Abstract
Objective
This study aimed to explore the influence of SLC22A1, PXR, ABCG2, ABCB1 and CYP3A5*3 genetic polymorphisms on imatinib mesylate (IM) pharmacokinetics in Asian patients with chronic myeloid leukemia (CML).
Patients and Methods
Healthy subjects belonging to three Asian populations (Chinese, Malay, Indian; n = 70 each) and CML patients (n = 38) were enrolled in a prospective pharmacogenetics study. Imatinib trough (C0h) and clearance (CL) were determined in the patients at steady state. Haplowalk method was applied to infer the haplotypes and generalized linear model (GLM) to estimate haplotypic effects on IM pharmacokinetics. Association of haplotype copy numbers with IM pharmacokinetics was defined by Mann-Whitney U test.
Results
Global haplotype score statistics revealed a SLC22A1 sub-haplotypic region encompassing three polymorphisms (rs3798168, rs628031 and IVS7+850C>T), to be significantly associated with IM clearance (p = 0.013). Haplotype-specific GLM estimated that the haplotypes AGT and CGC were both associated with 22% decrease in clearance compared to CAC [CL (*10−2 L/hr/mg): CAC vs AGT: 4.03 vs 3.16, p = 0.017; CAC vs CGC: 4.03 vs 3.15, p = 0.017]. Patients harboring 2 copies of AGT or CGC haplotypes had 33.4% lower clearance and 50% higher C0h than patients carrying 0 or 1 copy [CL (*10−2 L/hr/mg): 2.19 vs 3.29, p = 0.026; C0h (*10−6 1/ml): 4.76 vs 3.17, p = 0.013, respectively]. Further subgroup analysis revealed SLC22A1 and ABCB1 haplotypic combinations to be significantly associated with clearance and C0h (p = 0.002 and 0.009, respectively).
Conclusion
This exploratory study suggests that SLC22A1-ABCB1 haplotypes may influence IM pharmacokinetics in Asian CML patients.
Introduction
Chronic myeloid leukemia (CML) is a hematopoietic stem cell disorder, characterized by the presence of the Philadelphia chromosome (Ph) that results from a balanced reciprocal translocation between chromosomes 9 and 22. Functionally, this translocation results in the formation of the BCR-ABL gene which is then translated to the BCR-ABL protein with intrinsic tyrosine kinase activity that is critical to the development of CML [1]. Imatinib mesylate (IM), a selective inhibitor of the BCR-ABL tyrosine kinase, has been established as the first-line treatment option for CML since the remarkable success of the IRIS (International Randomized Study of Interferon vs. STI571) trial [2], which demonstrated the dramatic superiority of IM over interferon plus cytarabine - at 18 months, the rate of complete cytogenetic response (CCyR) in patients treated with IM was a remarkable 76% versus 15% for patients treated with interferon plus cytarabine [2].
However, despite significant progress in CML treatment, about 30–40% of the patients still fail to achieve major molecular response (MMR) at 18 months [3]. This is clinically important, since it is now recognized that there is an intimate link between early molecular response and long-term clinical outcome. Recently, an expanded 7 year follow-up study of the IRIS trial found that patients who achieved MMR by 18 months enjoyed remarkably durable responses, with no disease progression and 95% event-free survival at 7 years. The probability of loss of CCyR by 7 years was only 3% for patients in MMR at 18 months compared to 26% for patients with CCyR but not MMR [4]. In the words of the IRIS investigators, achieving an MMR may be a “safe haven that promises” favorable long-term outcomes in patients with CML [4].
Suboptimal response and eventual treatment failure may be associated with several factors including the presence of BCR-ABL mutations [5] as well as those associated with suboptimal therapeutic drug levels. These include poor medication compliance [6], drug-drug interactions [7], variable metabolizing enzyme activities [8], as well as different efflux and influx transporter activities [9]. Notably, higher plasma IM trough levels have recently been correlated with achievement of CCyR and MMR in Caucasians, with an optimum threshold approximately above 1000 ng/mL [3], [10]. These results have been replicated in studies on the Chinese [11], Japanese [12], Koreans [13], Israelis [14] and Jordanians [15]. However, there are some other studies which did not agree with this finding [16]–[18]. Although there may be good efficacy in escalating the dose of IM above the “standard” 400 mg in clinical practice so as to achieve therapeutic drug levels in vivo [19], the high inter-ethnic and inter-individual variability in IM plasma trough levels [3], [10], together with the fact that a significant proportion of patients with IM levels above 1000 ng/mL still do not achieve optimal response, suggest that there are other critical factors that limit IM efficacy.
One plausible hypothesis to explain the wide variability may be a result of disparities in population pharmacokinetics and pharmacogenetics. Differences in activity, expression levels as well as functional single nucleotide polymorphisms (SNPs) of both efflux transporters (ATP-binding cassette transporters, such as ABCB1 and ABCG2) and uptake transporters (solute carriers, such as hOCT-1 and OATP1A2) [20] of IM into leukemic cells have been suggested. Particularly, White et al. showed that in patients on IM doses less than 600 mg/day, 82% of those with low activity of hOCT-1, the major active influx pump for IM, failed to achieve MMR by 18 months, compared to only 17% of those with high hOCT-1 activity. Furthermore, the negative impact of low hOCT-1 activity may be overcome by escalating to high dose IM at 800 mg/day [9]. On the other hand, several studies focusing on the impact of SLC22A1 (encoding for hOCT-1) expression or polymorphisms on IM pharmacokinetics have revealed rather heterogeneous results. High expression levels have been associated with favorable response in some studies [6], [21], [22] but not others [9], [23]. An association between SLC22A1 SNPs and response to IM has also been observed in some reports [12], [24]–[26], whereas other studies failed to support such findings [27], [28].
To address these issues, we investigated if differences in SNP frequencies of efflux and influx transporters, as well as drug metabolizing enzymes, would be able to account for inter-ethnic and inter-individual variation in IM trough levels. In this exploratory study, the effects of 89 SLC22A1 SNPs identified on whole gene sequencing, as well as 11 candidate SNPs from ABCB1, ABCG2, CYP3A5 and PXR genes on IM pharmacokinetics were examined in a cohort of healthy subjects belonging to Asian populations from Singapore, consisting of Chinese, Malay and Indian ethnic groups as well as patients with Ph+ chronic phase CML.
Patients, Materials, and Methods
Ethics Statement
All participants provided written informed consent for the study, which was approved by the ethics review committee of the National Cancer Centre, Singapore.
Patients and Healthy Participants
A total of 210 healthy subjects belonging to Asian populations from Singapore, consisting of Chinese, Malay and Indian ethnic groups (n = 70 per group) were enrolled for this study. In addition, a total of 38 patients with Ph+ chronic phase CML (32 Chinese, 4 Malays, 2 Indians) who received IM at the Singapore General Hospital from January 2001 to September 2007 were prospectively recruited. Ethnicity of the subjects was confirmed by careful screening and verified against their National Registry Identification Cards. Prior to commencing therapy, all patients were required to have a complete blood count including the white cell count differential, as well as standard biochemistry. Baseline investigations also included bone marrow evaluation for morphology, conventional cytogenetic analysis by G-banding, and BCR-ABL fluorescence in situ hybridization (FISH) studies. At least 20 metaphases were assessed for cytogenetic analysis. Peripheral blood BCR-ABL/total ABL ratio was obtained by using quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR). Patient characteristics are summarized in Table 1.
Table 1. Demographic and clinicopathological information of patients with CML.
| Characteristics | n (%) |
| Total | 38 (100) |
| Ethnicity | |
| Chinese | 32 (84.2) |
| Malay | 4 (10.5) |
| Indian | 2 (5.3) |
| Gender | |
| Male | 24 (63.2) |
| Female | 14 (36.8) |
| Prior Interferon | |
| No | 23 (60.5) |
| Yes | 15 (39.5) |
| Imatinib dosage (mg) | |
| 400 | 32 (84.2) |
| 600 | 4 (10.5) |
| 300 | 2 (5.3) |
| Treatment response | |
| CCyR 6 months | |
| No | 13 (34.2) |
| Yes | 24 (63.2) |
| Unknown | 1 (2.6) |
| CCyR 12 months | |
| No | 6 (15.8) |
| Yes | 30 (78.9) |
| Unknown | 2 (5.3) |
| MMR 18 months | |
| No | 14 (36.8) |
| Yes | 14 (36.8) |
| Unknown | 10 (26.3) |
| Treatment failure | |
| No | 33 (86.8) |
| Yes | 5 (13.2) |
| Physical variables | Median (range) |
| Age (years) | 48 (23–79) |
| Body weight (kg) | 67.5 (47–135.5) |
| Height (cm) | 165.5 (145–180) |
| BMI (kg/m2) | 24.4 (17.9–49.5) |
| BSA (m2) | 1.8 (1.4–2.5) |
Abbreviations: CCyR, complete cytogenetic response; MMR, major molecular response; BMI, body mass index; BSA, body surface area.
Patient Monitoring and Response to Therapy
Patients were monitored regularly for treatment response on an outpatient basis. A complete hematological response was defined as normalization of peripheral blood count and disappearance of splenomegaly, if present, within 3 months. To evaluate for cytogenetic response, a repeat bone marrow biopsy was performed 6 monthly after treatment with IM. Cytogenetic response was defined using standard convention with respect to the percentage of Ph-positive cells in the bone marrow as complete (CCyR, 0%), partial (PCyR, 1–34%) and minor (35–90%). A <1% BCR-ABL/total ABL ratio obtained from RT-PCR was also taken to indicate CCyR [4]. A major cytogenetic response (MCyR) was defined as the sum of complete and partial cytogenetic responses. The quantification of peripheral blood BCR-ABL/total ABL ratio was repeated every 3 months after treatment with IM. According to the definition of the international scale [29], major molecular response (MMR) was defined as a ≤0.1% BCR-ABL/total ABL ratio. Treatment failure was defined as (1) inability to achieve any CyR at 6 months, (2) less than PCyR at 12 months or less than CCyR at 18 months, (3) disease relapse as indicated by a loss of CCyR or a confirmed one log increase of BCR-ABL/total ABL ratio, as well as (4) transformation to accelerated phase disease or blast crisis. In cases of treatment failure, tyrosine kinase domain mutation tests were indicated.
Pharmacogenetic Analysis
Genomic DNA was extracted from 3 ml of peripheral blood samples using the Gentra® Puregene® extraction kit (Qiagen Inc, Minneapolis, MN, USA) and stored at −80°C until analyzed. Pharmacogenetic analyses involved screening and genotyping of polymorphic variants in the entire SLC22A1 gene (UCSC RefSeq: NM_003057), spanning 40 kilobases, including 2 kilobases upstream and 1.5 kilobases downstream, on chromosome 6. Eleven candidate functional polymorphisms in ABCB1, ABCG2, CYP3A5 and PXR genes which were described in our previous publications [30]–[33] were also investigated. CYP3A4 polymorphisms were observed very rarely in Asian population in our previous studies [30], [34] and therefore not included in the present study. Data of healthy subjects from these publications is shown in Table S1. The PCR conditions employed are listed in Table S2. PCR amplicons were treated with Exonuclease I and Shrimp Alkaline Phosphatase before direct sequencing using the Applied Biosystems 3730 DNA Analyzer (Applied Biosystems Inc, CA, USA).
Pharmacokinetic Analysis
Peripheral blood samples were taken from the patients 24 hours following their last IM dose. Steady state plasma trough concentrations of IM (C0h) were estimated via a validated high performance liquid chromatography (HPLC) method that was modified from a previously published technique [35]. Briefly, after mixing with 100 ng of 4-hydroxy-benzophenone as an internal standard [36], 50 µL plasma was deproteinized with 100 µL of methanol. After vortex-mixing, the mixture was centrifuged at 10,000× g at 4°C for 10 min, and 20 µl of the supernatant was subject to HPLC analysis. Chromatographic separation was accomplished using a C8 column (150 mm×4.6 mm I.D., 5 µm XTerra RP-8, Waters, USA) with a mobile phase consisting of 20 mM potassium dihydrogen phosphate:acetonitrile (72∶28, v/v). The lower limit of quantitation was 7.8 ng/mL. The calibration curve was linear over a concentration range of 7.8–8000 ng/mL. The within-day and between-day coefficients of variation were less than 10%. Imatinib clearance (CL) was calculated by the formula IM dosage/(dosing interval*C0h), whereby the IM dosage ranged from 300–600 mg and dosing interval was 24 hours. The final parameters used in the analyses on C0h and CL were normalized by IM dosage.
Statistical Analysis
The Hardy-Weinberg equilibrium between the genotypes was assessed using Fisher’s exact test. The Chi-square test was employed to assess the differences in genotype and allele distributions among the different groups in healthy subjects. Linkage disequilibrium (LD) analysis and LD block constructions were carried out using the software Haploview ver. 4.2 (Broad Institute, MA, USA) and quantified by |D′| and rho-square (r2) values. Tag-SNPs were identified by using Tagger program in Haploview software. Haplowalk method [37], [38] was applied to infer the haplotypes and generalized linear model (GLM) to estimate the individual haplotypic effect on IM pharmacokinetics. Haplotype specific GLM (haplo.glm) method is available under “haplostats” package in R-software to analyze the influence of haplotypes on a given trait (binary or quantitative) in unrelated subjects where haplotypes are often ambiguous because of unknown linkage phase of the measured sites along a chromosome. To assign the haplotypes copy number for each patient, haplotype phasing was conducted using PLINK 1.02 v. Association of genotype and haplotype copy numbers with IM pharmacokinetics was evaluated by Mann-Whitney U or Kruskal-Wallis tests, as indicated. Based on our results, we assessed the combinatorial effect of SLC22A1 and ABCB1 haplotype profiles on the prediction of IM pharmacokinetics. Imatinib trough level association with clinico-demographic characteristics and response parameters was evaluated with Mann-Whitney U test for categorical data and Kendall Tau correlation test for continuous data. All tests were performed using SPSS statistics software ver. 18 (IBM, Chicago, IL, USA). All statistical evaluations were made assuming a two-sided test with significance level of 0.05 unless otherwise stated.
Results
Patient Demographics
The median age at diagnosis and BMI of the patients were 42 years (range: 16–69) and 24.4 kgm−2 (range: 17.9–49.5), respectively. All patients received IM for at least 18 months, starting at a daily dose of 400 mg. At the point of recruitment, 4 patients were on an escalated daily dose of 600 mg as they could not achieve major molecular response, and 2 were on an attenuated dose of 300 mg due to toxicity issues. Median follow-up time from the start of IM was 101.6 months (range 25.6–132.7). All patients achieved complete hematological response within 3 months of IM treatment. At 6 months, one patient had no CyR and was considered to have failed treatment, whereas the remainder had achieved minor CyR (n = 2), PCyR (n = 10) or CCyR (n = 24), or remained unknown (n = 1). At 12 months, no patients were identified to have achieved less than PCyR except for the one who had failed treatment earlier. At 18 months, all patients achieved CCyR except for 3 who were considered to have failed treatment. Of 28 patients with available data for molecular response at 18 months, 14 patients did not achieve MMR and were considered to have suboptimal response to IM treatment. In the course of follow-up post-IM therapy, one patient eventually relapsed at 56.5 months whilst another patient succumbed to blast crisis at 90.7 months. Upon further analysis, non-synonymous mutations were detected in the BCR-ABL kinase domain in these two patients - a double mutation at loci G250E and E255V in the former patient and a single mutation at Y253H in the latter patient. No mutations were detected in the remaining three patients. Overall, a total of five patients (13.2%) were considered to have failed treatment.
The median time from diagnosis to IM plasma level testing was 71.5 months, and the median time from the commencement of IM therapy to plasma level testing was 58.4 months. Trough IM plasma levels were highly variable, ranging from 625 to 5271 ng/mL, with mean (±SD) and median levels of 1959 (±1073) and 1719 ng/mL, respectively. For patients on the standard IM dose of 400 mg (n = 32), the mean (±SD) and median levels were 1899 (±1054) and 1395 ng/mL, respectively. Patients on the escalated dose of 600 mg (n = 4) had higher mean (±SD) and median levels of 2785 (±1093) and 2384 ng/mL, respectively, although this was not statistically significant. Notably, we observed that out of 34 patients on ≤400 mg dose of IM, 31 (94%) had an IM trough concentration of >1000 ng/mL (range: 679–5272 ng/mL) (Table 2). None of the patients with an IM trough of <1000 ng/mL failed treatment. The IM trough levels of the 5 patients who failed treatment were 2873, 1372, 1162, 1177 and 2633 ng/mL, respectively. IM trough levels were not significantly associated with patient clinico-demographic characteristics such as body weight, body mass index, body surface area, gender and age, nor with any of the response parameters (Table 2).
Table 2. Clinical correlates with imatinib trough in patients on 400 mg standard dose.
| Characteristics | n | IM Trough (ng/mL) | SLC22A1 haplotype profile | ||||
| Mean±SD | Median (range) | p-valuea | Shigh | Slow | p-valueb | ||
| Total | 32 | 1899±1054 | 1395 (678–5272) | – | – | – | |
| Gender | – | – | – | ||||
| Male | 19 | 1944±1137 | 1417 (678–5272) | 0.863 | 12 | 7 | 1.000 |
| Female | 13 | 1833±959 | 1365 (714–4219) | 8 | 5 | ||
| Treatment response | |||||||
| CCyR 6 months | |||||||
| No | 11 | 2017±800 | 1881 (1108–3278) | 0.505 | 8 | 3 | 0.466 |
| Yes | 19 | 1896±1226 | 1372 (678–5272) | 11 | 8 | ||
| CCyR 12 months | |||||||
| No | 4 | 2289±964 | 2359 (1161–3278) | 0.312 | 4 | 0 | 0.268 |
| Yes | 25 | 1857±1111 | 1372 (678–5272) | 15 | 10 | ||
| MMR 18 months | |||||||
| No | 11 | 1828±936 | 1506 (625–4219) | 0.260 | 7 | 4 | 0.080 |
| Yes | 10 | 1974±1375 | 1364 (678–4272) | 2 | 8 | ||
| Treatment failure | |||||||
| No | 28 | 1935±1090 | 1506 (678–5272) | 0.894 | 16 | 12 | 0.265 |
| Yes | 3 | 1802±933 | 1372 (1161–2873) | 3 | 0 | ||
| Continuous variables | Mean±SD | Median (range) | p –valuec | ||||
| Age (years) | 49±12 | 48 (23–79) | 0.486 | – | – | – | |
| Body weight (kg) | 67.5±15.6 | 65.5 (47–135.5) | 0.838 | – | – | – | |
| Height (cm) | 164.2±7.8 | 165 (145–180) | 0.183 | – | – | – | |
| BMI (kg/m2) | 25.1±5.7 | 24.4 (17.9–49.5) | 0.285 | – | – | – | |
| BSA (m2) | 1.8±0.2 | 1.7 (1.4–2.5) | 0.769 | – | – | – | |
Associations of clinico-demographic characteristics and response parameters with imatinib trough and SLC22A1 haplotypes were evaluated statistically. Abbreviations: CCyR, complete cytogenetic response; MMR, major molecular response; BMI, body mass index; BSA, body surface area.
p-values were calculated using aMann-Whitney, bFisher exact and cKendall Tau correlation tests, respectively.
SLC22A1 Polymorphisms in Healthy Asians
Complete screening of the SLC22A1 gene was first performed on healthy subjects of Chinese, Malay and Indian ethnicity. Table S1 summarizes the genotypic and allelic frequencies in healthy Asian subjects. A total of 89 SNPs were identified whereby 4 were located in the 5′-upstream region, 7 in the exonic region and 78 in the intronic region. Fig. S1 illustrates the SLC22A1 gene structure and locations of these SNPs. Fisher’s exact test showed that SLC22A1 genotype frequencies at all loci conformed to Hardy-Weinberg equilibrium except for a single intronic SNP IVS1+556G>A (rs62440864) in the Malay population (p<0.001). Of these 89 SNPs, 20 were found to be monomorphic in Chinese, 9 in Malays and 11 in Indians. Six SNPS [IVS2-1010T>G (rs614890), 1222A>G (rs628031), IVS8+4215T>C (rs654993), IVS8+5402T>C, IVS8-2964C>A (rs622342), and IVS8-1295C>A (rs650284)] were found to be highly polymorphic in all ethnic groups (variant allele frequency range, 52% to 85%). Statistically significant interethnic differences in the genotypic distributions among three ethnic groups were detected for 34% (n = 30) of the polymorphisms (Table S1, p<0.016).
LD and Tag-SNPs Analysis in Healthy Asians
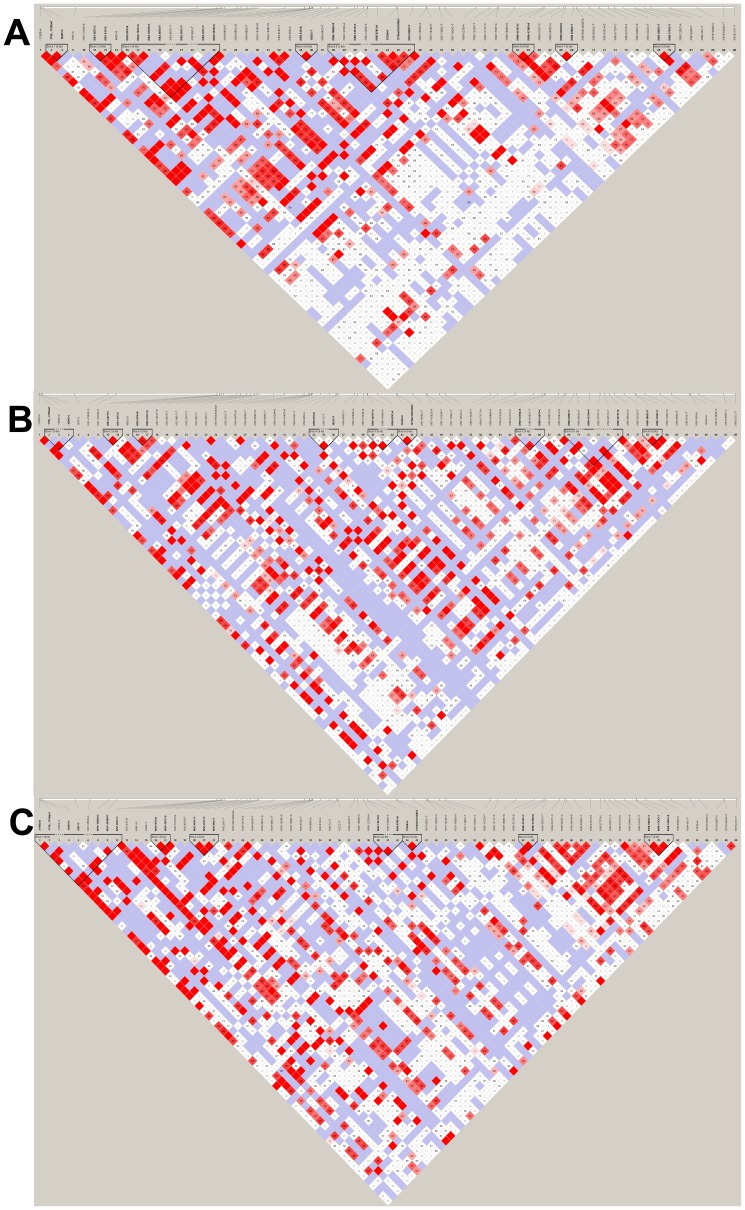
Pairwise LD analyses were performed between the 89 SNPs identified earlier for each of the ethnic groups. Pairwise LD matrices demonstrated moderate to strong linkage between several SNPs throughout the SLC22A1 gene region. Tight linkage was observed amongst the following SNPs across all ethnicities: IVS5-61G>A (rs2282142) with 1022C>T (rs2282143), IVS2-687G>T (rs3798175) and IVS4+1028A>G (rs3798170) (|D′| >0.90, r2>0.85); IVS2+97G>A (rs4646273) and IVS2+797C>G (rs4646274) with IVS4+1040T>G (rs3798169) and IVS7-1368C>T (rs1867350) (|D′| >0.94, r2>0.82); IVS2-461C>T (rs3798174) with IVS2-99C>T (rs3737088), IVS4+886C>G (rs3798172) and IVS7-1053C>T (rs7766568) (|D′| >0.90, r2>0.80); IVS2-257C>T (rs4646275) with IVS4+886C>G (rs3798172), IVS4-98G>A (rs4646276) and IVS7-1053C>T (rs7766568) (|D′| >0.90, r2>0.80); IVS2-1010T>G (rs614890) with IVS4+597A>G (rs594709) (|D′| = 1, r2>0.91) located in the region spanning introns 2–7; 5′-upstream region SNPs were in moderate linkage with intronic SNPs, -1756_-1755insT and -1620T>C (rs9457840) with IVS2-687G>T (rs3798175), IVS4+1028A>G (rs3798170) and IVS5-61G>A (rs2282142) (|D′| >0.90, r2>0.80); −1795G>A (rs6935207) with IVS1-207T>C (rs9457841) and IVS1-43T>G (rs4646272) (|D′| >0.80, r2>0.52); introns 8 and 9 polymorphisms, IVS8+1322G>T (rs644992) with IVS8+2698A>G (rs637841) (|D′| >0.96, r2>0.90); IVS8+5101T>C (rs7750592) with IVS8-4331A>G (rs9347386) (|D′| >0.91, r2>0.80) and IVS8-1803C>T (rs4709401) with IVS8-1793C>T (rs4709402) and IVS9+43C>T (rs2297374) (|D′| >0.92, r2>0.82). A detailed LD pattern is shown in Fig. 1 A–C where LD matrices are displayed by Chinese, Malay and Indian ethnic groups, respectively. Tag-SNPs were selected to represent the entire SLC22A1 region for each ethnic group by using pairwise tagging method in the Tagger program (Haploview 4.2, Broad institute USA) as described by de Bakker et al. [39]. A total of 24 tag-SNPs were selected to represent the 89 SNPs in all ethnicities, which included 2 5′-upstream, 2 exonic and 20 intronic SNPs (Table S1, see footnotes).
Figure 1. Linkage disequilibrium plots of SLC22A1 polymorphisms in healthy Asians.
Pairwise LD matrices represent moderate to strong linkage between SLC22A1 polymorphisms among (A) Chinese, (B) Malay and (C) Indian ethnic groups.
Pharmacogenetic Screening in CML Patients
Pharmacogenetic screening of the selected 24 SLC22A1 tag-SNPs, as well as candidate SNPs from ABCB1 [1236C>T (rs1128503), 2677G>T/A (rs2032582), 3435C>T (rs1045642)], ABCG2 421C>A (rs2231142), CYP3A5*3 (rs776746) and PXR [IVS2+55A>G (rs1464603), IVS2+78A>G (rs1464602), IVS6-17C>T (rs2276707), 1792A>G (rs3732359), 1944T>C (rs3732360), 2654T>C (rs3814058)] was performed in 38 CML patients. The genotypic and allelic frequencies of SLC22A1 tag-SNPs in the patient cohort were found to be similar to that observed in the healthy Chinese subjects as majority of patients were of Chinese descent. Candidate SNPs selected from ABCB1, ABCG2, CYP3A5 and PXR genes were present with high variant allele frequency ranging from 20% to 70% in the patients with CML which was also comparable with healthy subjects from the 3 ethnic groups (Table S1). None of the individual genotypes demonstrated a significant association with IM pharmacokinetic parameters (data not shown). Haplotype analysis have been recognized to be more robust than single marker analysis [40], [41] and these finding supported the hypothesis to look into multi-marker interactions to reveal the genotypic-phenotypic association. The 24 SLC22A1 tag-SNPs were considered for haplotypes and IM pharmacokinetic association analysis in the CML patients.
Haplotypic Effect of SLC22A1 Tag-SNPs on IM Pharmacokinetics
The haplotypic effect of SLC22A1 tag-SNPs on IM pharmacokinetics was evaluated using the haplowalk method. This method was adapted to identify a sub-haplotypic region that could best explain the variability in IM pharmacokinetic parameters. Global haplotype score statistics was applied to estimate the overall association of haplotypes and phenotypic traits [37], [38]. Our analysis revealed a sub-haplotypic region encompassing one exonic SNP [1222A>G (rs628031)] surrounded by two intronic SNPs [IVS6-878C>A (rs3798168) and IVS7+850C>T] that is significantly associated with IM clearance (p = 0.013, adjusted for gender, weight, age and dosing interval, Fig. S2 ). The haplowalk score statistic displays a global overview of the association of haplotypes in a particular region with phenotypic traits, and cannot be interpreted as a specific individual haplotypic effect. Therefore, we further employed the haplotype-specific GLM in order to estimate the individual haplotypic effects.
Haplotype-specific GLM estimates the regression coefficient associated with IM pharmacokinetics (Table 3). The high frequency haplotypes (IVS6-878C>A; 1222A>G; IVS7+850C>T) AGT and CGC (frequency: 41% and 36%, respectively) were significantly associated with a 22% decrease in IM clearance compared to the reference haplotype CAC (frequency: 22%) [CL (*10−2 L/hr/mg); CAC vs AGT: 4.03 vs 3.16, p = 0.017; CAC vs CGC: 4.03 vs 3.15, p = 0.017]. The SLC22A1 haplotype AGC was found to be present in only 1% of our CML patients, however the IM clearance was observed to be 1.7-fold higher than reference haplotype [CL (*10−2 L/hr/mg); CAC vs AGC: 4.03 vs 6.71, p = 0.04]. Similarly, haplotypes AGT and CGC were modestly associated with a 38% and 30% increase in C0h, respectively, as compared to reference haplotype CAC, although this was not statistically significant [C0h (*10−6 1/ml); CAC vs AGT: 3.13 vs 4.32; CAC vs CGC: 3.13 vs 4.07; p>0.05]. As both of these low clearance-associated haplotypes had similar impact on IM disposition, we examined the combinatorial effect of copy numbers of these two haplotypes (AGT and CGC) on IM pharmacokinetics. Two copies of haplotypes AGT or CGC were observed in 23 patients, 1 copy in 12 patients whereas 3 patients were not carrying any copies of AGT or CGC haplotypes. The patients were divided in two groups according to the number of copies of haplotypes AGT or CGC. The first group (n = 15) comprised of patients carrying 0 or 1 copy (Slow) and the second group (n = 23) comprised of patients carrying 2 copies of AGT or CGC haplotypes (Shigh). The copy numbers of AGT and CGC haplotypes were significantly associated with IM clearance and C0h (Fig. 2). Patients harboring Shigh had 33.4% lower clearance and 50% higher C0h than patients with Slow [median clearance (*10−2 L/hr/mg); Slow vs Shigh: 3.29 vs 2.19, p = 0.026; median C0h (*10−6 1/ml); Slow vs Shigh: 3.17 vs 4.76, p = 0.013].
Table 3. Influence of SLC22A1 haplotypes on imatinib clearance, CL and trough concentration, C0h using haplotype specific generalized linear model.
| Haplotypes | Haplotype frequency (%) | Mean ± SE | Fold change | p-value | ||||||
| IVS6 -878C>A | 1222A>G | IVS7+850C>T | CMLPatients | Healthy Chinese | CL(*10−2 L/hr/mg ) | C0h(*10−6 1/ml) | CL | C0h | CL | C0h |
| C | A | C | 22.3 | 25.8 | 4.03±0.53 | 3.13±1.06 | 1.00a | 1.00a | 0 | 0.005 |
| A | G | C | 1.30 | 0.90 | 6.71±1.28 | 0.60±2.56 | 1.66 | 0.192 | 0.044 | 0.328 |
| A | G | T | 40.7 | 38.7 | 3.16±0.35 | 4.32±0.70 | 0.784 | 1.38 | 0.017 | 0.097 |
| C | G | C | 35.5 | 30.2 | 3.15±0.35 | 4.07±0.71 | 0.781 | 1.30 | 0.017 | 0.192 |
Reference: Imatinib pharmacokinetics parameters values corresponding to haplotype CAC were used as reference to compare with other three haplotypes.
Figure 2. SLC22A1 haplotypes association with imatinib pharmacokinetics in Asian patients with CML (n = 38).
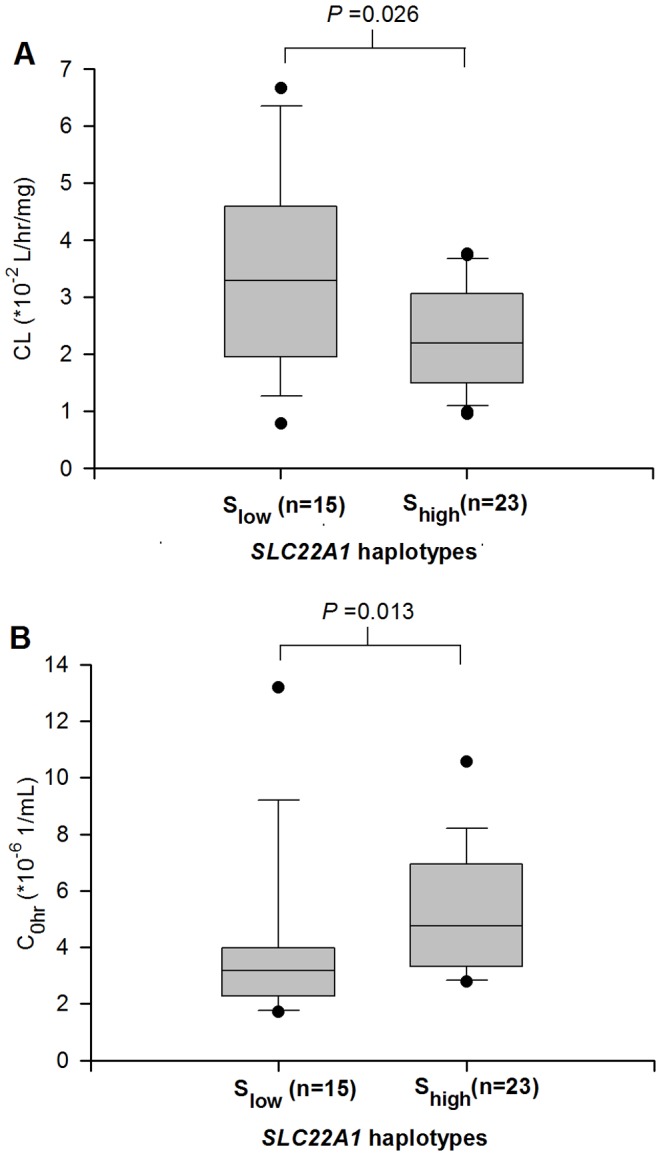
The patients were divided in two groups according to copy numbers of SLC22A1 haplotypes AGT and CGC, Slow (0 or 1 copy) and Shigh (2 copies). SLC22A1 haplotypes were significantly associated with imatinib (A) clearance, CL and (B) trough concentration, C0h. Patients harboring Shigh haplotypes had 33.4% lower clearance and 50% higher trough concentration than patients with Slow haplotypes.
Effect of a Combined SLC22A1-ABCB1 Haplotype Profile on IM Pharmacokinetics
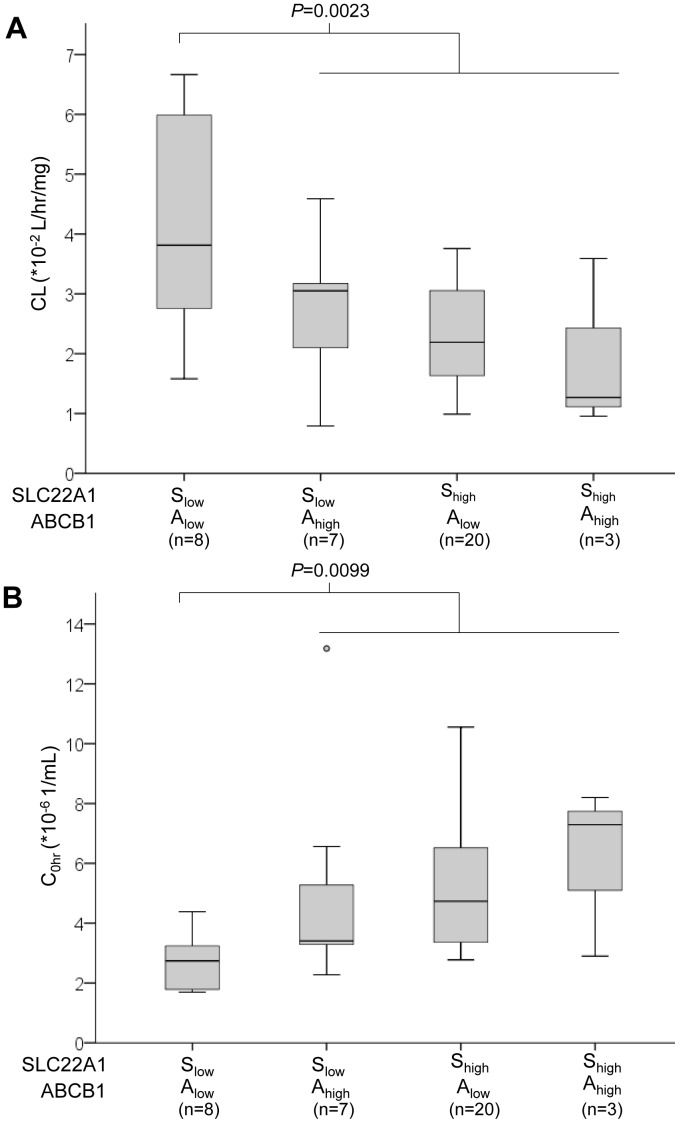
We explored if specific ABCB1 haplotypes could affect IM clearance and concentrations. Interestingly, although we did not find any direct correlations amongst ABCB1 haplotypes with clearance and C0h, we observed that patients harboring Shigh tended to carry either 0 or 1 copy of ABCB1 haplotypes (1236C>T; 267G>T/A; 3435C>T) TGT or TTT (Alow), rather than 2 copies (Ahigh) (p = 0.030). When stratified according to specific SLC22A1-ABCB1 combination haplotype profiles, a trend towards low clearance and high C0h was observed from Slow-Alow, Slow-Ahigh, Shigh-Alow to Shigh-Ahigh. Notably, the simultaneous possession of Slow with Alow was associated with a 73.2% higher clearance and 41.2% lower C0h than patients carrying other haplotype combinations [median CL (*10−2 L/hr/mg); Slow-Alow vs others: 3.81 vs 2.20, p = 0.0023; median C0h (*10−6 1/ml); Slow-Alow vs others: 2.74 vs 4.66, p = 0.0099] (Fig. 3). The combined SLC22A1-ABCB1 haplotype profile was not significantly associated with treatment response parameters.
Figure 3. SLC22A1 and ABCB1 haplotypes association with imatinib pharmacokinetics in Asian patients with CML (n = 38).
SLC22A1 and ABCB1 haplotypes were stratified according to specific combination haplotype profiles and associations were checked with imatinib (A) clearance, CL and (B) trough concentration, C0h. A trend towards low clearance and high C0h was observed from Slow-Alow, Slow-Ahigh, Shigh-Alow to Shigh-Ahigh. The simultaneous possession of Slow with Alow was associated with a 73.2% higher clearance and 41.2% lower trough concentration than patients carrying other haplotype combinations.
Discussion
Our results demonstrated wide inter-individual variability of trough IM plasma levels in Asian patients with CML, confirming previous reports on Caucasian [3], [6], [10], [16], Korean [13], Japanese [12], Chinese [11], [17] and other ethnic cohorts [14], [15], [18]. In addition, we not only observed that the mean IM trough concentration in our cohort of Asian patients with CML was significantly higher than that previously reported in studies on Caucasians [3], [6], [10], [14], [16], [18], 31 of 34 patients (94%) who received standard IM doses of 400 mg in fact had an IM trough concentration above the recommended threshold of >1000 ng/mL. This result is consistent with studies on the Chinese [11], [17], [42], Korean [13] and Jordanian [15] populations. These differences in IM levels may be attributed to several factors including poor medication compliance [6], intake of other drugs [7], gender as well as physical variables [3], [17]. In our study however, there was no evidence that any of the above factors could have accounted for the large inter-patient and inter-ethnic variability observed. Instead, the data indicated that the underlying reason may invoke intrinsic differences in cellular influx (e.g. hOCT1) and efflux (e.g. ABCB1) transporters [9]. hOCT1 is known to mediate the active transport of IM into primary CML cells [20], and is likely to be a major determinant of hepatocyte uptake for systemic clearance [43]. Low hOCT1 or high ABCB1 activity/expression in leukemic cells may result in reduced influx or increased efflux of drugs respectively, thereby reducing intracellular levels whilst increasing plasma levels. Simultaneously, the same effect on hepatocytes may impair drug uptake into the liver, leading to elevated plasma levels. Whilst it may be plausible that since some patients with CML achieve supra-therapeutic IM levels on standard 400 mg doses and may be considered for de-escalated regimens without aggravating their outcome [17], such observations may also reflect compromised cellular transport mechanisms resulting in high plasma levels and detrimentally-low intracellular levels.
In this exploratory study, we screened the entire SLC22A1 gene for polymorphic variation in three healthy Asian ethnic populations and further examined tag-SNPs in Asian CML patients. Our approach revealed a sub-haplotypic region encompassing 1 exonic SNP [1222A>G (rs628031)] surrounded by 2 intronic SNPs [IVS6-878C>A (rs3798168) and IVS7+850C>T] that is significantly associated with IM pharmacokinetics. The SLC22A1 gene was found to be highly polymorphic and displayed significant interethnic variations amongst healthy individuals of Chinese, Malay, and Indian ethnicity (Table S1, see footnotes). For example, the allelic frequency of the 5′-upstream polymorphism [−1795G>A (rs6935207)], was significantly higher amongst Chinese (0.57) as compared to Malays (0.39), Indians (0.30) and Caucasians (0.21) [26]. Interestingly, the majority of previously studied SLC22A1 coding SNPs in Caucasians [24], [25], [27] were absent in the Asian ethnic groups examined, and only 1222A>G (rs628031) was found in common amongst Caucasian, Japanese [44] and the current Asian cohort. Similarly, polymorphisms 181C>T (rs12208357) and 1260_1262delGAT (rs72552763) which have been linked to reduced transport activity of hOCT1 substrates in European-American groups [45], were not observed in Asians.
Amongst our cohort of Asian patients with CML, we were able to identify two groups based on unique haplotype profiles - one composed of those harboring 0 or 1 copy (Slow) and the other carrying 2 copies of AGT or CGC haplotypes (Shigh). Patients harboring Shigh had 50% higher IM trough level and 33.4% lower clearance than patients with Slow. The haplotype CGC was represented by the presence of the wild type allele of intronic polymorphisms IVS6-878C>A (rs3798168) and IVS7+850C>T, as well as the variant allele of the non-synonymous SNP 1222A>G (rs628031). On the other hand, the haplotype AGT was represented by the presence of the variant allele of all three polymorphisms. To the best of our knowledge, the intronic polymorphisms IVS6-878C>A (rs3798168) and IVS7+850C>T were not studied with phenotypic data previously. Although IM is a substrate of hOCT1 [20], it has been shown in previous single marker studies that 1222A>G (rs628031) does not alter the function of the hOCT1 protein [46] and does not affect response to IM therapy in CML patients [12], [24], [26], [27]. Our positive results from haplotype analysis is unsurprising, given that such an approach is known to be more robust than single marker analysis for identifying genomic regions enriched for phenotype-relevant casual variants [40], [41]. In the present study, a moderate to strong linkage pattern was observed amidst SLC22A1 variants in different ethnic groups. Particularly, strong linkage (|D′| >0.90) was detected amongst these three key variants in the Chinese population, which formed 84.2% of our patient cohort. Hence, the linkage effect of the region may have been the confounding factor in previous studies. The sub-haplotypic region is composed of two intronic and one coding SNP which suggests that the influence of haplotypes on IM pharmacokinetics could be mediated through interactions involving post-transcriptional modification. Taken together, out results highlight the importance of haplotype over single marker analysis to explain the variability in IM disposition and warrant further investigations in other ethnic groups.
Interestingly, we found that ABCB1 haplotypes may exert an indirect modulatory effect upon IM pharmacokinetics, depending on the specific SLC22A1 haplotype, with a trend towards high IM trough levels being observed from Slow-Alow, Slow-Ahigh, Shigh-Alow to Shigh-Ahigh (Fig. 3). Previous studies focusing only on genotype rather than haplotype profiles obtained heterogeneous results, with Gurney et al. reporting a lower IM clearance in individuals with the ABCB1 1236CC, 2677GG or 3435CC genotypes [47], Yamakawa et al. demonstrating higher clearance in Japanese patients with the 3435CC genotype [28], and others reporting a lack of any association [12], [48]. In agreement with our finding though, Dulucq et al. showed that both ABCB1 haplotype TTT as well as genotype 1236TT were correlated with high IM trough levels [49]. These inconsistencies may perhaps be explained by a functional dependency of ABCB1 on SLC22A1. In support of this thinking, it was noted in our study that patients with the SLC22A1 haplotype Shigh tended to possess the ABCB1 haplotype Alow, whilst a previous microarray analysis on a panel of leukemic cell lines revealed that SLC22A1 was inversely related with gene expression of ABCB1 [50]. This implies that specific ABCB1 and SLC22A1 haplotype profiles may affect IM levels by affecting their gene expression.
In conclusion, the present exploratory study comprehensively screened for SLC22A1 genetic variations and investigated its linkage and haplotype pattern in three distinct Asian populations, followed by its association with IM pharmacokinetics in CML patients. It is acknowledged that there are limitations in the current study and the results should be interpreted with care. Nonetheless, our report is the first to suggest that genetic polymorphisms and specific haplotypes in SLC22A1 and ABCB1 may contribute towards inter-individual and inter-ethnic variations in IM disposition in CML patients. The validation, as well as generalizability of results awaits future investigations in larger cohorts, including in other ethnic groups. More importantly, the functional and biological characterization of the haplotype variants ought to be further investigated.
Supporting Information
SLC22A1 gene structure and location of single nucleotide polymorphisms. Figure represents the 89 identified polymorphisms in the SLC22A1 gene. Four polymorphisms were located in the 5′-upstream region, 7 in the exonic region and 78 in the intronic region.
(TIF)
Haplowalk Analysis. Haplowalk analysis showing the sub-haplotypic region associated with imatinib clearance. P-values corresponding to polymorphisms are plotted on –log10 base. The sub-haplotypic region, significantly associated with imatinib clearance, is shown in blue dotted box. This region encompasses one exonic SNP (rs628031) surrounded by two intronic SNPs (rs3798168 and IVS7+850C>T).
(TIF)
Genotype and allele frequency of SLC22A1, ABCB1, ABCG2, CYP3A5 and PXR single nucleotide polymorphisms among Asian healthy subjects (Chinese, Malay and Indian, n = 70 each) and CML patients (n = 38). SLC22A1 24 Tag SNPs and statistically significant interethnic differences in the genotypic distributions among healthy subjects of three ethnic groups are indicated by table footnotes. Table also represents ABCB1, ABCG2, CYP3A5 and PXR gene polymorphism data from our previous publications [30]–[33].
(DOC)
Primer Sequences and PCR Conditions for Amplifications of SLC22A1 regions (UCSC RefSeq: NM_003057).
(DOC)
Acknowledgments
We would like to thank all subjects who have participated in this study.
Funding Statement
This project was supported by funds from the NMRC Institutional Grant (NMCCE10123 and NMPPG11122) and the Singhealth Research Fund (SRFIM07103) [http://www.nmrc.gov.sg/content/nmrc_internet/home.html, http://research.singhealth.com.sg/Pages/home.aspx]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rowley JD (1973) Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 243: 290–293. [DOI] [PubMed] [Google Scholar]
- 2. O’Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, et al. (2003) Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med 348: 994–1004. [DOI] [PubMed] [Google Scholar]
- 3. Larson RA, Druker BJ, Guilhot F, O’Brien SG, Riviere GJ, et al. (2008) Imatinib pharmacokinetics and its correlation with response and safety in chronic-phase chronic myeloid leukemia: a subanalysis of the IRIS study. Blood 111: 4022–4028. [DOI] [PubMed] [Google Scholar]
- 4. Hughes TP, Hochhaus A, Branford S, Müller MC, Kaeda JS, et al. (2010) Long-term prognostic significance of early molecular response to imatinib in newly diagnosed chronic myeloid leukemia: an analysis from the International Randomized Study of Interferon and STI571 (IRIS). Blood 116: 3758–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quintás-Cardama A, Kantarjian HM, Cortes JE (2009) Mechanisms of primary and secondary resistance to imatinib in chronic myeloid leukemia. Cancer Control 16: 122–131. [DOI] [PubMed] [Google Scholar]
- 6. Marin D, Bazeos A, Mahon FX, Eliasson L, Milojkovic D, et al. (2010) Adherence is the critical factor for achieving molecular responses in patients with chronic myeloid leukemia who achieve complete cytogenetic responses on imatinib. J Clin Oncol 28: 2381–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haouala A, Widmer N, Duchosal MA, Montemurro M, Buclin T, et al. (2011) Drug interactions with the tyrosine kinase inhibitors imatinib, dasatinib, and nilotinib. Blood 117: e75–e87. [DOI] [PubMed] [Google Scholar]
- 8. van Erp NP, Gelderblom H, Karlsson MO, Li J, Zhao M, et al. (2007) Influence of CYP3A4 inhibition on the steady-state pharmacokinetics of imatinib. Clin Cancer Res 13: 7394–7400. [DOI] [PubMed] [Google Scholar]
- 9. White DL, Saunders VA, Dang P, Engler J, Venables A, et al. (2007) Most CML patients who have a suboptimal response to imatinib have low OCT-1 activity: higher doses of imatinib may overcome the negative impact of low OCT-1 activity. Blood 110: 4064–4072. [DOI] [PubMed] [Google Scholar]
- 10. Picard S, Titier K, Etienne G, Teilhet E, Ducint D, et al. (2007) Trough imatinib plasma levels are associated with both cytogenetic and molecular responses to standard-dose imatinib in chronic myeloid leukemia. Blood 109: 3496–3499. [DOI] [PubMed] [Google Scholar]
- 11. Zhong JS, Meng FY, Xu D, Zhou HS, Dai M (2012) Correlation between imatinib trough concentration and efficacy in Chinese chronic myelocytic leukemia patients. Acta Haematol 127: 221–227. [DOI] [PubMed] [Google Scholar]
- 12. Takahashi N, Wakita H, Miura M, Scott SA, Nishii K, et al. (2010) Correlation between imatinib pharmacokinetics and clinical response in Japanese patients with chronic-phase chronic myeloid leukemia. Clin Pharmacol Ther 88: 809–813. [DOI] [PubMed] [Google Scholar]
- 13. Sohn SK, Oh SJ, Kim BS, Ryoo HM, Chung JS, et al. (2011) Trough plasma imatinib levels are correlated with optimal cytogenetic responses at 6 months after treatment with standard dose of imatinib in newly diagnosed chronic myeloid leukemia. Leuk Lymphoma 52: 1024–1029. [DOI] [PubMed] [Google Scholar]
- 14.Koren-Michowitz M, Volchek Y, Naparstek E, Gavish I, Levi I, et al.. (2012) Imatinib plasma trough levels in chronic myeloid leukaemia: results of a multicentre study CSTI571AIL11TGLIVEC. Hematol Oncol. [DOI] [PubMed]
- 15. Awidi A, Ayed AO, Bsoul N, Magablah A, Mefleh R, et al. (2010) Relationship of serum imatinib trough level and response in CML patients: long term follow-up. Leuk Res 34: 1573–1575. [DOI] [PubMed] [Google Scholar]
- 16. Forrest DL, Trainor S, Brinkman RR, Barnett MJ, Hogge DE, et al. (2009) Cytogenetic and molecular responses to standard-dose imatinib in chronic myeloid leukemia are correlated with Sokal risk scores and duration of therapy but not trough imatinib plasma levels. Leuk Res 33: 271–275. [DOI] [PubMed] [Google Scholar]
- 17. Li QB, Chen C, Chen ZC, Wang HX, Wu YL, et al. (2010) Imatinib plasma trough concentration and its correlation with characteristics and response in Chinese CML patients. Acta Pharmacol Sin 31: 999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Faber E, Friedecký D, Mičová K, Rožmanová S, Divoká M, et al. (2012) Imatinib trough plasma levels do not correlate with the response to therapy in patients with chronic myeloid leukemia in routine clinical setting. Ann Hematol 91: 923–929. [DOI] [PubMed] [Google Scholar]
- 19. Cortes JE, Egorin MJ, Guilhot F, Molimard M, Mahon FX (2009) Pharmacokinetic/pharmacodynamic correlation and blood-level testing in imatinib therapy for chronic myeloid leukemia. Leukemia 23: 1537–1544. [DOI] [PubMed] [Google Scholar]
- 20. Thomas J, Wang L, Clark RE, Pirmohamed M (2004) Active transport of imatinib into and out of cells: implications for drug resistance. Blood 104: 3739–3745. [DOI] [PubMed] [Google Scholar]
- 21.Crossman LC, Druker BJ, Deininger MWN, Pirmohamed M, Wang L, et al.. (2005) hOCT 1 and resistance to imatinib. Blood 106: 1133–4; author reply 1134. [DOI] [PubMed]
- 22. Nardinelli L, Sanabani SS, Didone A, Ferreira PDB, Serpa M, et al. (2012) Pretherapeutic expression of the hOCT1 gene predicts a complete molecular response to imatinib mesylate in chronic-phase chronic myeloid leukemia. Acta Haematol 127: 228–234. [DOI] [PubMed] [Google Scholar]
- 23. Zhang WW, Cortes JE, Yao H, Zhang L, Reddy NG, et al. (2009) Predictors of primary imatinib resistance in chronic myelogenous leukemia are distinct from those in secondary imatinib resistance. J Clin Oncol 27: 3642–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim DHD, Sriharsha L, Xu W, Kamel-Reid S, Liu X, et al. (2009) Clinical relevance of a pharmacogenetic approach using multiple candidate genes to predict response and resistance to imatinib therapy in chronic myeloid leukemia. Clin Cancer Res 15: 4750–4758. [DOI] [PubMed] [Google Scholar]
- 25. Bazeos A, Marin D, Reid AG, Gerrard G, Milojkovic D, et al. (2010) hOCT1 transcript levels and single nucleotide polymorphisms as predictive factors for response to imatinib in chronic myeloid leukemia. Leukemia 24: 1243–1245. [DOI] [PubMed] [Google Scholar]
- 26. Maffioli M, Camós M, Gaya A, Hernández-Boluda JC, Alvarez-Larrán A, et al. (2011) Correlation between genetic polymorphisms of the hOCT1 and MDR1 genes and the response to imatinib in patients newly diagnosed with chronic-phase chronic myeloid leukemia. Leuk Res 35: 1014–1019. [DOI] [PubMed] [Google Scholar]
- 27. White DL, Saunders VA, Dang P, Engler J, Hughes TP (2010) OCT-1 activity measurement provides a superior imatinib response predictor than screening for single-nucleotide polymorphisms of OCT-1. Leukemia 24: 1962–1965. [DOI] [PubMed] [Google Scholar]
- 28. Yamakawa Y, Hamada A, Nakashima R, Yuki M, Hirayama C, et al. (2011) Association of genetic polymorphisms in the influx transporter SLCO1B3 and the efflux transporter ABCB1 with imatinib pharmacokinetics in patients with chronic myeloid leukemia. Ther Drug Monit 33: 244–250. [DOI] [PubMed] [Google Scholar]
- 29.Hughes T (2006) ABL kinase inhibitor therapy for CML: baseline assessments and response monitoring. Hematology Am Soc Hematol Educ Program 211–218. [DOI] [PubMed]
- 30. Chowbay B, Cumaraswamy S, Cheung YB, Zhou Q, Lee EJD (2003) Genetic polymorphisms in MDR1 and CYP3A4 genes in Asians and the influence of MDR1 haplotypes on cyclosporin disposition in heart transplant recipients. Pharmacogenetics 13: 89–95. [DOI] [PubMed] [Google Scholar]
- 31. Balram C, Zhou Q, Cheung YB, Lee EJD (2003) CYP3A5*3 and *6 single nucleotide polymorphisms in three distinct Asian populations. Eur J Clin Pharmacol 59: 123–126. [DOI] [PubMed] [Google Scholar]
- 32. Jada SR, Lim R, Wong CI, Shu X, Lee SC, et al. (2007) Role of UGT1A1*6, UGT1A1*28 and ABCG2 c.421C>A polymorphisms in irinotecan-induced neutropenia in Asian cancer patients. Cancer Sci 98: 1461–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sandanaraj E, Lal S, Selvarajan V, Ooi LL, Wong ZW, et al. (2008) PXR pharmacogenetics: association of haplotypes with hepatic CYP3A4 and ABCB1 messenger RNA expression and doxorubicin clearance in Asian breast cancer patients. Clin Cancer Res 14: 7116–7126. [DOI] [PubMed] [Google Scholar]
- 34. Zhou Q, Sparreboom A, Tan EH, Cheung YB, Lee A, et al. (2005) Pharmacogenetic profiling across the irinotecan pathway in Asian patients with cancer. Br J Clin Pharmacol 59: 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Velpandian T, Mathur R, Agarwal NK, Arora B, Kumar L, et al. (2004) Development and validation of a simple liquid chromatographic method with ultraviolet detection for the determination of imatinib in biological samples. J Chromatogr B Analyt Technol Biomed Life Sci 804: 431–434. [DOI] [PubMed] [Google Scholar]
- 36. Oostendorp RL, Beijnen JH, Schellens JHM, Tellingen OV (2007) Determination of imatinib mesylate and its main metabolite (CGP74588) in human plasma and murine specimens by ion-pairing reversed-phase high-performance liquid chromatography. Biomed Chromatogr 21: 747–754. [DOI] [PubMed] [Google Scholar]
- 37. Karami S, Brennan P, Rosenberg PS, Navratilova M, Mates D, et al. (2009) Analysis of SNPs and haplotypes in vitamin D pathway genes and renal cancer risk. PloS one 4: e7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA (2002) Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 70: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de Bakker PIW, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, et al. (2005) Efficiency and power in genetic association studies. Nat Genet 37: 1217–1223. [DOI] [PubMed] [Google Scholar]
- 40. Purcell S, Daly MJ, Sham PC (2007) WHAP: haplotype-based association analysis. Bioinformatics 23: 255–256. [DOI] [PubMed] [Google Scholar]
- 41. Scherag A, Jarick I, Grothe J, Biebermann H, Scherag S, et al. (2010) Investigation of a genome wide association signal for obesity: synthetic association and haplotype analyses at the melanocortin 4 receptor gene locus. PloS one 5: e13967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Y, Zhou L, Dutreix C, Leroy E, Yin Q, et al. (2008) Effects of imatinib (Glivec) on the pharmacokinetics of metoprolol, a CYP2D6 substrate, in Chinese patients with chronic myelogenous leukaemia. Br J Clin Pharmacol 65: 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang L, Brett CM, Giacomini KM (1998) Role of organic cation transporters in drug absorption and elimination. Annu Rev Pharmacol Toxicol 38: 431–460. [DOI] [PubMed] [Google Scholar]
- 44. Chen L, Takizawa M, Chen E, Schlessinger A, Segenthelar J, et al. (2010) Genetic polymorphisms in organic cation transporter 1 (OCT1) in Chinese and Japanese populations exhibit altered function. J Pharmacol Exp Ther 335: 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shu Y, Brown C, Castro RA, Shi RJ, Lin ET, et al. (2008) Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin Pharmacol Ther 83: 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, et al. (2007) Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest 117: 1422–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gurney H, Wong M, Balleine RL, Rivory LP, McLachlan AJ, et al. (2007) Imatinib disposition and ABCB1 (MDR1, P-glycoprotein) genotype. Clin Pharmacol Ther 82: 33–40. [DOI] [PubMed] [Google Scholar]
- 48. Gardner ER, Burger H, van Schaik RH, van Oosterom AT, de Bruijn EA, et al. (2006) Association of enzyme and transporter genotypes with the pharmacokinetics of imatinib. Clin Pharmacol Ther 80: 192–201. [DOI] [PubMed] [Google Scholar]
- 49. Dulucq S, Bouchet S, Turcq B, Lippert E, Etienne G, et al. (2008) Multidrug resistance gene (MDR1) polymorphisms are associated with major molecular responses to standard-dose imatinib in chronic myeloid leukemia. Blood 112: 2024–2027. [DOI] [PubMed] [Google Scholar]
- 50. Hu S, Franke RM, Filipski KK, Hu C, Orwick SJ, et al. (2008) Interaction of imatinib with human organic ion carriers. Clin Cancer Res 14: 3141–3148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SLC22A1 gene structure and location of single nucleotide polymorphisms. Figure represents the 89 identified polymorphisms in the SLC22A1 gene. Four polymorphisms were located in the 5′-upstream region, 7 in the exonic region and 78 in the intronic region.
(TIF)
Haplowalk Analysis. Haplowalk analysis showing the sub-haplotypic region associated with imatinib clearance. P-values corresponding to polymorphisms are plotted on –log10 base. The sub-haplotypic region, significantly associated with imatinib clearance, is shown in blue dotted box. This region encompasses one exonic SNP (rs628031) surrounded by two intronic SNPs (rs3798168 and IVS7+850C>T).
(TIF)
Genotype and allele frequency of SLC22A1, ABCB1, ABCG2, CYP3A5 and PXR single nucleotide polymorphisms among Asian healthy subjects (Chinese, Malay and Indian, n = 70 each) and CML patients (n = 38). SLC22A1 24 Tag SNPs and statistically significant interethnic differences in the genotypic distributions among healthy subjects of three ethnic groups are indicated by table footnotes. Table also represents ABCB1, ABCG2, CYP3A5 and PXR gene polymorphism data from our previous publications [30]–[33].
(DOC)
Primer Sequences and PCR Conditions for Amplifications of SLC22A1 regions (UCSC RefSeq: NM_003057).
(DOC)