Abstract
Background
Toll like receptor 4 (TLR4) has been related to inflammation and beta-amyloid deposition in Alzheimer's disease (AD) brain. No study has explored the association between haplotype-tagging single nucleotide polymorphisms (htSNPs) of TLR4 and AD risk previously and ApoE e4 status alone showed low sensitivity in identifying late-onset AD (LOAD) patients.
Methods
A total of 269 LOAD patients were recruited from three hospitals in northern Taiwan (2007–2010). Controls (n = 449) were recruited from elderly health checkup and volunteers of the hospital during the same period of time. Five common (frequency≥5%) TLR4 htSNPs were selected to assess the association between TLR4 polymorphisms and the risk of LOAD in the Chinese ethnic population.
Results
Homozygosity of TLR4 rs1927907 was significantly associated with an increased risk of LOAD [TT vs. CC: adjusted odds ratio (AOR) = 2.45, 95% confidence interval (CI) = 1.30–4.64]. After stratification, the association increased further in ApoE e4 non-carriers (AOR = 3.07) and in hypertensive patients (AOR = 3.60). Haplotype GACGG was associated with a decreased risk of LOAD (1 vs. 0 copies: AOR = 0.59, 95% CI = 0.36–0.96; 2 vs. 0 copies: AOR = 0.31, 95% CI = 0.14–0.67) in ApoE e4 non-carriers. ApoE e4 status significantly modified this association (p interaction = 0.01). These associations remained significant after correction for multiple tests.
Conclusions
Sequence variants of TLR4 were associated with an increased risk of LOAD, especially in ApoE e4 non-carriers and in hypertensive patients. The combination of TLR4 rs1927907 and ApoE e4 significantly increased the screening sensitivity in identifying LOAD patients from 0.4 to 0.7.
Introduction
Alzheimer's Disease (AD) is the most common cause of dementia, and is characterized by accumulation of extracellular senile plaques (composed by amyloid-β peptide, Aβ), intracellular neurofibrillary tangles (containing hyperphosphorylated tau protein), and degenerating neurons [1]. In US elderlies (age 65 or older), the incidence of AD was 1.4% in 1995 and estimated to be 4.6% in 2050 [2]; it was also the sixth leading cause of death in 2009 [3]. In Taiwan elderlies, the prevalence of dementia is 1.7% to 4.3% [4]; however, AD is not the top 10 leading cause of death probably due to under-diagnosis and under-reported. As population aging quickly in most of developed countries, dementia has become an important health issue worldwide.
Beta amyloid (Aβ) load has been related to AD pathogenesis via its role in triggering the innate immune response. Toll like receptors (TLRs) recognize various pathogens infection and damaged host cells, which lead to the subsequent inflammation responses [5]. Toll-like receptor 4 (TLR4) expresses on the surface of microglia in the central nervous system and acts as the binding receptor of lipopolysaccharide (LPS) and Aβ [6], [7]. Aβ deposition increases the expression and activation of TLR4, which facilitates the uptake and clearance of Aβ in AD pathogenesis [8], [9], [10]. An animal study also showed that the Aβ load was greater in mutant TLR4 AD mice than that of wild-type mice [11]. These reflect that TLR4 may be an important susceptibility gene to AD via its role in innate immunity.
Two TLR4 missense polymorphisms (Asp299Gly, equivalent to rs4986790; Thr399Ile, equivalent to rs4986791) have been related to a blunted inflammatory response to LPS [12]. Asp299Gly may affect the production of pro-inflammatory cytokines [13] and has been associated with an increased risk of AD in two Italian studies [14], [15]. However, both Thr399Ile and Asp299Gly are very rare in Chinese [16] and other populations (Indonesian, Papuan, Trio Indians in Surinam) [17]. In addition, TLR4/11367, a SNP in the 3′ untranslated region (UTR), has been associated with an increased risk of AD in a Chinese population [18]. In summary, few studies have assessed the association between TLR4 polymorphisms and the risk of AD, and only 1 or 2 TLR4 SNPs were explored previously.
TLR4 plays an important role in inflammation and age-related diseases [19]. Past studies relating TLR4 polymorphisms to AD risk have been limited to few coding SNPs in TLR4 (Table S1). In addition, apolipoprotein E (ApoE) e4 is an important risk factor of AD but has low sensitivity (about 0.4) in screening late-onset (aged 65 or older) AD (LOAD) patients [20]. Therefore, this study was aimed to identify genetic marker(s) supplementary to ApoE e4 for identifying LOAD. We used a systematic approach to select TLR4 haplotype-tagging single nucleotide polymorphisms (htSNPs) and explored their associations with AD risk. Vascular risk factors [e.g., hypertension, hypercholesteremia, and type 2 diabetes mellitus (DM)] have been related to the pathogenesis of dementia [21], [22], [23]. This study further explored how these factors and ApoE e4 status modify the association above.
Materials and Methods
Study Population
This was a case-control study. A total of 294 LOAD cases were recruited from the neurology clinics of three teaching hospitals in northern Taiwan from 2007 to 2010. Healthy controls (n = 503) were recruited from elderly health checkup and volunteers of the hospital during the same period of time. All participants were Taiwanese aged 65 years or older. Participants with the history of the following diseases were excluded: depression, Parkinson's disease, hemorrhagic or ischemic stroke, cerebral infarction, or organic brain tumors.
A questionnaire was administered to collect information on demography, vascular risk factors (hypertension, hypercholesteremia, and type 2 DM), and life style (cigarette smoking and alcohol consumption, etc.). For each participant, a blood sample was collected in a tube containing sodium EDTA. After centrifugation, genomic DNA was extracted from buffy coat by QuickGene-Mini80 system (Fujifilm, Tokyo, Japan) and then stored in a −80°C freezer. After further exclusion of participants without blood samples, a total of 269 cases and 449 controls were included for data analyses.
Ethics Statement
The study protocol has been approved by the Institutional Review Boards of National Taiwan University Hospital, En Chu Kong Hospital, and Cardinal Tien Hospital. Written informed consent was obtained from each study participant. The consent from the legal guardian/next of kin was obtained when patients had serious cognitive impairment.
Dementia Evaluation
At each hospital, potential dementia cases were diagnosed by a neurologist. Mini-Mental State Examination was used to evaluate their cognitive function. The diagnosis of probable dementia was evaluated by Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition [24]. Head magnetic resonance imaging and computed tomography were taken to exclude participants with organic lesions. Diagnosis of AD was based on National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer's Disease and Related Disorders Association Alzheimer's Criteria [25]. The cognitive function of controls was assessed by using Short Portable Mental Status Questionnaire [26] to exclude participants with possible dementia.
SNP Selection and Genotyping Assay
Common (frequency≥5%) TLR4 SNPs were identified from the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov) using genotype data of Han Chinese in Beijing, China (CHB). Haploview (http://www.broadinstitute.org/haploview/haploview) was used to define haplotype block by applying the modified Gabriel algorithm [27], [28]. TagSNP program [29] was used to identify htSNPs among these common SNPs.
ApoE e4 status was determined using the assay developed by Chapman et al. [30]. Genotypes of TLR4 htSNPs were determined by Taqman Assay (Applied Biosystems Inc., CA, USA) with genotyping success rate greater than 95% for each SNP. Quality control samples were obtained from 5% of internal samples in duplicates and genotyped together with all other samples, and the concordance rate was 100%.
Statistical Analysis
For each SNP, the Hardy-Weinberg equilibrium (HWE) test was performed among controls to examine possible genotyping error and selection bias. Haplotype frequencies were estimated by utilizing the expectation-maximization algorithm. To control for the confounding effect of age, frequency matching was used to match cases and controls on age within an interval of 5 years. The conditional logistic regression models were performed to estimate SNP- and haplotype-specific odds ratios (OR) and 95% confidence intervals (CI) for LOAD adjusting for age, gender, education, and ApoE e4 status. Type I error for multiple tests was controlled by Bonferroni corrections [31].
This study further explored how ApoE e4 status and vascular risk factors (type 2 DM, hypertension, and hypercholesteremia) modified the association between TLR4 polymorphisms and the risk of LOAD by using the likelihood ratio test. Stratified analyses were performed by these vascular risk factors to assess the association between TLR4 polymorphisms and the risk of LOAD. SAS version 9.2 (SAS Institute, Cary, NC) was used for statistical analyses and all statistical tests were two-sided.
Statistical power for genetic main effect (SNPs and haplotypes) and gene-risk factor interaction are calculated by QUANTO (http://hydra.usc.edu/gxe/_vti_bin/shtml.dll/request.htm) and PGA (http://dceg.cancer.gov/bb/tools/pga) programs using 269 cases and 449 controls and a disease prevalence of 5%. For SNP analysis, with disease allele frequency of 26%, the power to detect a relative risk of 2.45 (codominant model with 2df) is around 0.94 (alpha = 0.05). For haplotype analysis, with disease haplotype frequency of 0.36, the power to detect a relative risk of 0.64 is about 78% (alpha = 0.05). Regarding gene-gene interaction, disease allele frequency of 0.26 and 0.23 for two SNPs, respectively, the power to detect an interaction relative risk of 3.4 is about 70% (alpha = 0.05). For gene-environment interaction, with disease allele frequency of 0.26, environmental exposure in the population of 0.48, the power to detect an interaction relative risk of 3.6 is about 95% (alpha = 0.05).
Results
Characteristics of Study Population
This study included 269 incident LOAD cases and 449 controls. As compared with controls, LOAD cases were older (79.8 vs. 73.2 years old); included more females (64% vs. 52%); had a lower education level (≤6 years: 51% vs. 11%), fewer with the history of hypertension (39% vs. 54%) or with hypercholesteremia (18% vs. 30%), and more ApoE e4 carriers (39% vs. 15%, Table 1). The distributions of body mass index at age 40 s, cigarette smoking, alcohol consumption, and type 2 DM were similar between LOAD cases and controls.
Table 1. Characteristics of the study population.
| Variables | LOAD N = 269 | Control N = 449 | p |
| Age (mean±SD) | 79.8±6.3 | 73.2±5.8 | <0.001* |
| Female (%) | 172 (64) | 234 (52) | 0.002* |
| Education (%) | <0.001* | ||
 6 years 6 years |
136 (51) | 51 (11) | |
| 6–12years | 91 (34) | 179 (40) | |
| >12 years | 39 (15) | 217 (48) | |
| BMI at age 40 s, kg/m2(mean±SD) | 22.6±3.1 | 22.4±2.8 | 0.43 |
| Cigarette smoking (%) | 62 (23) | 76 (17) | 0.05 |
| Alcohol consumption (%) | 32 (12) | 49 (11) | 0.69 |
| Type 2 DM (%) | 50 (17) | 63 (13) | 0.11 |
| Hypertension (%) | 104 (39) | 241 (54) | <0.001* |
| Hypercholesteremia (%) | 49 (18) | 133 (30) | 0.001* |
| ApoE e4 carriers (%) | 105 (39) | 66 (15) | <0.001* |
p value<0.05 was obtained by comparing LOAD cases and controls.
Abbreviations: LOAD, late-onset Alzheimer's disease; SD, standard deviation; BMI, body mass index; DM, diabetes mellitus; ApoE e4, apolipoprotein E e4.
TLR4 Polymorphisms and LOAD Risk
Five TLR4 htSNPs (rs1927911, rs11536879, rs1927907, rs11536889, and rs7873784) were genotyped. The minor allele frequency (MAFs) of the five SNPs among controls ranged from 11% to 41%, which were similar to the MAFs of CHB genotype data from the International HapMap Project (7% to 36%, Table 2). All TLR4 SNPs were in HWE among controls. For each SNP, the genotype frequencies were similar between cases and controls. Participants carrying two copies of variant SNP3 (rs1927907) had a significantly increased risk of LOAD [TT vs. CC: adjusted OR (AOR) = 2.45, 95% CI = 1.30–4.64, p = 0.004 Table 3]. This association remained significant after Bonferroni correction (α = 0.05/5). Significant association was also observed for SNP3 under the assumption of additive model (AOR = 1.36), which did not remain significant after Bonferroni correction.
Table 2. Characteristics of TLR4 haplotype-tagging SNPs.
| SNP name | Nucleotide change | Location | rs no. | HapMap CHB | Controls | LOAD Cases | ||
| MAF | MAF | HWE p | MAF | HWE p | ||||
| SNP1 | G→A | Intron | rs1927911 | 0.36 | 0.41 | 0.37 | 0.46 | <0.01 |
| SNP2 | A→G | Intron | rs11536879 | 0.09 | 0.13 | 0.63 | 0.14 | 0.92 |
| SNP3 | C→T | Intron | rs1927907 | 0.20 | 0.26 | 0.30 | 0.32 | <0.01 |
| SNP4 | G→C | 3′ UTR | rs11536889 | 0.22 | 0.22 | 0.41 | 0.22 | 0.89 |
| SNP5 | G→C | 3′ UTR | rs7873784 | 0.07 | 0.11 | 0.87 | 0.11 | 0.27 |
Abbreviations: UTR, untranslated region; CHB, Han Chinese in Beijing, China; HWE p, p value for Hardy–Weinberg equilibrium test; LOAD, late-onset Alzheimer's disease; MAF, minor allele frequency; SNP, single nucleotide polymorphism.
Table 3. Association between TLR4 SNPs and LOAD risk.
| Co-dominant model# | Additive model | |||||||||
| 0 copies | 1 copy | 2 copies | ||||||||
| Case/control | AOR | Case/control | AOR (95% CI) | p | Case/control | AOR (95% CI) | p | AOR (95% CI) | p | |
| SNP1 | 92/161 | 1.00 | 105/208 | 1.00 (0.65–1.54) | 0.47 | 69/80 | 1.33 (0.80–2.22) | 0.22 | 1.14 (0.88–1.47) | 0.33 |
| SNP2 | 196/335 | 1.00 | 61/100 | 1.13 (0.72–1.78) | 0.21 | 5/9 | 0.43 (0.10–1.94) | 0.24 | 0.98 (0.66–1.45) | 0.90 |
| SNP3 | 133/242 | 1.00 | 84/155 | 1.00 (0.65–1.52) | 0.05 | 43/32 | 2.45 (1.30–4.64) * | 0.004 | 1.36 (1.03–1.80) | 0.03 |
| SNP4 | 164/274 | 1.00 | 90/145 | 1.34 (0.89–2.03) | 0.46 | 13/24 | 1.16 (0.45–2.97) | 0.99 | 1.22 (0.88–1.70) | 0.24 |
| SNP5 | 206/341 | 1.00 | 48/86 | 0.97 (0.59–1.58) | 0.70 | 5/5 | 0.64 (0.11–3.75) | 0.63 | 0.93 (0.60–1.44) | 0.74 |
All models were adjusted for age, gender, education, and ApoE e4 status.
Abbreviations: LOAD, late-onset Alzheimer's disease; AOR, adjusted odds ratio; CI, confidence interval; SNP, single nucleotide polymorphism.
0 copies, wild type; 1 copy, heterozygotes; 2 copies, homozygous variants.
Numbers in bold indicates statistically significant findings (p<α = 0.05).
Additive model is assessing the association between number of variant allele and LOAD.
The result remained significant (2 copies of variant SNP3, p = 0.004) after controlling for type I error by using Bonferroni correction (α = 0.05/5).
Five common (frequency≥5%) htSNPs spanning TLR4 formed one haplotype block, which was determined by modified Gabriel et al. algorithm [27], [28] (Figure 1). Four common haplotypes with a cumulative frequency of 90% in controls were identified in TLR4; two of them were excluded from statistical analysis due to no controls carrying 2 copies of their corresponding haplotypes (data not shown). Participants carrying 1 copy of HAP1 (GACGG) had a significantly decreased risk of LOAD (AOR = 0.64, 95% CI = 0.42–0.97, Table 4) as compared with those carrying 0 copies of HAP1. This association did not reach statistical significance after Bonferroni correction. HAP3 was not associated with the risk of LOAD.
Figure 1. TLR4 linkage disequilibrium (LD) plot.
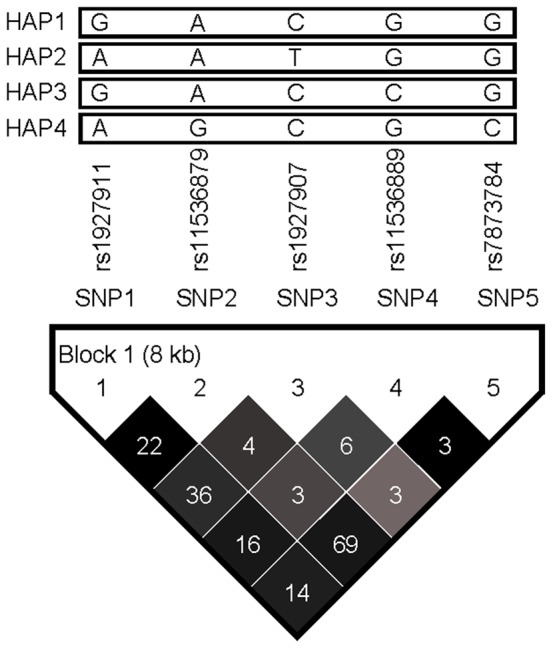
This plot was generated by Haploview program using data from this study. Five common (frequency≥0.05) htSNPs formed one block. The SNP name, e.g., SNP1, SNP2, etc., indicated five htSNPs genotyped in this study. Four common haplotypes were identified. The level of pairwise r2, which indicated the association degree between two SNPs in the LD block, was shown in the cell of the LD structure in numeric. The level of pair-wise D', which indicated the strength of LD between two SNPs, was shown in the LD structure in gray scale.
Table 4. Association between TLR4 haplotypes and LOAD.
| Prevalence in controls, % | Co-dominant modela | p interaction | ||||||
| 0 copies | 1 copy | 2 copies | ||||||
| Case/Control | AOR | Case/Control | AOR (95% CI) | Case/Control | AOR (95% CI) | |||
| HAP1 (GACGG) | 36.1 | 134/185 | 1.00 | 102/205 | 0.64 (0.42–0.97) | 33/59 | 0.60 (0.33–1.09) | NA |
| ApoE e4 carriers | ||||||||
| No | 83/155 | 1.00 | 64/173 | 0.59 (0.36–0.96) | 16/53 | 0.31 (0.14–0.67) * | 0.01 | |
| Yes | 50/29 | 1.00 | 38/31 | 0.78 (0.35–1.72) | 17/6 | 1.93 (0.54–6.82) | ||
| Hypertension | ||||||||
| No | 82/89 | 1.00 | 66/92 | 0.74 (0.41–1.32) | 17/26 | 0.57 (0.23–1.40) | 0.59 | |
| Yes | 52/95 | 1.00 | 36/113 | 0.55 (0.29–1.04) | 16/33 | 0.75 (0.32–1.73) | ||
| Hypercholesteremia | ||||||||
| No | 107/132 | 1.00 | 83/139 | 0.72 (0.45–1.17) | 27/43 | 0.68 (0.34–1.34) | 0.65 | |
| Yes | 26/51 | 1.00 | 17/66 | 0.34 (0.12–0.98) | 6/16 | 0.29 (0.07–1.25) | ||
| Type 2 DM | ||||||||
| No | 107/161 | 1.00 | 88/176 | 0.68 (0.43–1.08) | 24/49 | 0.61 (0.31–1.21) | 0.92 | |
| Yes | 26/23 | 1.00 | 14/28 | 0.43 (0.13–1.38) | 9/10 | 0.51 (0.14–1.89) | ||
| HAP3 (GACCG) | 20.3 | 177/290 | 1.00 | 82/136 | 1.36 (0.89–2.10) | 10/23 | 1.08 (0.39–2.98) | NA |
| ApoE e4 carriers | ||||||||
| No | 102/245 | 1.00 | 56/116 | 1.84 (1.11–3.06) | 5/20 | 1.46 (0.43–4.98) | 0.11 | |
| Yes | 74/44 | 1.00 | 26/19 | 0.70 (0.30–1.65) | 5/3 | 0.63 (0.11–3.55) | ||
| Hypertension | ||||||||
| No | 108/135 | 1.00 | 52/57 | 1.67 (0.91–3.09) | 5/15 | 0.95 (0.25–3.57) | 0.52 | |
| Yes | 69/154 | 1.00 | 30/79 | 1.08 (0.56–2.07) | 5/8 | 1.17 (0.24–5.84) | ||
| Hypercholesteremia | ||||||||
| No | 144/202 | 1.00 | 64/94 | 1.27 (0.78–2.09) | 9/17 | 1.17 (0.37–3.66) | 0.87 | |
| Yes | 31/86 | 1.00 | 17/42 | 1.91 (0.69–5.30) | 1/6 | 1.25 (0.10–16.30) | ||
| Type 2 DM | ||||||||
| No | 142/246 | 1.00 | 67/120 | 1.29 (0.80–2.09) | 10/19 | 1.51 (0.54–4.22) | NA | |
| Yes | 34/42 | 1.00 | 15/16 | 1.86 (0.60–5.82) | 0/4 | NA | ||
Abbreviations: LOAD, late-onset Alzheimer's disease; AOR, adjusted odds ratio; CI, confidence interval; DM, diabetes mellitus; HAP, haplotype; NA, not applicable; SNP, single nucleotide polymorphism; ApoE e4, apolipoprotein E e4.
All models were adjusted for age, gender, education, and ApoE e4 status.
Minor alleles were underlined.
Numbers in bold indicates statistically significant findings (p<α = 0.05).
0 copies, wild type; 1 copy, heterozygotes; 2 copies, homozygous variants.
The result remained significant (2 copies of HAP1, p = 0.003) after controlling for type I error by using Bonferroni correction (α = 0.05/4).
Effect Modification by ApoE e4 Status
ApoE e4 carriers was associated with a significantly increased risk of LOAD (AOR = 5.05, 95% CI = 3.20–7.97) as compared with ApoE e4 non-carriers. After stratified by ApoE e4 status, SNP3 was significantly associated with an increased LOAD risk [TT vs. CC: AOR = 3.07, 95% CI = 1.49–6.33, p = 0.004, Table 5] among ApoE e4 non-carriers, which remained statistically significant after Bonferroni correction (α = 0.05/5). However, no significant association was observed for SNP3 in ApoE e4 carriers. Similar findings were observed for SNP4 among ApoE e4 non-carriers (GC vs. GG: AOR = 1.82, 95% CI = 1.11–2.96, Table 5).
Table 5. Association between TLR4 SNPs and LOAD risk by ApoE e4 status.
| Co-dominant modela | p interaction | ||||||
| 0 copies | 1 copy | 2 copies | |||||
| Case/Control | AOR | Case/Control | AOR (95% CI) | Case/Control | AOR (95% CI) | ||
| SNP1 | |||||||
| Non-carriers | 56/138 | 1.00 | 63/181 | 1.00 (0.60–1.66) | 42/62 | 1.50 (0.81–2.78) | 0.70 |
| Carriers | 36/22 | 1.00 | 42/27 | 1.07 (0.47–2.45) | 27/17 | 1.13 (0.43–2.99) | |
| SNP2 | |||||||
| Non-carriers | 126/287 | 1.00 | 33/84 | 0.92 (0.53–1.59) | 1/6 | 0.32 (0.03–3.10) | 0.35 |
| Carriers | 70/47 | 1.00 | 28/15 | 1.75 (0.71–4.36) | 4/3 | 0.75 (0.11–5.30) | |
| SNP3 | |||||||
| Non-carriers | 75/210 | 1.00 | 54/127 | 1.24 (0.75–2.05) | 29/28 | 3.07 (1.49–6.33) * | 0.17 |
| Carriers | 58/31 | 1.00 | 30/27 | 0.54 (0.24–1.23) | 13/4 | 1.49 (0.37–5.96) | |
| SNP4 | |||||||
| Non-carriers | 94/232 | 1.00 | 61/122 | 1.82 (1.11–2.96) | 7/21 | 1.50 (0.49–4.61) | 0.06 |
| Carriers | 70/41 | 1.00 | 29/22 | 0.66 (0.29–1.50) | 6/3 | 0.63 (0.11–3.53) | |
| SNP5 | |||||||
| Non-carriers | 132/293 | 1.00 | 24/72 | 0.78 (0.43–1.44) | 1/3 | 0.63 (0.06–7.21) | 0.28 |
| Carriers | 74/47 | 1.00 | 24/13 | 1.63 (0.62–4.27) | 4/2 | 0.82 (0.08–8.16) | |
All models were adjusted for age, gender, and education.
Abbreviations: LOAD, late-onset Alzheimer's disease; AOR, adjusted odds ratio; CI, confidence interval; SNP, single nucleotide polymorphism; ApoE e4, apolipoprotein E e4.
Numbers in bold indicates statistically significant findings (p<α = 0.05).
0 copies, wild type; 1 copy, heterozygotes; 2 copies, homozygous variants.
The result remained significant (2 copies of variant SNP3 in AopE e4 non-carriers, p = 0.004) after controlling for type I error by using Bonferroni correction (α = 0.05/5).
For TLR4 haplotypes, ApoE e4 status significantly modified the association between TLR4 HAP1 and the risk of LOAD (p interaction = 0.01, Table 4). After stratified by ApoE e4 status, ApoE e4 non-carriers carrying 1 or 2 copies of HAP1 had a decreased risk of LOAD [1 vs. 0 copies: AOR = 0.59, 95% CI = 0.36–0.96; 2 vs. 0 copies: AOR = 0.31, 95% CI = 0.14–0.67, p = 0.003]. These associations remained statistically significant after Bonferroni correction (α = 0.05/4).
Effect Modification by Vascular Risk Factors
Vascular risk factors (hypertension, type 2 DM, and hypercholesteremia) did not significantly modify the association between TLR4 polymorphisms and the risk of LOAD. After stratification by these vascular risk factors, significant associations were observed in some subgroups as detailed below.
Hypertensive patients showed a decreased the risk of LOAD (AOR = 0.41, 95% CI = 0.28–0.61). After stratification, hypertensive patients carrying homozygosity of SNP3 had a significantly increased risk of LOAD (TT vs. CC: AOR = 3.60, 95% CI = 1.47–8.84, Table 6). After stratified by type 2 DM, non-DM patients carrying homozygosis SNP3 was associated with an increased LOAD risk (TT vs. CC: AOR = 2.34, 95% CI = 1.15–4.77, p = 0.002). These associations remained statistically significant after Bonferroni correction (α = 0.05/5). After stratification by hypercholesteremia, no significant association was observed (data not shown). None of the vascular risk factors significantly modified the association between TLR4 haplotypes (HAP1 and HAP3) and the risk of LOAD; stratified analyses did not show significant association in the subgroups after Bonferroni correction (Table 4).
Table 6. Association between TLR4 SNPs and LOAD risk by hypertension status.
| Co-dominant modela | p interaction | ||||||
| 0 copies | 1 copy | 2 copies | |||||
| Case/Control | AOR | Case/Control | AOR (95% CI) | Case/Control | AOR (95% CI) | ||
| SNP1 | |||||||
| No | 50/78 | 1.00 | 68/87 | 1.37 (0.74–2.53) | 44/42 | 1.15 (0.55–2.38) | 0.18 |
| Yes | 42/83 | 1.00 | 37/121 | 0.64 (0.34–1.22) | 25/37 | 1.39 (0.65–2.98) | |
| SNP2 | |||||||
| No | 117/157 | 1.00 | 40/45 | 1.30 (0.69–2.47) | 4/4 | 0.31 (0.03–3.45) | 0.76 |
| Yes | 79/178 | 1.00 | 21/54 | 0.95 (0.47–1.91) | 1/5 | 0.47 (0.05–4.59) | |
| SNP3 | |||||||
| No | 80/109 | 1.00 | 57/74 | 1.04 (0.58–1.86) | 24/15 | 1.75 (0.67–4.57) | 0.57 |
| Yes | 53/133 | 1.00 | 27/80 | 0.78 (0.40–1.54) | 19/17 | 3.60 (1.47–8.84) * | |
| SNP4 | |||||||
| No | 103/128 | 1.00 | 53/61 | 1.51 (0.84–2.72) | 7/15 | 1.07 (0.31–3.73) | 0.86 |
| Yes | 61/145 | 1.00 | 37/84 | 1.22 (0.66–2.26) | 6/9 | 1.07 (0.24–4.85) | |
| SNP5 | |||||||
| No | 123/159 | 1.00 | 33/40 | 1.17 (0.59–2.32) | 3/1 | 0.51 (0.01–22.48) | 0.59 |
| Yes | 83/182 | 1.00 | 15/45 | 0.73 (0.34–1.55) | 2/4 | 0.74 (0.07–7.70) | |
All models were adjusted for age, gender, education, and ApoE e4 status.
Abbreviations: LOAD, late-onset Alzheimer's disease; AOR, adjusted odds ratio; CI, confidence interval; SNP, single nucleotide polymorphism.
Numbers in bold indicates statistically significant findings(p<α = 0.05).
0 copies, wild type; 1 copy, heterozygotes; 2 copies, homozygous variants.
The result remained significant (2 copies of variant SNP3 in hypertensive persons, p = 0.002) after controlling for type I error by using Bonferroni correction (α = 0.05/5).
Before stratification, hypertensive patients showed a decreased the risk of LOAD (AOR = 0.41, 95% CI = 0.28–0.61).
Discussion
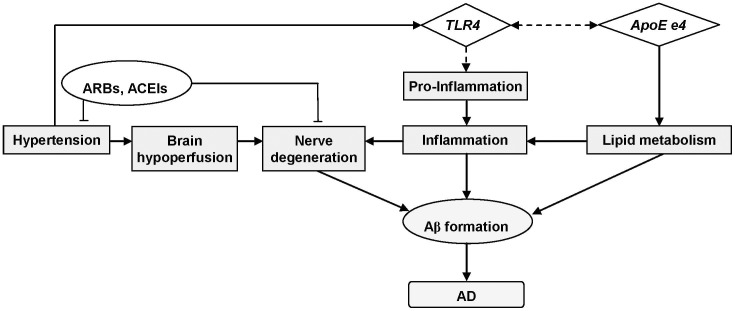
This study is the first to assess the association between five TLR4 htSNPs and LOAD risk. These htSNPs captured abundant genetic information in TLR4 gene and were representative of Chinese ethnic group. We found that homozygosity rs1927907 (SNP3) was significantly associated with an increased risk of LOAD, which has not been explored previously. rs1927907 is an intronic SNP and may affect LOAD risk via regulating the alternative splicing and the subsequent protein production [32]. Sequence variants of TLR4 may enhance the production of pro-inflammatory molecules and cytokine that leads to an increased risk of LOAD (Figure 2). Although five htSNPs are located in one linkage disequilibrium (LD) block (Figure 1), the pairwise correlations between TLR4 htSNPs were not strong (most of pairwise r2<0.40). This may explain the sole significant association for rs1927907 (SNP3) but not for other TLR4 htSNPs in the same block. Only two TLR4 SNPs (Asp299Gly and TLR4/11367) were explored for AD risk in previous studies (Table S1). However, they were not included in this study because Asp299Gly is very rare in Chinese population. As for TLR4/11367, no sufficient information (e.g., rs number or probe/primer sequence) is available for it from the only previous study [18] or the public domain for us to perform genotyping. We further stratification our findings by two important risk factors of LOAD, gender and age, and found that homozygosity of SNP3 were significantly associated with increased risk of LOAD in females and in older people (age ≦75), respectively (Table S2 and S3). This is consistent with our understanding of LOAD that is more common in people with advancing age as the accumulation of Aβ (http://www.alz.org/alzheimers_disease_causes_risk_factors.asp) and in females due to the loss of mitochondria protection as aging [33], respectively. Previous GWASs [34], [35], [36], [37], [38] and a recent meta-analysis for GWASs of AD [39] did not identify TLR4 as a susceptibility gene for AD. This may be due to different ethnic groups explored between this study (Asian) and most previous studies (Whites). In addition, a gene with true association with AD may not be selected in the exploratory stage (stage 1) of multi-stage GWAS because of its moderate effect and a marginal p value (false negative). This phenomenon has been observed by other study [40] and may also explain why TLR4 is not in the list of susceptibility gene identified from GWASs for AD.
Figure 2. Postulated pathway of TLR4 and factors involved in the pathogenesis of Alzheimer's disease.
Solid lines indicated pathways that have been well documented; dotted lines indicated speculated pathways. Abbreviations: Aβ, beta amyloid; ARBs, angiotensin receptor blockers; ACEIs, angiotensin-converting enzyme inhibitors; ApoE e4, apolipoprotein E e4; AD, Alzheimer's disease.
As for TLR4 haplotypes, 1 copy of HAP1 was associated with a decreased risk of LOAD (Table 4). HAP1 GACGG was composed of five major alleles, which may explain the protective effect of HAP1. The association between 2 copies of HAP1 and LOAD was not significant probably due to relatively small sample size.
ApoE e4 is a well-known risk factor of AD. This study found rs1927907 (SNP3) and HAP1 were significantly associated with an increased risk of LOAD in ApoE e4 non-carriers and the association remained statistically significant after correction for multiple tests by using Bonferroni correction. Both previous [20] and this study observed a low sensitivity (about 0.40) by using ApoE e4 to screen LOAD (i.e., only 40% of LOAD cases can be identified). In contrast, TLR4 SNP3 has better sensitivity (0.51). Therefore, simultaneous testing by using both ApoE e4 and TLR4 SNP3 can significantly increase the screening sensitivity of LOAD to 0.7. In addition, the population attributable risk of SNP3 is even higher than that of ApoE e4 (0.39 vs. 0.37). Therefore, TLR4 SNP3 may be used for screening LOAD together with ApoE e4.
Hypertension is associated with brain-hypoperfusion and neurodegeneration, which are associated with an increased AD risk in some studies [21], [22], [23]. In contrast, some studies reported that anti-hypertension drugs (e.g., angiotensin receptor blockers or angiotensin-converting enzyme inhibitors) may provide neuroprotective effects, e.g., reduce neuronal damage and slow progression of AD [41], [42]. This may explain the protective effect of hypertension on LOAD risk in our study. However, the protective effect no longer exists when a hypertensive patient carrying homozygosity of rs1927907 (SNP3). It is possible that the protective effect from hypertensive medications is not strong enough to offset the elevated AD risk from rs1927907 (SNP3) polymorphisms and hypertension itself. Alternatively, hypertension may lead to the injury of tissue wall, which exposed the cardiovascular system to pathogens and danger signals (endogenous ligand) and then activate TLRs [43]. Elevated expression of TLR4 has been observed in AD patients and has been associated with Aβ deposition [9]. Furthermore, a recent study found that angiotensin-converting enzyme inhibitors in therapeutic dose have no effect on TLR4 expression [44]. In summary, these may explain the increased AD risk in hypertensive patients carrying variant TLR4 via elevated inflammation responses (Figure 2).
This study had some strength. First, five TLR4 htSNPs were identified for Chinese population to predict LOAD risk, especially among ApoE e4 “non-carriers”. Clinically, this is helpful to identify more people with elevated risk of LOAD by simultaneously using TLR4 SNP3 and ApoE e4. Second, the selection of five representative htSNPs captured abundant genetic information in TLR4 gene (r2 = 0.95) as compared with the limited genetic information captured by the single TLR4 SNP (Asp299Gly, r2<0.01) in two White studies [13], [14]. Importantly, the associations between TLR4 polymorphisms (rs1927907 and HAP1 GACGG) and LOAD risk remained significant after Bonferroni correction. Therefore, these significant findings are unlikely due to chance. In addition, high false positive rate, which prevents identifying SNPs associated with the outcome because of their moderate p values in the exploratory stage, has been observed in genome-wide association studies [40]. This study used a systematic approach to select TLR4 htSNPs and may thus resolve this issue. Furthermore, brain imaging helped us to exclude other diseases with similar presentation as AD.
This study had some limitations. A self-report questionnaire was used to collect information on vascular risk factors (e.g., hypertension, hypercholesteremia, and type 2 DM). Because these diseases/conditions are major health issues, participants' recall of disease/condition diagnosis and their awareness of these diseases/conditions tend to be accurate [45], [46], [47]. Therefore, information bias should not be a concern. In addition, cases were older than controls, which may lead to overestimate our findings. To solve this issue, frequency matching on age with a 5-year interval was used to compare cases with controls within the same age stratum.
This study, for the first time, found a strong association between TLR4 polymorphisms (rs1927907 and HAP1 GACGG) and LOAD risk, especially among ApoE e4 “non-carriers”. These associations remained significant after correction for multiple tests. The majority (about 60%) of AD patients was ApoE e4 non-carriers and the sensitivity of ApoE e4 has been low for LOAD. Therefore, the combination of ApoE 4e and TLR4 rs1927907 can significantly increase the sensitivity from 40% (use ApoE e4 alone) to 70%. Future large studies are warranted to explore the role of TLR4 polymorphisms with expression data and levels of pro-inflammatory mediators on the risk of LOAD in multi-ethnic groups.
Supporting Information
Previous studies relating TLR4 polymorphisms to AD risk.
(DOCX)
Association between TLR4 SNPs and LOAD risk by gender.
(DOCX)
Association between TLR4 SNPs and LOAD risk by age status.
(DOCX)
Acknowledgments
The authors gratefully acknowledge Dr. Wen-Chung Lee for statistical consultation.
Funding Statement
Funding for the study was provided by National Science Council grants 96-2314-B-002-197 and 97-2314-B-002-168-MY3. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Querfurth HW, LaFerla FM (2010) Alzheimer's disease. N Engl J Med 362: 329–344. [DOI] [PubMed] [Google Scholar]
- 2. Hebert LE, Beckett LA, Scherr PA, Evans DA (2001) Annual incidence of Alzheimer disease in the United States projected to the years 2000 through 2050. Alzheimer Dis Assoc Disord 15: 169–173. [DOI] [PubMed] [Google Scholar]
- 3. Kochanek KD, Xu J, Murphy SL, Minino AM, Kung HC (2011) Deaths: Preliminary Data for 2009. National Vital Statistics Reports 59: 1–51. [PubMed] [Google Scholar]
- 4. Fuh JL, Wang SJ (2008) Dementia in Taiwan: past, present, and future. Acta Neurol Taiwan 17: 153–161. [PubMed] [Google Scholar]
- 5. Akira S, Uematsu S, Takeuchi O (2006) Pathogen recognition and innate immunity. Cell 124: 783–801. [DOI] [PubMed] [Google Scholar]
- 6. Streit WJ (2004) Microglia and Alzheimer's disease pathogenesis. J Neurosci Res 77: 1–8. [DOI] [PubMed] [Google Scholar]
- 7. Streit WJ, Miller KR, Lopes KO, Njie E (2008) Microglial degeneration in the aging brain–bad news for neurons? Front Biosci 13: 3423–3438. [DOI] [PubMed] [Google Scholar]
- 8. Tahara K, Kim HD, Jin JJ, Maxwell JA, Li L, et al. (2006) Role of toll-like receptor signalling in Abeta uptake and clearance. Brain 129: 3006–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walter S, Letiembre M, Liu Y, Heine H, Penke B, et al. (2007) Role of the toll-like receptor 4 in neuroinflammation in Alzheimer's disease. Cell Physiol Biochem 20: 947–956. [DOI] [PubMed] [Google Scholar]
- 10. Udan ML, Ajit D, Crouse NR, Nichols MR (2008) Toll-like receptors 2 and 4 mediate Abeta(1–42) activation of the innate immune response in a human monocytic cell line. J Neurochem 104: 524–533. [DOI] [PubMed] [Google Scholar]
- 11. Jin JJ, Kim HD, Maxwell JA, Li L, Fukuchi K (2008) Toll-like receptor 4-dependent upregulation of cytokines in a transgenic mouse model of Alzheimer's disease. J Neuroinflammation 5: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arbour NC, Lorenz E, Schutte BC, Zabner J, Kline JN, et al. (2000) TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet 25: 187–191. [DOI] [PubMed] [Google Scholar]
- 13. Balistreri CR, Caruso C, Listi F, Colonna-Romano G, Lio D, et al. (2011) LPS-mediated production of pro/anti-inflammatory cytokines and eicosanoids in whole blood samples: biological effects of +896A/G TLR4 polymorphism in a Sicilian population of healthy subjects. Mech Ageing Dev 132: 86–92. [DOI] [PubMed] [Google Scholar]
- 14. Minoretti P, Gazzaruso C, Vito CD, Emanuele E, Bianchi M, et al. (2006) Effect of the functional toll-like receptor 4 Asp299Gly polymorphism on susceptibility to late-onset Alzheimer's disease. Neurosci Lett 391: 147–149. [DOI] [PubMed] [Google Scholar]
- 15. Balistreri CR, Grimaldi MP, Chiappelli M, Licastro F, Castiglia L, et al. (2008) Association between the polymorphisms of TLR4 and CD14 genes and Alzheimer's disease. Curr Pharm Des 14: 2672–2677. [DOI] [PubMed] [Google Scholar]
- 16. Hang J, Zhou W, Zhang H, Sun B, Dai H, et al. (2004) TLR4 Asp299Gly and Thr399Ile polymorphisms are very rare in the Chinese population. J Endotoxin Res 10: 238–240. [DOI] [PubMed] [Google Scholar]
- 17. Ferwerda B, McCall MB, Alonso S, Giamarellos-Bourboulis EJ, Mouktaroudi M, et al. (2007) TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc Natl Acad Sci U S A 104: 16645–16650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang LZ, Yu JT, Miao D, Wu ZC, Zong Y, et al. (2011) Genetic association of TLR4/11367 polymorphism with late-onset Alzheimer's disease in a Han Chinese population. Brain Res 1381: 202–207. [DOI] [PubMed] [Google Scholar]
- 19. Balistreri CR, Colonna-Romano G, Lio D, Candore G, Caruso C (2009) TLR4 polymorphisms and ageing: implications for the pathophysiology of age-related diseases. J Clin Immunol 29: 406–415. [DOI] [PubMed] [Google Scholar]
- 20. Wang M, Jia J (2010) The interleukin-6 gene −572C/G promoter polymorphism modifies Alzheimer's risk in APOE epsilon 4 carriers. Neurosci Lett 482: 260–263. [DOI] [PubMed] [Google Scholar]
- 21. Roman GC (2005) Vascular dementia prevention: a risk factor analysis. Cerebrovasc Dis 20 Suppl 2: 91–100. [DOI] [PubMed] [Google Scholar]
- 22. Stozicka Z, Zilka N, Novak M (2007) Risk and protective factors for sporadic Alzheimer's disease. Acta Virol 51: 205–222. [PubMed] [Google Scholar]
- 23. Fillit H, Nash DT, Rundek T, Zuckerman A (2008) Cardiovascular risk factors and dementia. Am J Geriatr Pharmacother 6: 100–118. [DOI] [PubMed] [Google Scholar]
- 24. American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders. [Google Scholar]
- 25. McKhann G, Drachman D, Folstein M, Katzman R, Price D, et al. (1984) Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 34: 939–944. [DOI] [PubMed] [Google Scholar]
- 26. Pfeiffer E (1975) A short portable mental status questionnaire for the assessment of organic brain deficit in elderly patients. J Am Geriatr Soc 23: 433–441. [DOI] [PubMed] [Google Scholar]
- 27. Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. (2002) The structure of haplotype blocks in the human genome. Science 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- 28. Chen YC, Giovannucci E, Lazarus R, Kraft P, Ketkar S, et al. (2005) Sequence variants of Toll-like receptor 4 and susceptibility to prostate cancer. Cancer Res 65: 11771–11778. [DOI] [PubMed] [Google Scholar]
- 29. Stram DO, Leigh Pearce C, Bretsky P, Freedman M, Hirschhorn JN, et al. (2003) Modeling and E-M estimation of haplotype-specific relative risks from genotype data for a case-control study of unrelated individuals. Hum Hered 55: 179–190. [DOI] [PubMed] [Google Scholar]
- 30. Chapman J, Estupinan J, Asherov A, Goldfarb LG (1996) A simple and efficient method for apolipoprotein E genotype determination. Neurology 46: 1484–1485. [DOI] [PubMed] [Google Scholar]
- 31. Holm S (1979) A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics 6: 65–70. [Google Scholar]
- 32. Liao PY, Lee KH (2010) From SNPs to functional polymorphism: The insight into biotechnology applications. Biochem Eng J 49: 149–158. [Google Scholar]
- 33. Vina J, Lloret A (2010) Why women have more Alzheimer's disease than men: gender and mitochondrial toxicity of amyloid-beta peptide. J Alzheimers Dis 20 Suppl 2: S527–533. [DOI] [PubMed] [Google Scholar]
- 34. Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, et al. (2008) A genome-wide association study for late-onset Alzheimer's disease using DNA pooling. BMC Med Genomics 1: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lambert JC, Heath S, Even G, Campion D, Sleegers K, et al. (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nature Genetics [DOI] [PubMed] [Google Scholar]
- 36. Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, et al. (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nature Genetics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, et al. (2010) Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303: 1832–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beecham GW, Martin ER, Li YJ, Slifer MA, Gilbert JR, et al. (2009) Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. Am J Hum Genet 84: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kamboh MI, Demirci FY, Wang X, Minster RL, Carrasquillo MM, et al. (2012) Genome-wide association study of Alzheimer's disease. Transl Psychiatry 2: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zheng SL, Liu W, Wiklund F, Dimitrov L, Balter K, et al. (2006) A comprehensive association study for genes in inflammation pathway provides support for their roles in prostate cancer risk in the CAPS study. Prostate 66: 1556–1564. [DOI] [PubMed] [Google Scholar]
- 41. Hajjar IM, Keown M, Lewis P, Almor A (2008) Angiotensin converting enzyme inhibitors and cognitive and functional decline in patients with Alzheimer's disease: an observational study. Am J Alzheimers Dis Other Demen 23: 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li NC, Lee A, Whitmer RA, Kivipelto M, Lawler E, et al. (2010) Use of angiotensin receptor blockers and risk of dementia in a predominantly male population: prospective cohort analysis. BMJ 340: b5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bomfim GF, Dos Santos RA, Oliveira MA, Giachini FR, Akamine EH, et al. (2012) Toll-like receptor 4 contributes to blood pressure regulation and vascular contraction in spontaneously hypertensive rats. Clin Sci (Lond) 122: 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eissler R, Schmaderer C, Rusai K, Kuhne L, Sollinger D, et al. (2011) Hypertension augments cardiac Toll-like receptor 4 expression and activity. Hypertens Res 34: 551–558. [DOI] [PubMed] [Google Scholar]
- 45. St Sauver JL, Hagen PT, Cha SS, Bagniewski SM, Mandrekar JN, et al. (2005) Agreement between patient reports of cardiovascular disease and patient medical records. Mayo Clin Proc 80: 203–210. [DOI] [PubMed] [Google Scholar]
- 46. Okura Y, Urban LH, Mahoney DW, Jacobsen SJ, Rodeheffer RJ (2004) Agreement between self-report questionnaires and medical record data was substantial for diabetes, hypertension, myocardial infarction and stroke but not for heart failure. J Clin Epidemiol 57: 1096–1103. [DOI] [PubMed] [Google Scholar]
- 47. El Fakiri F, Bruijnzeels MA, Hoes AW (2007) No evidence for marked ethnic differences in accuracy of self-reported diabetes, hypertension, and hypercholesterolemia. J Clin Epidemiol 60: 1271–1279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Previous studies relating TLR4 polymorphisms to AD risk.
(DOCX)
Association between TLR4 SNPs and LOAD risk by gender.
(DOCX)
Association between TLR4 SNPs and LOAD risk by age status.
(DOCX)