degradative enzymes and receptors that could contribute to nutrient acquisition and thus to bacterial survival (5, 76, 78). Proteases such as the P. aeruginosa aminopeptidase can be both external and active when associated with OMVs. As a result, they are positioned to release into the environment amino acids that may be critical to bacterial growth (5). PQS is a highly hydrophobic molecule associated with P. aeruginosa OMVs, and PQS binds iron, which is essential for bacterial viability and often limiting in biological environments (17, 54). Similar to siderophores, OMVs carrying PQS could enable bacteria to scavenge iron. PQS-Fe-OMVs could then be absorbed into the OM by fusion, or the OMVs could release PQS-bound iron in the vicinity of the cell. OMVs can also acquire electrons from the surrounding environment (24).
Biofilm OMVs
Biofilm OMVs have been studied most extensively for P. aeruginosa and Helicobacter pylori (69, 86). Although significantly smaller than bacteria, OMVs contribute a majority (52 %) of the LPS present in P. aeruginosa biofilms and are associated with the entire biofilm matrix (69). Biofilm-derived OMVs can be separated into two distinct populations based on their density. The high-density biofilm OMVs are slightly larger and associate with fimbriae and pili fragments. The low-density biofilm OMVs are the same size as OMVs from planktonic cultures; however, these OMVs have more LPS and less protein than their planktonic counterparts.
The addition of OMVs to a H. pylori culture stimulated biofilm production (86). This result suggests that vesicles enable biofilms to form and that their presence in biofilms is not solely the result of their entrapment in the matrix. Given the above mentioned studies on DNA association with OMVs, and that exogenous addition of DNA to cells stimulates biofilm production (81), it is possible that OMVs play a role in signaling biofilm production via surface-associated DNA.
Beyond a role in nucleation, OMVs can mediate interactions within and external to the biofilm. OMVs may mediate communication by OMV-associated quorum-signaling molecules (54). Further, biofilm OMVs could serve to bind and/or inactivate harmful molecules (such as antibiotics, complement, and antibodies) and thereby prevent damage to bacteria within the biofilm. By their contributions to nucleation, communication, nutrient acquisition, and defense, OMVs are key multifunctional elements of biofilms, and further studies of these abundant biofilm components are warranted.
OMVs in Bacterial Pathogenesis
The survival-related functions of OMVs described above are likely important to the ability of bacterial pathogens to cause disease. OMVs can shield infecting bacteria from the host immune response, enable survival in the stressful environment inside the host during infection, and help acquire nutrients in an iron- and nutrient-scarce host environment. However, OMVs also have specific virulence-associated activities. OMVs can contain, and sometimes be enriched for, active toxins, and they can deliver these toxins into host cells using a variety of mechanisms (23, 37, 40). The stressful environment encountered during infection can cause OMV upregulation, with substantial downstream consequences: Increased production of OMVs is accompanied by increased toxicity and activation of both innate and adaptive immune responses. Thus, OMVs likely play a variety of key roles in the various stages of bacterial infections. For a recent, detailed review of the role of OMVs in the host-pathogen interactions, see Reference 17a.
REGULATION OF OUTER MEMBRANE VESICLE PRODUCTION
Although no known growth condition or mutation results in the absence of OMV formation, they can affect the levels of vesiculation. Examining the regulation of vesicle production provides some insight into the mechanism of OMV biogenesis. Quantitation of OMVs in these studies usually relies on the quantitation of protein in sedimented OMVs from a culture supernatant, but it should be mentioned that some conditions or mutations dramatically affect the OM protein-to-lipid ratio; therefore, quantitation of lipids in the OMV pellet (e.g., using FM4–64, a lipophilic fluorescent dye, as described in Reference 56) can and often should also be taken into account when quantifying vesicle production.
Growth Conditions
Growth conditions can have a drastic effect on OMV production. For example, high growth temperatures can increase the number of OMVs produced (38). However, this effect is likely indirect because increased growth temperature affects many processes. For example, at high temperatures more proteins are denatured, activating stress response pathways that influence vesiculation, but also membranes become more fluid, enabling the OM to bulge and release OMVs. Higher growth temperatures also increase the rate of cell division, with concomitant increased rates of cell wall growth and turnover. Indeed, cells growing at exponential phase produce more OMVs and more OMVs are produced at division septa, possibly because of the increased peptidoglycan turnover that occurs during cell division (38, 58).
As discussed above, nutrient availability affects vesiculation. Depending on the species of bacteria, this response can depend on opposite signals. Upregulation of OMV production for Lysobacter occurs in response to low nutrition, whereas for Pseudomonas fragi it occurs in response to the availability of nutrients in meat media (76, 79). Enzymes contained within OMVs might degrade a distant, immobile nutrient, thereby releasing soluble nutrients into the environment.
External conditions that generate envelope stress elicit a strong vesiculation response in E. coli (57; A. Manning & M.J. Kuehn, unpublished data). These treatments may increase vesiculation indirectly by activating envelope stress response pathways or directly by disrupting the peptidoglycan layer or the proteins linking the OM to the peptidoglycan.
Genetic Control
Several genes are involved in OMV production based on vesiculation changes that occur when the gene is mutated or knocked-out. Genetic mutants must be carefully analyzed to reveal whether they change only vesiculation levels and not simply reduce membrane integrity. For example, mutations of components of the Tol/Pal-envelope-spanning complex caused vesiculation to increase sharply, and this result has been interpreted to mean that the Tol/Pal system was a master regulator of vesiculation (7, 14, 30). However, defects in this system cause considerable leakiness of the OM (45, 50). Because other mutations result in hypervesiculation without concomitant loss of membrane integrity (56), it is not likely that the Tol/Pal system is the sole mechanistic basis for OMV production.
A study of two closely related strains of Lysobacter has provided a compelling example of OMV regulation at the genetic level (79). Strains XL1 and XL2 have distinct OMV production phenotypes, even though XL2 arose from subculturing XL1. Strain XL1 produced a higher basal level of OMVs and responded much more dramatically to changes in nutrient availability. For both strains, only low levels of OMVs were observed in nutrient-rich media, but when grown in poor media, XL1 produced up to 100-fold-more OMVs whereas XL2 did not. Thus, for XL2, the ability to vesiculate was unaffected, but the regulation of OMV production was disrupted. Because XL2 did not result from mutagenic treatment, it can be assumed that it was nearly genetically identical to XL1. The identity of the genes that differ between these two strains could provide valuable insight into OMV regulation.
A transposon mutant screen of E. coli was performed to identify genes responsible for vesiculation (56). Although this screen was not saturating, it did identify a number of mutations that altered vesiculation levels up to 100-fold. Two of the identified hypovesiculating mutants had mutations in genes encoding putative cell-envelope-localized proteins (ypjA and nlpA). A larger number of hypervesiculating mutants were identified, several of which also displayed compromised OM integrity. Increased OM proteins and lipids in the supernatants of these mutants could be the result of OM disintegration rather than a regulated process of generating OMVs.
Transposon mutations in several genes involved in the SigmaE stress response pathway caused vesiculation phenotypes with little to no membrane integrity defects. The SigmaE pathway responds to misfolded OM proteins by upregulating many genes, including those encoding periplasmic chaperones and proteases (64, 66). Vesiculation increased when the SigmaE pathway was activated (57). This effect could be due either to a specific Sigma E-regulated protein or to a general pressure increase in the envelope caused by increased protein content. The transposon screen also identified hypervesiculation mutants with significantly lower SigmaE activities (57). These might also be explained by increased pressure on the OM: Without SigmaE-regulated proteases and chaperones, misfolded proteins would accumulate in the periplasm.
Whether accumulation of envelope proteins could increase vesiculation was tested directly. Vesiculation of E. coli increased when genes encoding either nonnative or native periplasmic proteins were overexpressed (57). Thus, the response was due to the overabundance of periplasmic material, since this effect was dose dependent, not limited to endogenous proteins, and not due to SigmaE activation.
The vesiculation screen also identified a gene, nlpI, that encodes an OM-anchored lipoprotein. The null mutation of nlpI did not alter SigmaE levels or cause compromised membrane integrity but did cause hypervesiculation (56). The role of nlpI in vesiculation has not yet been elucidated, although it appears to also play a role in cell division (62).
A role for OM-peptidoglycan links in vesiculation has been suggested from the early studies and has gained more support recently. Deleting or truncating OmpA, an abundant protein linking the OM and peptidoglycan layer, results in increased vesiculation in E. coli, Salmonella, and Vibrio cholerae (14, 72, 73). It also was recently shown that a small RNA of V. cholera, VrrA, can block the expression of OmpA and that the presence of VrrA correlated with the amount of OMVs produced (72). The increase in vesiculation observed with VrrA over-expression was similar to that seen in an ompA mutant. The sRNA mechanism to downregulate OmpA is conserved across gram-negative species, with homologues of VrrA found in E. coli, Salmonella, Yersinia pestis, and Klebsiella pneumoniae (3), suggesting a common mechanism for upregulating vesiculation. VrrA and the E. coli homolog MicA are regulated by the SigmaE system, possibly explaining how vesiculation is increased in response to SigmaE-activating envelope stress (72, 77). Further evidence for the role of OM-peptidoglycan links in vesiculation was presented in a study using truncation mutants of several other abundant OM-peptidoglycan-linking proteins in Salmonella (14). The truncations caused vesiculation to increase to varying extents, further showing that these linkages may be critical in regulating vesiculation.
ANALYSIS OF OUTER MEMBRANE VESICLE CONTENT
The content of OMVs from various species has been characterized using protein mass spectroscopy (MS). Compositional data from lipid and SDS-PAGE profiles, as well as comparisons between bacterial fractions and OMVs, have long been used to identify the source of OMVs; however, this method results in conflicting conclusions (31). Proteomic data must be carefully interpreted. In some studies, vesicles from chemically or mechanically treated bacteria were analyzed. These treatments do not produce native OMVs in protein content or reflect their biogenesis (see Figure 1) (20). Also, whereas MS is extremely sensitive, it is unable to determine relative abundance in the sample. As a result, the OMV compositional data is easily misinterpreted. For instance, many MS studies of OMVs report they include cytoplasm owing to the presence of cytoplasmic components (44, 46). Because the MS studies do not provide information on the abundance of identified proteins, it is possible that the cytoplasmic proteins are the result of small amounts of contamination. Quantitative comparative techniques such as two-dimensional difference gel electrophoresis have been used to circumvent these problems (5).
Mass Spectroscopy of OMVs
Challenges in the MS proteomic study of E. coli OMVs are exemplified by two studies. In the first study, Lee et al. (46) identified OMV proteins from a stationary-phase wild-type culture. They found a large variety of cytoplasmic proteins, suggesting that E. coli OMVs regularly contain cytoplasmic material. However, cytoplasmic proteins may actually represent only a small portion of OMV protein. In the second study, Berlanda Scorza et al. (6) characterized OMVs from a Tol/Pal mutant of E. coli. The authors took great care to examine highly purified OMVs and, by collecting OMVs from a log phase culture, to avoid contaminations from lysed cells. Cytoplasmic or inner membrane proteins were not found, and they argue that Lee et al. found different results owing to contamination from cell lysis. Although it appears that Berlanda Scorza et al. analyzed pure OMVs, it is not clear that the Tol/Pal mutant OMVs are the same as wild-type OMVs, despite the authors finding similar protein profiles by SDS-PAGE. Because Tol/Pal mutants cause general membrane instability (45, 50) and because natural OMVs have enriched/excluded OM content (described above), it is likely that the protein content of OMVs purified from mutants could differ in subtle but important ways from the protein content of wild-type OMVs. Nevertheless, most proteomic studies concur that the inner membrane is excluded from OMVs, supporting EM observations of a single lipid bilayer (5, 41). How cytoplasmic proteins could enter OMVs without lysis or being accompanied by inner membrane is a feat hard to imagine. However, current analyses cannot rule out the possibility that OMVs include some inner membrane and cytoplasmic content, including DNA In order to use the power of MS proteomics to deduce OMV origins or biogenesis by OMV content, carefully purified native OMVs should be studied (see Figure 1 and References 32 and 39 for a detailed method).
Differential Packaging of OMV Cargo
As described above, certain proteins and lipids are enriched in OMVs whereas others are excluded (5, 37, 80). This consistent population of enriched and excluded OM and periplasmic components in OMVs supports a specific vesicle biogenesis mechanism and not simply a random blebbing of the OM. However, it should also be considered that OMVs shed by a single bacterial strain could have heterogeneous content owing to multiple mechanisms of biogenesis. This might confound the detection of more examples of differential cargo packaging.
Preferential packaging and exclusion of cargo into OMVs can help to reveal the mechanism of OMV biogenesis. For instance, the finding that Lpp is relatively excluded from OMVs (31, 80) means that the mechanism must account for the absence of this abundant envelope protein from the OMV budding site. Because Lpp bridges the OM and peptidoglycan, it is obvious that the lack of such a component would promote OMV development. As mentioned above, P. aeruginosa OMVs primarily contain B-band LPS; however, the OM contains both A-band and B-band LPS, with the B-band being the minor OM constituent (35, 48). B-band LPS is more highly charged than A-band LPS; therefore, it was proposed that B-band LPS localized to a small area on the cell surface, and the close proximity of like charges forced the membrane to curve outward, thereby increasing the distance between the charged LPS chains. This localized curvature could then be resolved by release as an OMV. Less obvious is what would cause the clustering of B-band LPS in the OM to initiate these events. Similarly, it is not understood how overexpressed mimics of misfolded OM proteins increase vesiculation and become enriched in the secreted OMVs (57). It is possible that a gathering of such periplasmic proteins (possibly complexed with an OM component) causes outward pressure on the OM and leads to OMV formation.
COMPARISON WITH OTHER EXAMPLES OF MEMBRANE BUDDING
Our understanding of the mechanism of OMV biogenesis is assisted by studies of similar outward budding and fission processes in other biological cells. Vesicles are made with the egress of enveloped viruses and during the development of archaeal vesicular bodies and multivesicular bodies of the endosomal sorting complex required for transport (ESCRT) pathway in mammalian cells.
Viral Budding
Enveloped virus budding offers numerous examples of how small fragments of membrane can be selectively released from the cell. This process involves three key aspects that are found in bacterial vesiculation: membrane bulging, packaging of specific components inside the bud, and membrane fission to release the virion.
Influenza virus release is a particularly well-studied process that presents several similarities to OMV release. Unlike many envelope viruses, the size and shape of the influenza virion are not determined by an underlying nucleocapsid; so, much like OMVs, a range of sizes are observed (27). The size of the influenza virus probably depends on membrane proteins, and similarly, OMV size may be modulated by OM proteins.
The budding and release of influenza depend on three proteins (the transmembrane proteins NA and M2, and the membrane-associated protein M1) as well as lipid rafts in the membrane. Lipid rafts concentrate critical viral proteins such as NA, which is needed for proper virus release (60). Whereas the majority of the viral envelope is derived from lipid rafts, the bud scission site appears to be devoid of lipid rafts (4). It is thought that the lipid raft, which is more viscous than the rest of the membrane, is not sufficiently fluid to support membrane fission. These data demonstrate how lipid composition can play a key role in initiating budding through protein recruitment. Similarly, OMV formation may depend on a heterogeneous patch of OM lipids or proteins, and the observed variation in the size of OMVs, even for a single cell, may be due to varying sizes of the membrane patches.
M1 plays a more direct role in budding (11). M1 remains associated with the inner leaflet of the plasma membrane and likely helps concentrate the viral proteins NA and HA through interactions with other envelope proteins (2, 60). M1 also creates a striking asymmetry between membrane leaflets that could generate membrane curvature (60). A similar process may occur for OMVs in which membrane proteins may be predominately recruited at one side of the OM, leading to a change in membrane curvature that induces OMV budding. M1 also binds to ribonucleoproteins and is crucial to packaging the viral genome into the virus (85). The enrichment of particular OMV cargo might be similar, owing to a protein that interacts with both enriched proteins and lipids and proteins involved in OMV formation. Finally, M1 appears to play a role in the fission step, as M1 mutants produced longer virus particles (11, 61). That a single protein can be important to multiple budding steps gives precedent that OM vesiculation may not require a large set of proteins.
Bud release typically requires an active step. The release of influenza virus is an ATP-dependent process that is inefficient; only around 10% of the fully budded viral particles are released (60). It is not clear if this is caused by a lack of ATP in the host cell late in infection or a general inefficiency of the scission process. Similarly, filoviruses with defects in viral release form long buds at the membrane of infected cells, indicating that viral scission is an important step that does not occur randomly (28). This stage of the process contrasts with OMV budding: OMV release appears to be a rapid, efficient process (8).
ESCRT-Mediated Budding by Eukaryotes and Archaea
The ESCRT pathway creates vesicles by an outward budding mechanism, and therefore it is, at least in principle, related to the OMV biogenesis pathway. The ESCRT pathway is conserved in most taxa of eukaryotes and was also discovered to exist in archaea (18, 71, 82).
In eukaryotes, the ESCRT pathway is responsible for budding of intracellular multi-vesicular bodies. In this process, four proteins, ESCRT-0, -I, -II, and -III, are successively recruited to vesicle budding sites (82). The vesicular bodies are released by a fission event involving a ring of proteins that forms at the interface between the bud and the membrane. The polymerized ring constricts by eliminating proteins from the ring, forming progressively smaller rings, and eventually pinching off the neck of the budding vesicle. Identifying conserved bacterial proteins homologous to the ESCRT proteins or at least having features analogous to those of ESCRT proteins, such as the presence of lipid binding domains, might aid the search for the OMV biogenesis pathway.
ESCRT proteins are also involved in cargo recruitment to the newly forming vesicle (71, 82). However, cargo sorting into multivesicular bodies is ubiquitin dependent (71), and thus this process would not be conserved in bacteria. However, there may be another, as yet undiscovered, tag in bacteria that takes the place of ubiquitin.
The ESCRT-III proteins are the final ESCRT proteins recruited to the sites of budding. They have an inactive monomeric form and an active multimeric lattice form (59). It is not clear what role this lattice plays. It is not proposed that the ESCRT proteins themselves create membrane curvature, but instead that they recruit curvature-inducing components. For example, it is suggested that such membrane curvature in yeast is induced by the interaction between ESCRT-0 and phosphoinositides with Ent3 and Ent5, which have ENTH (epsin N-terminal homology) domains (19). A search for analogs in gram-negative bacteria might identify major players in the vesiculation pathway. In archaea, a cell division pathway similar to ESCRT-III mediates external vesicle formation (18). This conservation between domains increases the likelihood that bacteria possess a similar system.
IF-BAR-Mediated Budding
Another form of outward budding in eukaryotes is through the use of inverted F-BAR (IF-BAR) proteins. These proteins create curvature in membranes both in vitro and in vivo (26, 68). IF-BAR proteins are crescent-shaped proteins with charged residues located on the convex side. These residues are thought to interact with the charged head groups of the membrane and provide a scaffold for the initial budding (68). Also, domains on the edges of the protein seem to recruit phosphatidylserine, which is concentrated at those sites (68). Proteins involved in OMV formation could, similarly, specifically interact with certain lipids of the OM, causing the observed enrichment of particular lipids seen in OMVs.
OUTER MEMBRANE VESICLE BIOGENESIS
In proposing how OM vesiculation occurs, we must consider both the cumulative OMV data and the mechanistic principles detailed in this review. A gram-negative species, strain, or mutant that does not produce OMVs has never been identified. This makes it likely (but not certain) that vesiculation is an essential, and possibly conserved, process. It has been established that vesicle biogenesis is regulated, it does not result from or cause fissures in the OM, it leads to heterogeneous packaging of proteins and lipids, and it cannot require ATP or other energy sources directly at the site of budding. In sum, OM vesiculation likely results from nonexclusive events that depend on physiological circumstances or species (Figure 3).
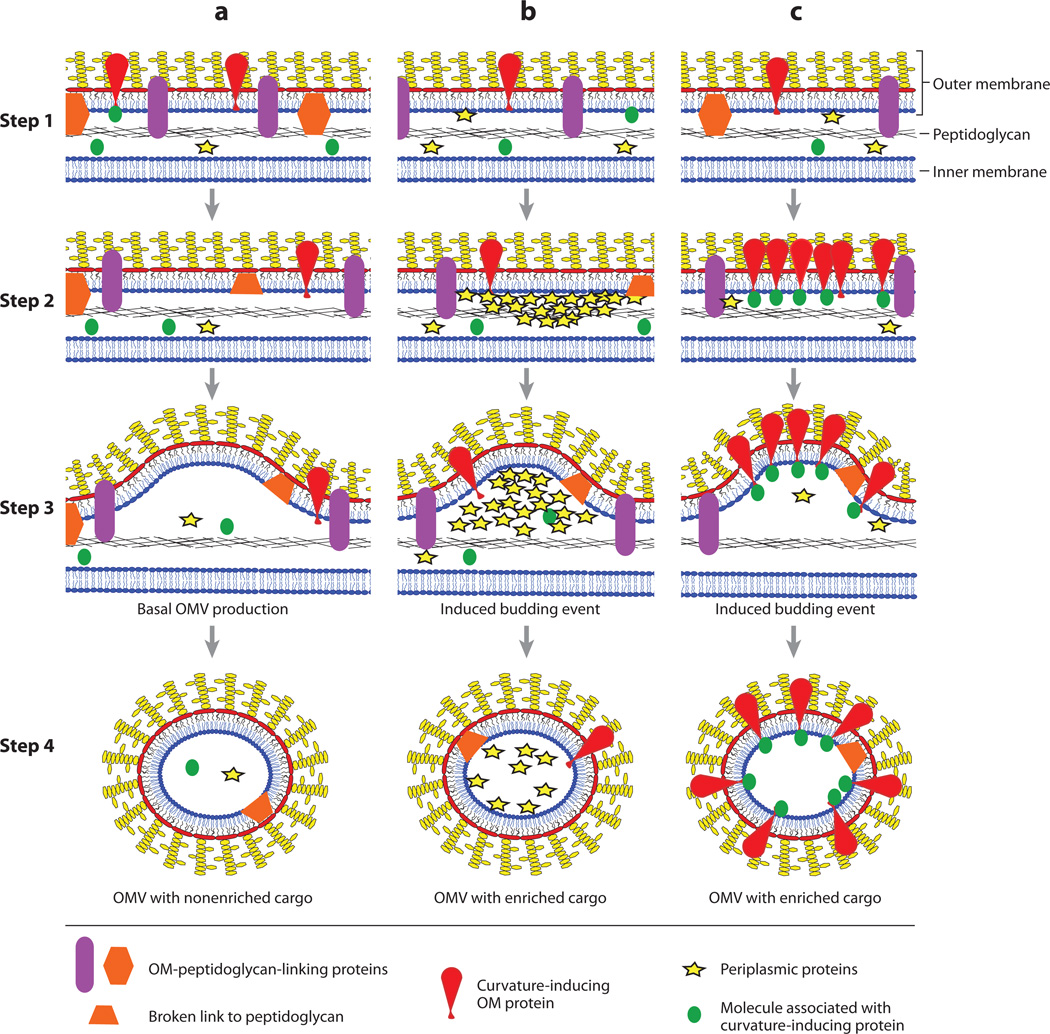
Figure 3.
Events in outer membrane vesicle (OMV) biogenesis. Step 1: Unbudded gram-negative envelope. The overall homogenous distribution of envelope proteins, including outer membrane (OM)-peptidoglycan-linking proteins (purple ovals or orange hexagons), does not lead to significant areas of OM unlinked from peptidoglycan. Steps 2 and 3: Initial stages of vesiculation. In various areas, links between the OM and the peptidoglycan are lost, either by movement of the linking protein (purple ovals) or by breaking the connections directly (orange half hexagons). This could be sufficient for basal OMV production (column a). Gathering of periplasmic proteins ( yellow stars) (column b) and/or accumulation of curvature-inducing OM proteins (red wedges) (column c) could induce additional budding events. Step 4: Released OMVs. OMVs that form without an inducing force (column a) may not have particularly enriched cargo. If OMVs are formed because of localized pressure on the OM, then the proteins causing the pressure (yellow stars) would be enriched in the OMVs (column b). If curvature-inducing molecules (red wedges) are involved, then these molecules and molecules associated with them (green circles) would also be enriched in the OMVs (column c). In all cases, inactive or cleaved OM-peptidoglycan-bridging proteins may be present in the OMVs. These multiple budding mechanisms are not exclusive, so OMVs released in a culture could be a mixed population and would make cargo enrichment difficult to detect.
Triggering OM Bulging
The first step toward creating an OMV is outward bulging of the OM. This event implies that buds form in areas where proteins linking the OM to the peptidoglycan layer are absent (Figure 3). This could occur if the peptidoglycan is disrupted, such as with antibiotics or autolysins (35), and could result in the release of OMVs containing peptidoglycan fragments and portions of OM-peptidoglycan bridging proteins.
Although this model accounts for the increased vesiculation that occurs during envelope disruption, it is not likely the main mode of vesiculation. Vesiculation can be increased over 100-fold without affecting membrane integrity (56), a feat hard to imagine if the peptidoglycan layer was heavily degraded. Other envelope events could trigger OMV budding. For example, OM-peptidoglycan links could be moved or targeted for destruction (Figure 3). Evidence exists to support both models, as the linking protein OmpA is depleted but present in OMVs and Lpp is excluded (31, 51, 58, 80). Simply relocating the links would provide the benefit of maintaining overall membrane integrity, as there would always be some level of OM-peptidoglycan links present, even if vesiculation were highly upregulated.
However, simply removing OM-peptidoglycan links cannot suffice for the mechanism of OMV formation because it does not take into account that overexpressed and misfolded periplasmic proteins can cause budding and that specific proteins are enriched/excluded in OMVs. Proteins could gather at the inner surface of the OM first, creating pressure on the membrane, but not result in budding until OM-peptidoglycan links are removed from the area (Figure 3). This would increase the odds of a bud forming when the links are removed, and allow vesiculation to be upregulated by the accumulation of periplasmic material as well as by removal of OM-peptidoglycan-linking proteins. Specific gathering of particular proteins at these sites would enrich these proteins in the OMVs (32, 37, 57).
Alternative to or in addition to periplasmic turgor pressure, curvature-inducing molecules could cause membrane bulging, as mentioned for viral, archaeal, and eukaryotic processes (Figure 3). If, upon gathering of curvature-inducing molecules, the underlying OM-peptidoglycan links were broken, then the membrane could bud. This would result in the enrichment of these molecules in OMVs, such as observed with B-band LPS of P. aeruginosa. Also, the curvature-inducing molecules could interact with specific periplasmic proteins and cause enrichment of particular OMV cargo. In each scenario, vesiculation would be regulated by the aggregation/dispersion of envelope proteins.
Fission
It is often proposed that OMVs release from the cell when the bud grows to the point at which membrane curvature forces separation. However, it is not clear that this actually happens. OMVs are found in a variety of sizes, indicating that there is not a critical threshold of curvature that precipitates OMV release, and viral particles do not spontaneously release, as discussed above. Further, OMVs from Shewanella spp. remain tethered to the cell by a thin membrane wire (24). If scission spontaneously resulted from high membrane curvature, these tethers would be unlikely to occur. Many membrane fission processes are energy dependent, but the gram-negative bacterial envelope does not have a direct energy source such as ATP or NADPH, so it is not clear what would drive fission. One possibility is that energy is provided from the cytoplasm via an inner membrane conduit. Alternatively, the required energy is stored by the folding of membrane-associated proteins and released by their conformational changes, as occurs for the membrane fusion step of many viruses.
CONCLUSIONS
Rather than a simple, passive barrier, the OM of gram-negative bacteria is a dynamic region of the cell. Whereas some aspects of the envelope have been well studied, understanding OMV production has only recently received attention. Studies of OMVs have established that vesiculation is a common, regulated process of gram-negative bacteria in which envelope proteins, including membrane-bound proteins, lipids, quorum-signaling molecules, and even DNA, can be secreted in a protected and often concentrated form that allows targeted delivery. OMVs play a number of important roles in the growth and survival of gram-negative bacteria. In addition, OMVs are a stress response to both internal stresses such as protein misfolding and external stresses such as phage infections (51, 57). Vesiculation is regulated by growth conditions, by induction of certain stress response pathways, and by overexpression of periplasmic proteins (38, 57). Also, vesiculation seems to be regulated at the genetic level, as single-gene mutations and closely related strains affect the levels of OMVs released (56).
It is likely that OMV biogenesis involves several key features: breaking the contacts between the OM and the peptidoglycan wall, inducing localized membrane curvature, enriching/excluding particular proteins, and releasing OMVs. The study of membrane dynamics and vesicle formation in eukaryotes and archaea will likely provide significant guidance toward identifying the bacterial components that are critical to OMV biogenesis.
FUTURE DIRECTIONS
The study of OMVs will continue to elucidate general features of gram-negative bacteria and allow the application of this knowledge to other uses. OMV studies have already revealed the nonhomogenous distribution of protein and lipid in the bacterial envelope. An important breakthrough in finding the mechanism of vesiculation will be the identification of conserved envelope components that are directly involved in OMV formation and are common to all gram-negative bacteria. Based on the discovery that OMV production is essential in specific, stressful growth conditions, synthetic genetic analysis might be used to identify which proteins are involved in the mechanism of OM vesiculation. Ultimately, it is possible that a novel antibiotic could be designed to target this conserved, essential, and virulence-associated process and thereby inhibit bacterial growth and pathogenesis. In addition, although already successful, OMV-based vaccines can be optimized further. Several studies have shown that vesicle vaccines can elicit a strong, lasting immune response in animal models (47); however, these studies rely on artificially created vesicles that may not be effective in presenting the most efficacious antigens. Further understanding of OMV biogenesis will help in the production of OMVs enriched in ideal antigens, thus creating more effective vaccines and improving human health.
SUMMARY POINTS.
OMVs are naturally occurring structures derived from the OM of gram-negative bacteria.
OMVs are a defined secretory pathway, releasing a cohort of insoluble as well as enclosed or associated soluble molecules with a distinct composition.
OMVs can deliver secreted molecules to specific targets in a protected, active, concentrated form.
OMVs mediate bacterial envelope stress, survival, colonization, biofilm nucleation and maintenance, virulence, and transformation.
Vesiculation is a regulated but poorly defined process that is likely conserved among gram-negative bacteria and shares common principles with outward budding systems of other biological kingdoms.
OMV biogenesis results from a heterogenous distribution of envelope components.
Vesiculation is probably regulated through the steps that initiate budding, although membrane scission is not necessarily a spontaneous event.
ACKNOWLEDGMENTS
The authors’ research program is supported by grants from the National Institutes of Health R01AI064464 and R01AI079068.
Glossary
- Vesicle biogenesis
the mechanism of OMV budding and release
- Outer membrane vesicle (OMV)
spherical structure derived from the OM of gram-negative bacteria
- LPS
lipopolysaccharide
- OM
outer membrane
- Vesiculation
the process of making natural OMVs
- MS
mass spectroscopy
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Alaniz RC, Deatherage BL, Lara JC, Cookson BT. Membrane vesicles are immunogenic facsimiles of Salmonella typhimurium that potently activate dendritic cells, prime B and T cell responses, and stimulate protective immunity in vivo. J. Immunol. 2007;179:7692–7701. doi: 10.4049/jimmunol.179.11.7692. [DOI] [PubMed] [Google Scholar]
- 2.Ali A, Avalos RT, Ponimaskin E, Nayak DP. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 2000;74:8709–8719. doi: 10.1128/jvi.74.18.8709-8719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 4.Barman S, Nayak DP. Lipid raft disruption by cholesterol depletion enhances influenza A virus budding from MDCK cells. J. Virol. 2007;81:12169–12178. doi: 10.1128/JVI.00835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauman SJ, Kuehn MJ. Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect. 2006;8:2400. doi: 10.1016/j.micinf.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berlanda Scorza F, Doro F, Rodriguez-Ortega MJ, Stella M, Liberatori S, et al. Proteomics characterization of outer membrane vesicles from the extraintestinal pathogenic Escherichia coli DeltatolR IHE3034 mutant. Mol. Cell Proteomics. 2008;7:473–485. doi: 10.1074/mcp.M700295-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Bernadac A, Gavioli M, Lazzaroni JC, Raina S, Lloubes R. Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 1998;180:4872–4878. doi: 10.1128/jb.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 1999;181:4725–4733. doi: 10.1128/jb.181.16.4725-4733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop DG, Work E. An extracellular glycolipid produced by Escherichia coli grown under lysinelimiting conditions. Biochem. J. 1965;96:567–576. doi: 10.1042/bj0960567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- 11.Burleigh LM, Calder LJ, Skehel JJ, Steinhauer DA. Influenza A viruses with mutations in the m1 helix six domain display a wide variety of morphological phenotypes. J. Virol. 2005;79:1262–1270. doi: 10.1128/JVI.79.2.1262-1270.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciofu O, Beveridge TJ, Kadurugamuwa J, Walther-Rasmussen J, Hoiby N. Chromosomal betalactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2000;45:9–13. doi: 10.1093/jac/45.1.9. [DOI] [PubMed] [Google Scholar]
- 13.Collins RF, Beis K, Dong C, Botting CH, McDonnell C, et al. The 3D structure of a periplasmspanning platform required for assembly of group 1 capsular polysaccharides in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2007;104:2390–2395. doi: 10.1073/pnas.0607763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deatherage BL, Lara JC, Bergsbaken T, Rassoulian Barrett SL, Lara S, Cookson BT. Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 2009;72:1395–1407. doi: 10.1111/j.1365-2958.2009.06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorward DW, Garon CF. DNA is packaged within membrane-derived vesicles of gram-negative but not gram-positive bacteria. Appl. Environ. Microbiol. 1990;56:1960–1962. doi: 10.1128/aem.56.6.1960-1962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorward DW, Garon CF, Judd RC. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J. Bacteriol. 1989;171:2499–2505. doi: 10.1128/jb.171.5.2499-2505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubern JF, Diggle SP. Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol. Biosyst. 2008;4:882–888. doi: 10.1039/b803796p. [DOI] [PubMed] [Google Scholar]
- 17a.Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ettema TJ, Bernander R. Cell division and the ESCRT complex: a surprise from the archaea. Commun. Integr. Biol. 2009;2:86–88. doi: 10.4161/cib.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eugster A, Pecheur EI, Michel F, Winsor B, Letourneur F, Friant S. Ent5p is required with Ent3p and Vps27p for ubiquitin-dependent protein sorting into the multivesicular body. Mol. Biol. Cell. 2004;15:3031–3041. doi: 10.1091/mbc.E03-11-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ferrari G, Garaguso I, Adu-Bobie J, Doro F, Taddei AR, et al. Outer membrane vesicles from group B Neisseria meningitidis delta gna33 mutant: proteomic and immunological comparison with detergent-derived outer membrane vesicles. Proteomics. 2006;6:1856–1866. doi: 10.1002/pmic.200500164. Compares proteomic profiles of detergent-derived OMVs with natural OMVs and definitively shows that detergent-derived OMVs were different than their natural counterparts.
- 21.Fiocca R, Necchi V, Sommi P, Ricci V, Telford J, et al. Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J. Pathol. 1999;188:220–226. doi: 10.1002/(SICI)1096-9896(199906)188:2<220::AID-PATH307>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 22.Furuta N, Takeuchi H, Amano A. Entry of Porphyromonas gingivalis outer membrane vesicles into epithelial cells causes cellular functional impairment. Infect. Immun. 2009;77:4761–4770. doi: 10.1128/IAI.00841-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gankema H, Wensink J, Guinee PA, Jansen WH, Witholt B. Some characteristics of the outer membrane material released by growing enterotoxigenic Escherichia coli. Infect. Immun. 1980;29:704–713. doi: 10.1128/iai.29.2.704-713.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorby Y, McLean J, Korenevsky A, Rosso K, El-Naggar MY, Beveridge TJ. Redox-reactive membrane vesicles produced by Shewanella. Geobiology. 2008;6:232–241. doi: 10.1111/j.1472-4669.2008.00158.x. [DOI] [PubMed] [Google Scholar]
- 25.Grenier D, Mayrand D. Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect. Immun. 1987;55:111–117. doi: 10.1128/iai.55.1.111-117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, et al. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–104. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. USA. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harty RN. No exit: targeting the budding process to inhibit filovirus replication. Antiviral Res. 2009;81:189–197. doi: 10.1016/j.antiviral.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellman J, Loiselle PM, Zanzot EM, Allaire JE, Tehan MM, et al. Release of gram-negative outermembrane proteins into human serum and septic rat blood and their interactions with immunoglobulin in antiserum to Escherichia coli J5. J. Infect. Dis. 2000;181:1034–1043. doi: 10.1086/315302. [DOI] [PubMed] [Google Scholar]
- 30.Henry T, Pommier S, Journet L, Bernadac A, Gorvel JP, Lloubes R. Improved methods for producing outer membrane vesicles in gram-negative bacteria. Res. Microbiol. 2004;155:437–446. doi: 10.1016/j.resmic.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 31. Hoekstra D, van der Laan JW, de Leij L, Witholt B. Release of outer membrane fragments from normally growing Escherichia coli. Biochim. Biophys. Acta. 1976;455:889–899. doi: 10.1016/0005-2736(76)90058-4. Performed the first thorough analysis of OMVs to determine their origin.
- 32.Horstman AL, Kuehn MJ. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 2000;275:12489–12496. doi: 10.1074/jbc.275.17.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inagaki S, Onishi S, Kuramitsu HK, Sharma A. Porphyromonas gingivalis vesicles enhance attachment, and the leucine-rich repeat BspA protein is required for invasion of epithelial cells by “Tannerella forsythia”. Infect. Immun. 2006;74:5023–5028. doi: 10.1128/IAI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadurugamuwa JL, Beveridge TJ. Virulence factors are released from Pseudomonas aeruginosa in association withmembrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J. Bacteriol. 1995;177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kadurugamuwa JL, Beveridge TJ. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J. Bacteriol. 1996;178:2767–2774. doi: 10.1128/jb.178.10.2767-2774.1996. Shows that OMVs are capable of fusing with other cell membranes and delivering contents directly to the interior of a target cell.
- 36.Kadurugamuwa JL, Beveridge TJ. Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other gram-negative bacteria. Microbiology. 1999;145(Pt 8):2051–2060. doi: 10.1099/13500872-145-8-2051. [DOI] [PubMed] [Google Scholar]
- 37.Kato S, Kowashi Y, Demuth DR. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb. Pathog. 2002;32:1–13. doi: 10.1006/mpat.2001.0474. [DOI] [PubMed] [Google Scholar]
- 38.Katsui N, Tsuchido T, Hiramatsu R, Fujikawa S, Takano M, Shibasaki I. Heat-induced blebbing and vesiculation of the outer membrane of Escherichia coli. J. Bacteriol. 1982;151:1523–1531. doi: 10.1128/jb.151.3.1523-1531.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kesty NC, Kuehn MJ. Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles. J. Biol. Chem. 2004;279:2069–2076. doi: 10.1074/jbc.M307628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kesty NC, Mason KM, Reedy M, Miller SE, Kuehn MJ. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 2004;23:4538–4549. doi: 10.1038/sj.emboj.7600471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knox KW, VeskM,Work E. Relation between excreted lipopolysaccharide complexes and surface structures of a lysine-limited culture of Escherichia coli. J. Bacteriol. 1966;92:1206–1217. doi: 10.1128/jb.92.4.1206-1217.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koch AL. The biophysics of the gram-negative periplasmic space. Crit. Rev. Microbiol. 1998;24:23–59. doi: 10.1080/10408419891294172. [DOI] [PubMed] [Google Scholar]
- 43.Kolling GL, Matthews KR. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl. Environ. Microbiol. 1999;65:1843–1848. doi: 10.1128/aem.65.5.1843-1848.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon SO, Gho YS, Lee JC, Kim SI. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol. Lett. 2009;297:150–156. doi: 10.1111/j.1574-6968.2009.01669.x. [DOI] [PubMed] [Google Scholar]
- 45.Lazzaroni JC, Germon P, Ray MC, Vianney A. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol. Lett. 1999;177:191–197. doi: 10.1111/j.1574-6968.1999.tb13731.x. [DOI] [PubMed] [Google Scholar]
- 46.Lee EY, Bang JY, Park GW, Choi DS, Kang JS, et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics. 2007;7:3143–3153. doi: 10.1002/pmic.200700196. [DOI] [PubMed] [Google Scholar]
- 47.Lee SR, Kim SH, Jeong KJ, Kim KS, Kim YH, et al. Multi-immunogenic outer membrane vesicles derived from an MsbB deficient Salmonella enterica serovar Typhimurium mutant. J. Microbiol. Biotechnol. 2009;19:1271–1279. [PubMed] [Google Scholar]
- 48.Li Z, Clarke AJ, Beveridge TJ. A major autolysin of Pseudomonas aeruginosa : subcellular distribution, potential role in cell growth and division and secretion in surface membrane vesicles. J. Bacteriol. 1996;178:2479–2488. doi: 10.1128/jb.178.9.2479-2488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z, Clarke AJ, Beveridge TJ. Gram-negative bacteria produce membrane vesicles which are capable of killing other bacteria. J. Bacteriol. 1998;180:5478–5483. doi: 10.1128/jb.180.20.5478-5483.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Llamas MA, Ramos JL, Rodriguez-Herva JJ. Mutations in each of the tol genes of Pseudomonas putida reveal that they are critical for maintenance of outer membrane stability. J. Bacteriol. 2000;182:4764–4772. doi: 10.1128/jb.182.17.4764-4772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loeb MR, Kilner J. Release of a special fraction of the outer membrane from both growing and phage T4-infected Escherichia coli B. Biochim. Biophys. Acta. 1978;514:117–127. doi: 10.1016/0005-2736(78)90081-0. [DOI] [PubMed] [Google Scholar]
- 52.Loeb MR, Kilner J. Effect of growth medium on the relative polypeptide composition of cellular outer membrane and released outer membrane material in Escherichia coli. J. Bacteriol. 1979;137:1031–1034. doi: 10.1128/jb.137.2.1031-1034.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, et al. Interaction of quorum signals with outer membrane lipids: insights into prokaryoticmembrane vesicle formation. Mol. Microbiol. 2008;69:491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437:422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 55.MayrandD,Grenier D. Biological activities of outer membrane vesicles. Can. J. Microbiol. 1989;35:607–613. doi: 10.1139/m89-097. [DOI] [PubMed] [Google Scholar]
- 56. McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J. Bacteriol. 2006;188:5385–5392. doi: 10.1128/JB.00498-06. Shows that vesiculation is independent of membrane instability.
- 57.McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mug-Opstelten D, Witholt B. Preferential release of new outer membrane fragments by exponentially growing Escherichia coli. Biochim. Biophys. Acta. 1978;508:287–295. doi: 10.1016/0005-2736(78)90331-0. [DOI] [PubMed] [Google Scholar]
- 59.Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, et al. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 60.Nayak DP, Balogun RA, Yamada H, Zhou ZH, Barman S. Influenza virus morphogenesis and budding. Virus Res. 2009;143:147–161. doi: 10.1016/j.virusres.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nayak DP, Hui EK, Barman S. Assembly and budding of influenza virus. Virus Res. 2004;106:147–165. doi: 10.1016/j.virusres.2004.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohara M, Wu HC, Sankaran K, Rick PD. Identification and characterization of a new lipoprotein, NlpI, in Escherichia coli K-12. J. Bacteriol. 1999;181:4318–4325. doi: 10.1128/jb.181.14.4318-4325.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Post DM, Zhang D, Eastvold JS, Teghanemt A, Gibson BW, Weiss JP. Biochemical and functional characterization of membrane blebs purified from Neisseria meningitidis serogroup B. J. Biol. Chem. 2005;280:38383–38394. doi: 10.1074/jbc.M508063200. [DOI] [PubMed] [Google Scholar]
- 64.Raivio TL. Envelope stress responses and gram-negative bacterial pathogenesis. Mol. Microbiol. 2005;56:1119–1128. doi: 10.1111/j.1365-2958.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- 65.Renelli M, Matias V, Lo RY, Beveridge TJ. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology. 2004;150:2161–2169. doi: 10.1099/mic.0.26841-0. [DOI] [PubMed] [Google Scholar]
- 66.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rothfield L, Pearlman-Kothencz M. Synthesis and assembly of bacterial membrane components. A lipopolysaccharide-phospholipid-protein complex excreted by living bacteria. J. Mol. Biol. 1969;44:477–492. doi: 10.1016/0022-2836(69)90374-x. [DOI] [PubMed] [Google Scholar]
- 68.Saarikangas J, Zhao H, Pykalainen A, Laurinmaki P, Mattila PK, et al. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr. Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 69.Schooling SR, Beveridge TJ. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 2006;188:5945–5957. doi: 10.1128/JB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schooling SR, Hubley A, Beveridge TJ. Interactions of DNA with biofilm-derived membrane vesicles. J. Bacteriol. 2009;191:4097–4102. doi: 10.1128/JB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shields SB, Oestreich AJ, Winistorfer S, Nguyen D, Payne JA, et al. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J. Cell Biol. 2009;185:213–224. doi: 10.1083/jcb.200811130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song T, Mika F, Lindmark B, Liu Z, Schild S, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol. 2008;70:100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sonntag I, Schwarz H, Hirota Y, Henning U. Cell envelope and shape of Escherichia coli : multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J. Bacteriol. 1978;136:280–285. doi: 10.1128/jb.136.1.280-285.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stock JB, Rauch B, Roseman S. Periplasmic space in Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 1977;252:7850–7861. [PubMed] [Google Scholar]
- 75.Tan TT, Morgelin M, Forsgren A, Riesbeck K. Haemophilus influenzae survival during complementmediated attacks is promoted by Moraxella catarrhalis outer membrane vesicles. J. Infect. Dis. 2007;195:1661–1670. doi: 10.1086/517611. [DOI] [PubMed] [Google Scholar]
- 76.Thompson SS, Naidu YM, Pestka JJ. Ultrastructural localization of an extracellular protease in Pseudomonas fragi by using the peroxidase-antiperoxidase reaction. Appl. Environ. Microbiol. 1985;50:1038–1042. doi: 10.1128/aem.50.4.1038-1042.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Udekwu KI,Wagner EG. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res. 2007;35:1279–1288. doi: 10.1093/nar/gkl1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vasilyeva NV, Tsfasman IM, Suzina NE, Stepnaya OA, Kulaev IS. Secretion of bacteriolytic endopeptidase L5 of Lysobacter sp. XL1 into the medium by means of outer membrane vesicles. FEBS J. 2008;275:3827–3835. doi: 10.1111/j.1742-4658.2008.06530.x. [DOI] [PubMed] [Google Scholar]
- 79.Vasilyeva NV, Tsfasman IM, Suzina NE, Stepnaya OA, Kulaev IS. Outer membrane vesicles of Lysobacter sp. Dokl. Biochem. Biophys. 2009;426:139–142. doi: 10.1134/s1607672909030041. [DOI] [PubMed] [Google Scholar]
- 80.Wensink J, Witholt B. Outer-membrane vesicles released by normally growing Escherichia coli contain very little lipoprotein. Eur. J. Biochem. 1981;116:331–335. doi: 10.1111/j.1432-1033.1981.tb05338.x. [DOI] [PubMed] [Google Scholar]
- 81.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:148. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 82.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 83.Work E, Knox KW, Vesk M. The chemistry and electron microscopy of an extracellular lipopolysaccharide from Escherichia coli. Ann. N. Y. Acad. Sci. 1966;133:438–449. doi: 10.1111/j.1749-6632.1966.tb52382.x. [DOI] [PubMed] [Google Scholar]
- 84.Yaron S, Kolling GL, Simon L, Matthews KR. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 2000;66:4414–4420. doi: 10.1128/aem.66.10.4414-4420.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ye Z, Liu T, Offringa DP, McInnis J, Levandowski RA. Association of influenza virus matrix protein with ribonucleoproteins. J. Virol. 1999;73:7467–7473. doi: 10.1128/jvi.73.9.7467-7473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yonezawa H, Osaki T, Kurata S, Fukuda M, Kawakami H, et al. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol. 2009;9:197. doi: 10.1186/1471-2180-9-197. [DOI] [PMC free article] [PubMed] [Google Scholar]