Vesiclepedia is a community-annotated compendium of molecular data on extracellular vesicles.
Abstract
Extracellular vesicles (EVs) are membraneous vesicles released by a variety of cells into their microenvironment. Recent studies have elucidated the role of EVs in intercellular communication, pathogenesis, drug, vaccine and gene-vector delivery, and as possible reservoirs of biomarkers. These findings have generated immense interest, along with an exponential increase in molecular data pertaining to EVs. Here, we describe Vesiclepedia, a manually curated compendium of molecular data (lipid, RNA, and protein) identified in different classes of EVs from more than 300 independent studies published over the past several years. Even though databases are indispensable resources for the scientific community, recent studies have shown that more than 50% of the databases are not regularly updated. In addition, more than 20% of the database links are inactive. To prevent such database and link decay, we have initiated a continuous community annotation project with the active involvement of EV researchers. The EV research community can set a gold standard in data sharing with Vesiclepedia, which could evolve as a primary resource for the field.
Introduction
A growing body of research has implicated extracellular vesicles (EVs), membraneous sacs released by a variety of cells, in diverse physiological and patho-physiological conditions [1]–[9]. They can be detected in body fluids including blood plasma, urine, saliva, amniotic fluid, breast milk, and pleural ascites [10]–[13], and contain proteins, lipids, and RNA representative of the host cell [14]–[18]. Though a definitive categorization is yet to be achieved [19], EVs can be broadly classified into three main classes, based on the mode of biogenesis: (i) ectosomes (also referred to as shedding microvesicles), (ii) exosomes, and (iii) apoptotic bodies (ABs) (see Box 1).
Box 1. Categories of EVs Based on the Mode of Biogenesis
Ectosomes or shedding microvesicles: Ectosomes are large EVs ranging between 50–1,000 nm in diameter [1]. They are shed from cells by outward protrusion (or budding) of a plasma membrane (PM) followed by fission of their membrane stalk [3],[5]. Ectosomes are released by a variety of cells including tumour cells, polymorphonuclear leucocytes, and aging erythrocytes [5]. The expression of phosphatidylserine (PS) on the membrane surface has been shown to be one of the characteristic features of ectosomes [1],[5].
Exosomes: Exosomes are small membranous vesicles of endocytic origin ranging from 40–100 nm in diameter [1],[42]. The density of exosomes varies from 1.10–1.21 g/ml and the commonly found markers of exosomes are Alix, TSG101, tetraspanins, and heat shock proteins [10]. The biogenesis of exosomes begins with the internalisation of molecules via endocytosis [42]. Once internalised, endocytosed molecules are either recycled to the PM or trafficked to multivesicular bodies (MVBs) [3]. The “exocytic” fate of MVBs results in their exocytic fusion with the PM, resulting in the release of intraluminal vesicles into the extracellular microenvironment as exosomes [43].
Apoptotic bodies: ABs are released from fragmented apoptotic cells and are 50–5,000 nm in diameter [1]. ABs are formed about during the process of programmed cell death or apoptosis, and represent the fragments of dying cells [3]. Similar to ectosomes, the expression of PS on the membrane surface has been shown to be a key characteristic of ABs [1],[5].
Recent studies have highlighted the role of EVs in intercellular communication [20]–[22], vaccine and drug delivery [23]–[25], and suggested a potential role in gene vector therapy [26] and as disease biomarkers [27]. More than three decades of research has advanced our basic understanding of these extracellular organelles and has generated large amounts of multidimensional data [14],[17]. Whilst most of the data are presented in the context of the biological findings/technical development and are mentioned in the inline text of the published article, a vast majority are often placed as supplementary information or not provided [28],[29]. Importantly, none of the molecular data in published articles is easily searchable [28]. With the immense interest in EVs and advances in high-throughput techniques, the data explosion will only increase. An online compendium of heterogeneous data will help the biomedical community to exploit the publicly available datasets and accelerate biological discovery [30].
ExoCarta and Need for an EV Database
Existing databases are not comprehensive. For example, ExoCarta (http://www.exocarta.org), a database for molecular data (proteins, RNA, and lipids) identified in exosomes, catalogs only exosomal studies (as reported by the authors) [31]. Described initially in 2009 [32], the database has been visited by more than 16,000 unique users [33]. However, only exosomal studies (as reported by the authors) are catalogued in ExoCarta. With the confusion in terminologies and inefficiency of the purification protocols to clearly segregate each class of EVs [1],[19], it is critical to build a repository with data from all classes of EVs to understand more about the molecular repertoire of the various classes of EVs and their biological functions. This was the rationale for starting the Vesiclepedia online compendium for EVs.
Vesiclepedia
Vesiclepedia (http://www.microvesicles.org) is a manually curated compendium that contains molecular data identified in all classes of EVs, including AB, exosomes, large dense core vesicles, microparticles, and shedding microvesicles. The main criterion for manual curation was the presence of these vesicles in the extracellular microenvironment (EVs) as approved by the investigators who undertook the research. At this juncture, the EVs are named as per the curated article or submitting author, as the nomenclature is yet to be standardized [19]. Vesiclepedia was built using ZOPE, an open source content management system. Python a portable, interpreted, object oriented programming language was used in the three-tier system to connect the web interface with a MySQL database. Users can query or browse through proteins, lipids, and RNA molecules identified in EVs. Selecting a gene of interest directs the user to a gene/molecule page with information on the gene, its external references to other primary databases, experiment description of the study that identified the molecule, gene ontology based annotations, protein-protein interactions, and a graphical display of such network with relevance to molecules identified in EVs. Gene ontology annotations of molecular functions, biological process, and subcellular localization were retrieved from Entrez Gene [34] and mapped onto the proteins/mRNA identified in EVs. Under the experiment description, the sample source including the tissue name or cell line name, EV isolation procedures, and floatation gradient density as reported in the study are provided to the users. EV proteins are mapped onto their protein physical interactors along with the protein interaction identification method and PubMed identifier. Protein-protein interaction data was obtained from HPRD [35],[36], BioGRID [37], and Human Proteinpedia [38].
Database Issues and Community Annotation
Though biological databases are indispensable resources for effective scientific research, it has to be noted that more than 20% of the database links are non-existent after their initial publication [39]–[41]. More than 50% of the databases are never updated reducing their usability [39], primarily due to the lack of continuous funding to maintain and update these resources. At this juncture, funding for databases is largely non-existent in many parts of the world. To overcome funding-related limitations and to keep the database updated, it is essential to involve the scientific community in annotating the data. Community annotation will significantly ease the burden of the curators who maintain and update the databases. Whilst community annotation is the permanent solution to keep the database updated, it seldom happens without a clear and transparent mechanism. In addition, the system has to ensure continuous deposition of data and “not just once” uploads. It has to be noted that data annotation can be regulated at two levels: (i) principal investigators voluntarily contributing data and (ii) peer-reviewed journals mandating data deposition before publication. Currently available community annotation tools don't have a continuous data deposition arrangement with an investigator. Additionally, only few journals mandate the deposition of data to public repositories before acceptance of a manuscript. To this end, we have initiated a community annotation project through Vesiclepedia that involves members of the EV research community (53 laboratories from 20 countries: Table 1).
Table 1. Vesiclepedia statistics.
| Statistics | n |
| EV studies | 341 |
| Protein entries | 35,264 |
| mRNA entries | 18,718 |
| miRNA entries | 1,772 |
| Lipid molecules | 342 |
| Participating laboratories | 53 |
| Countries | 20 |
Community annotation via Vesiclepedia happens through the founding members who agree to the conditions listed in Box 2. All of the members are listed in the credits page (http://www.microvesicles.org/credits).
Box 2. Conditions to Become a Member of Vesiclepedia Community Annotation
1, Agree to submit datasets pertaining to EVs to Vesiclepedia before or after publication on a continuous basis
2, When reviewing articles, mandate/request investigators to submit datasets on EVs to Vesiclepedia
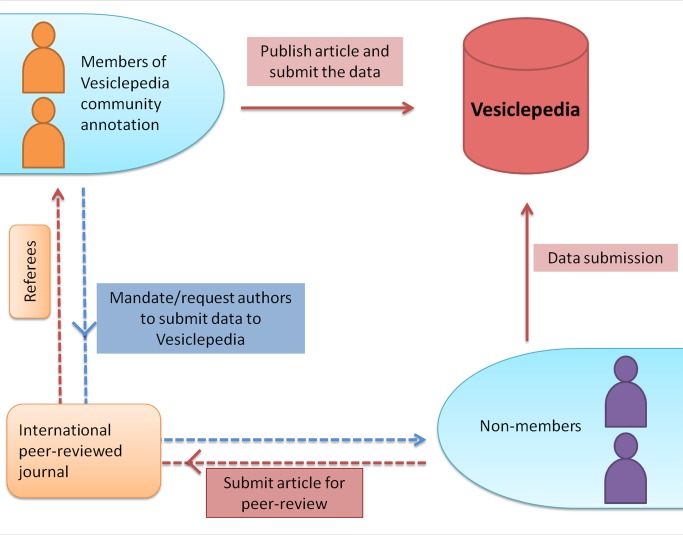
On the basis of the agreement of community participation, members will submit their data automatically to Vesiclepedia before or after publication (Figure 1). Non-members submitting their research findings for peer-review through international journals might find the Vesiclepedia members as referees who will request/mandate the authors to submit the data to Vesiclepedia. By instituting this mechanism the datasets will be continuously deposited to Vesiclepedia. However, a non-member can also be appointed as a referee in which case the data might not be submitted to Vesiclepedia. The Vesiclepedia-data capture team will work along with the researchers to make the data submission as easy as possible. Detailed information on the format of data required for submission is provided in the Vesiclepedia webpage (http://www.microvesicles.org/data_submission). Currently, Vesiclepedia comprises 35,264 protein, 18,718 mRNA, 1,772 miRNA, and 342 lipid entries (Table 1). All of these data were obtained from 341 independent studies that were published over the past several years.
Figure 1. A schematic of Vesiclepedia community annotation.
Based on the agreement of community participation, members will submit their data automatically to Vesiclepedia before and after publication. Non-members submitting their research findings for peer-review through international journals might find some of the Vesiclepedia members as referees who will request/mandate the authors to submit the data to Vesiclepedia. Alternatively, a non-member can also be appointed as a referee in which case the data might not be submitted to Vesiclepedia. A non-member can also submit data directly to Vesiclepedia.
Conclusions and Future Directions
ExoCarta will be active even after the release of Vesiclepedia and will become a primary resource for high-quality exosomal datasets. Data deposited to ExoCarta can also be accessed through Vesiclepedia; however, only high quality exosomal datasets deposited to Vesiclepedia can be accessed through ExoCarta. With the launch of Vesiclepedia, we expect to have an organised data deposition mechanism. We expect active participation from the EV research community, along with the addition of new members and numerous heterogeneous datasets. All datasets submitted by EV researchers will be listed in the credits page along with the investigator details.
Abbreviations
- AB
apoptotic body
- EV
extracellular vesicle
Funding Statement
This work was supported by a NH&MRC fellowship (1016599) and LIMS fellowship to SM. Part of tool development for data upload into Vesiclepedia was funded by the eResearch Office at La Trobe University with programming support from the Victorian eResearch Strategic Initiative (VeRSI). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Thery C, Ostrowski M, Segura E (2009) Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 9: 581–593. [DOI] [PubMed] [Google Scholar]
- 2. Keller S, Sanderson MP, Stoeck A, Altevogt P (2006) Exosomes: from biogenesis and secretion to biological function. Immunol Lett 107: 102–108. [DOI] [PubMed] [Google Scholar]
- 3. Mathivanan S, Ji H, Simpson RJ (2010) Exosomes: Extracellular organelles important in intercellular communication. J Proteomics 73: 1907–1920. [DOI] [PubMed] [Google Scholar]
- 4. Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ (2006) Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia 20: 1487–1495. [DOI] [PubMed] [Google Scholar]
- 5. Cocucci E, Racchetti G, Meldolesi J (2009) Shedding microvesicles: artefacts no more. Trends Cell Biol 19: 43–51. [DOI] [PubMed] [Google Scholar]
- 6. Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G (2002) The biogenesis and functions of exosomes. Traffic 3: 321–330. [DOI] [PubMed] [Google Scholar]
- 7. van Niel G, Porto-Carreiro I, Simoes S, Raposo G (2006) Exosomes: a common pathway for a specialized function. J Biochem (Tokyo) 140: 13–21. [DOI] [PubMed] [Google Scholar]
- 8. Johnstone RM (2006) Exosomes biological significance: A concise review. Blood Cells Mol Dis 36: 315–321. [DOI] [PubMed] [Google Scholar]
- 9. Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, et al. (2006) Alzheimer's disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci U S A 103: 11172–11177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simpson RJ, Lim JW, Moritz RL, Mathivanan S (2009) Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics 6: 267–283. [DOI] [PubMed] [Google Scholar]
- 11. Lasser C, Alikhani VS, Ekstrom K, Eldh M, Paredes PT, et al. (2011) Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Admyre C, Johansson SM, Qazi KR, Filen JJ, Lahesmaa R, et al. (2007) Exosomes with immune modulatory features are present in human breast milk. J Immunol 179: 1969–1978. [DOI] [PubMed] [Google Scholar]
- 13. Mincheva-Nilsson L, Baranov V (2010) The role of placental exosomes in reproduction. Am J Reprod Immunol 63: 520–533. [DOI] [PubMed] [Google Scholar]
- 14. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, et al. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654–659. [DOI] [PubMed] [Google Scholar]
- 15. Thery C, Zitvogel L, Amigorena S (2002) Exosomes: composition, biogenesis and function. Nat Rev Immunol 2: 569–579. [DOI] [PubMed] [Google Scholar]
- 16. Al-Nedawi K, Meehan B, Rak J (2009) Microvesicles: messengers and mediators of tumor progression. Cell Cycle 8: 2014–2018. [DOI] [PubMed] [Google Scholar]
- 17. Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, et al. (2010) Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics 9: 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, et al. (2008) Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 10: 619–624. [DOI] [PubMed] [Google Scholar]
- 19. Simpson RJ, Mathivanan S (2012) Extracellular microvesicles: the need for internationally recognised nomenclature and stringent purification criteria. J Proteomics Bioinform 5: ii–ii. [Google Scholar]
- 20. Hood JL, San RS, Wickline SA (2011) Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res 71: 3792–3801. [DOI] [PubMed] [Google Scholar]
- 21. Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, et al. (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 18: 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, et al. (2011) Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, et al. (2011) Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 29: 341–345. [DOI] [PubMed] [Google Scholar]
- 24. Sun D, Zhuang X, Xiang X, Liu Y, Zhang S, et al. (2010) A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther 18: 1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lakhal S, Wood MJ (2010) Exosome nanotechnology: an emerging paradigm shift in drug delivery: exploitation of exosome nanovesicles for systemic in vivo delivery of RNAi heralds new horizons for drug delivery across biological barriers. Bioessays 33: 737–741. [DOI] [PubMed] [Google Scholar]
- 26. Maguire CA, Balaj L, Sivaraman S, Crommentuijn MH, Ericsson M, et al. (2012) Microvesicle-associated AAV vector as a novel gene delivery system. Mol Ther 20: 960–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, et al. (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10: 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Santos C, Blake J, States DJ (2005) Supplementary data need to be kept in public repositories. Nature 438: 738. [DOI] [PubMed] [Google Scholar]
- 29. Mathivanan S, Ahmed M, Ahn NG, Alexandre H, Amanchy R, et al. (2008) Human Proteinpedia enables sharing of human protein data. Nat Biotechnol 26: 164–167. [DOI] [PubMed] [Google Scholar]
- 30. Vizcaino JA, Foster JM, Martens L (2010) Proteomics data repositories: providing a safe haven for your data and acting as a springboard for further research. J Proteomics 73: 2136–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mathivanan S, Fahner CJ, Reid GE, Simpson RJ (2012) ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res 40: D1241–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mathivanan S, Simpson RJ (2009) ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 21: 4997–5000. [DOI] [PubMed] [Google Scholar]
- 33. Simpson RJ, Kalra H, Mathivanan S (2012) ExoCarta as a resource for exosomal research. Journal of Extracellular Vesicles 1: 18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maglott D, Ostell J, Pruitt KD, Tatusova T (2007) Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 35: D26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mishra GR, Mathivanan S, Kumaran K, Kannabiran N, Suresh S, et al. (2006) Human protein reference database–2006 update. Nucleic Acids Res 34: D411–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, et al. (2009) Human Protein Reference Database–2009 update. Nucleic Acids Res 37: D767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, et al. (2008) The BioGRID Interaction Database: 2008 update. Nucleic Acids Res 36: D637–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mathivanan S, Pandey A (2008) Human Proteinpedia as a resource for clinical proteomics. Mol Cell Proteomics 7: 2038–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wren JD (2008) URL decay in MEDLINE–a 4-year follow-up study. Bioinformatics 24: 1381–1385. [DOI] [PubMed] [Google Scholar]
- 40. Ducut E, Liu F, Fontelo P (2008) An update on Uniform Resource Locator (URL) decay in MEDLINE abstracts and measures for its mitigation. BMC Med Inform Decis Mak 8: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wren JD (2004) 404 not found: the stability and persistence of URLs published in MEDLINE. Bioinformatics 20: 668–672. [DOI] [PubMed] [Google Scholar]
- 42. Fevrier B, Raposo G (2004) Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol 16: 415–421. [DOI] [PubMed] [Google Scholar]
- 43. Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, et al. (1996) B lymphocytes secrete antigen-presenting vesicles. J Exp Med 183: 1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]